Abstract
Genome-wide association studies have identified a number of signals for both Type 2 Diabetes and related quantitative traits. For the majority of loci, the transition from association signal to mutational mechanism has been difficult to establish. Glucokinase (GCK) regulates glucose storage and disposal in the liver where its activity is regulated by glucokinase regulatory protein (GKRP; gene name GCKR). Fructose-6 and fructose-1 phosphate (F6P and F1P) enhance or reduce GKRP-mediated inhibition, respectively. A common GCKR variant (P446L) is reproducibly associated with triglyceride and fasting plasma glucose levels in the general population. The aim of this study was to determine the mutational mechanism responsible for this genetic association. Recombinant human GCK and both human wild-type (WT) and P446L-GKRP proteins were generated. GCK kinetic activity was observed spectrophotometrically using an NADP+-coupled assay. WT and P446L-GKRP-mediated inhibition of GCK activity and subsequent regulation by phosphate esters were determined. Assays matched for GKRP activity demonstrated no difference in dose-dependent inhibition of GCK activity or F1P-mediated regulation. However, the response to physiologically relevant F6P levels was significantly attenuated with P446L-GKRP (n = 18; P ≤ 0.03). Experiments using equimolar concentrations of both regulatory proteins confirmed these findings (n = 9; P < 0.001). In conclusion, P446L-GKRP has reduced regulation by physiological concentrations of F6P, resulting indirectly in increased GCK activity. Altered GCK regulation in liver is predicted to enhance glycolytic flux, promoting hepatic glucose metabolism and elevating concentrations of malonyl-CoA, a substrate for de novo lipogenesis, providing a mutational mechanism for the reported association of this variant with raised triglycerides and lower glucose levels.
INTRODUCTION
A recent genome-wide association (GWA) scan identified GCKR as a potential locus for modulating triglyceride and fasting plasma glucose (fpg) levels (1). GCKR encodes glucokinase regulatory protein (GKRP), and following fine mapping attributed the signal to the common non-synonymous SNP rs1260326 (c.1403 C > T, p.P446L, MAF 34%) (2). Subsequent population-based studies have replicated the association of this variant with triglyceride and fpg levels. Individuals homozygous for the risk allele (L) have on average a 0.15 mmol/l increase in triglyceride and a 0.06 mmol/l reduction in fpg levels compared with individuals with two copies of the wild-type (WT) allele (P) (2–4). The mutational mechanism behind this genetic association is currently unknown. However, unlike many signals arising from GWA studies, there is a non-synonymous SNP in a strong biological candidate gene driving the genetic association, thus facilitating functional studies. Even with such tractable variants, identifying the mutational mechanism for common risk alleles presents a challenge due to their small physiological effects. It is therefore essential that the ‘correct’ assays are selected and performed if these small differences in protein function are to be observed.
Glucokinase (GCK) is a key regulator of glucose storage and disposal in the liver. GKRP regulates GCK activity competitively with respect to the substrate glucose (5,6). GKRP action is in turn controlled by the phosphate esters fructose 6- and fructose 1-phosphate (F6P and F1P), which compete with each other for binding, and enhance or inhibit the action of the regulatory protein, respectively (7–9).
There is some controversy in the literature as to whether GKRP also regulates GCK in pancreatic β-cells (10–12). The vast majority of studies state that GCKR is not expressed in rodent β-cells (11,12). However, there is evidence from one study that an alternatively spliced GCKR variant is expressed in the β-cells of rodents and represents the major isoform in this tissue (10). To date, no studies have been reported which investigate GCKR expression in human pancreatic islets.
Biological evidence to support the association of these phenotypes with the GCKR SNP comes from both cellular and rodent models (13,14). Counter intuitively, Slosberg et al. over expressed GCKR in the human liver cell line HepG2 and showed an increase in GCK activity and expression at the protein level. From this, they suggested that GKRP as well as inhibiting GCK activity, also has a paradoxical role in extending GCK half-life by binding to and stabilizing the enzyme, thus protecting it from degradation (14). This finding is supported by data from both homo- and heterozygous gkrp knockout mice which display a marked reduction in gck expression and reduced GCK enzymatic activity under saturating glucose concentrations (11). At least one study has also shown that in vivo over-expression of gck in rat hepatocytes led to a drop in plasma glucose levels with a corresponding increase in circulating triglycerides (13). Therefore, findings from both groups suggested that (i) GKRP expression has a direct affect on GCK activity and (ii) there is a link between GCK activity and plasma glucose/circulating triglyceride levels.
A number of studies investigating the interaction of GCK and its regulatory protein have been reported but due to technical difficulties in purifying human recombinant GKRP, all of these have used the rat isoform (9,15,16). One of these studies investigated the effect of rodent P446L-GKRP on GCK activity and reported no differences in the inhibitory ability of the variant protein or its response to phosphate esters compared with WT (9). However, despite their high sequence homology (∼88%), there are important differences in the regulation of rat and human GKRP (7,17). First, human regulatory protein inhibits GCK even in the absence of F6P, an affect not observed with the same concentration of rat GKRP. Secondly, human regulatory protein was shown to have a higher affinity for this phosphate ester compared with the rodent protein (7).
The aims of this study were therefore, first, to investigate the affect of both human WT and P446L-GKRP on human GCK activity and secondly, to determine whether their regulation by phosphate esters was altered. Finally, we aimed to establish whether GCKR is expressed in human pancreatic islets, and if so, to look at this expression relative to that of GCK.
RESULTS
WT and P446L-GKRP both inhibit GCK activity
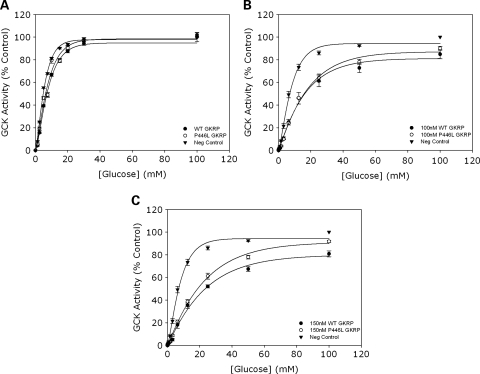
To assess the inhibitory function of our purified recombinant human WT and P446L-GKRP proteins following heterologous expression in Escherichia coli, one unit of each regulatory protein was used in assays to study competitive inhibition of GCK activity over a glucose concentration range (0–100 mm). Figure 1A demonstrates that GCK activity is inhibited with both GKRPs (each data point being plotted as a percentage of the negative control in which GKRP was absent and glucose concentration was 100 mm), establishing that both WT and P446L proteins are functional. These results also show no difference between the ability of WT and variant regulatory protein to inhibit GCK in the absence of the phosphate esters F1P and F6P.
Figure 1.
(A) Competitive inhibition of GCK using equivalent activities of WT and P446L-GKRP (n = 24). Competitive inhibition of 10 m U/ml GCK by one GKRP unit (WT=black circles, P446L=white circles, negative control=black triangles) was observed over a glucose concentration range of 0–100 mm. Each data point was plotted as a percentage of the negative control in which GKRP was absent and glucose concentration equaled 100 mm. Statistical analysis revealed no difference between the intrinsic inhibitory capacity of WT and P446L-GKRP. (B and C) Competitive inhibition of GCK using equimolar concentrations of WT and P446L-GKRP (n = 3). GCK activity was assayed in the presence of 0, 100 and 150 nm WT (black circles) and P446L (white circles) GKRP, over a glucose concentration range of 0–100 mm. Each data point was plotted as a percentage of the negative control in which GKRP was absent (black triangles) and glucose concentration equaled 100 mm. At 100 nm GKRP, there was statistically no difference between the responses of the two regulatory proteins (although it was noted that the variance of this data was larger than seen for other experiments and n = 3). At 150 nm GKRP, statistical analysis showed the intrinsic inhibitory capacity of P446L-GKRP to be significantly lower than that of the WT regulatory protein at [glucose] over 25 mm (P ≤ 0.03).
To further compare GKRP activity, competitive inhibition assays were repeated at fixed concentrations of both GKRP proteins (n = 3) as shown in Figure 1B (100 nm GKRP) and C (150 nm GKRP). These data also demonstrate no difference between the two regulatory proteins at 100 nm. However, at 150 nm, P446L-GKRP is less inhibitory than WT at concentrations of 25 mm glucose and above (P ≤ 0.03). Ki (inhibitor dissociation constants) were calculated from Dixon plots of 1/GCK activity versus [GKRP] for the equimolar concentration experiments (Supplementary Material, Fig. S1). The Ki values did not significantly differ between the two regulatory proteins (24.0 ± 40.7 nm for WT versus 23.1 ± 40.8 nm for P446L-GKRP; mean ± SEM; P = 1).
Our data are consistent with GKRP acting as a competitive inhibitor of GCK activity (18). The KcatGCK (maximal specific activity of GCK) remained unchanged in the presence of 0–150 nm regulatory protein (51.8 ± 1.6 to 51.9 ± 6.9 for WT versus 46.8 ± 0.7 to 53.4 ± 3.6 for P446L-GKRP; n = 3), and the S0.5 (glucose concentration needed for half maximal rate) was raised with increasing concentrations of both GKRPs (8.7 ± 0.2 to 16.8 ± 1.3 for WT versus 8.0 ± 0.3 to 18.2 ± 1.1 for P446L-GKRP).
F1P increases GCK activity with both WT and P446L-GKRP
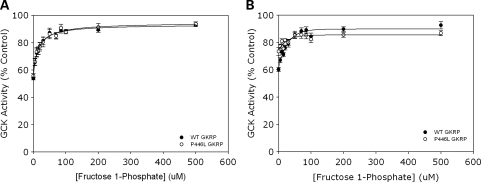
Data obtained using comparable activities of regulatory protein demonstrated the expected reduction in GKRP-mediated inhibition of GCK activity across a F1P concentration range of 0–500 µm (Fig. 2A). This GKRP-mediated inhibition was decreased by the same extent for both WT and P446L-GKRP (53.7 ± 1.1 to 92.8 ± 1.2% GCK activity for WT versus 55.7 ± 1.5 to 93.6 ± 1.4% GCK activity with P446L; means ± SEM; n = 18; P > 0.1).
Figure 2.
(A and B) F1P negatively modulates both WT and P446L-GKRP-mediated inhibition of GCK, leading indirectly to an increase in enzyme activity. Inhibition of 10 m U/ml GCK by (A) one GKRP unit, 5 mm glucose (n = 18) and (B) 100 nm GKRP, 10 mm glucose (n = 3) was observed over 0–500 µm F1P (black circles=WT, white circles=P446L). GCK activity was plotted as a percentage of that obtained in the absence of either regulatory protein. Statistical analysis revealed no difference in response of WT and P446L-GKRP to F1P up to and including 500 µm F1P (P ≥ 0.1). Kiapparent from Dixon plots of [F1P] versus 1/GCK activity did not significantly differ between the two regulatory proteins (A) 31.2 ± 4.3 µm for WT versus 24.9 ± 2.0 µm for P446L-GKRP, (B) 54.0 ± 10.8 µm for WT versus 46.3 ± 4.2 µm for P446L-GKRP; mean ± SEM; P ≥ 0.1.
Experiments using fixed concentrations of both WT and variant regulatory protein confirmed that there was no difference in F1P-mediated inhibition of the two GKRPs at concentrations up to and including 500 µm of phosphate ester (92.5 ± 2.6% for WT versus 90.1 ± 1.8% for P446L; n = 9; P = 0.2), despite the initial difference in intrinsic inhibitory capacity at 0 µm F1P (Fig. 2B).
It has been suggested previously that F1P causes an increase in GCK activity by promoting dissociation of the GKRP:GCK complex (7). Therefore the ‘apparent’ inhibitor dissociation constant (Kiapparent) would be expected to increase upon addition of F1P, as regulatory protein dissociation from the enzyme is promoted. In our studies, Kiapparent from Dixon plots of 1/GCK activity versus [F1P] reflected this. Also, these calculated Kiapparent values did not significantly differ between the two regulatory proteins when using either comparable activity (24.5 ± 2.0 µm for WT versus 31.2 ± 4.3 µm for P446L-GKRP; P = 0.1) or equimolar concentrations (54.0 ± 10.8 µm for WT versus 46.3 ± 4.2 µm for P446L-GKRP; P = 0.5), suggesting a similar dissociation of both regulatory proteins from GCK in response to F1P. This was also corroborated by observing the [F1P] that caused reversal of GCK inhibition by half (i.e. 75% enzyme activity), which was ∼15 µm F1P for both GKRPs in the matched activity experiments, and ∼15 and 20 µm F1P for P446L and WT-GKRP, respectively, in the comparable concentration experiments.
Differences in F6P-mediated regulation of P446L-GKRP compared with WT
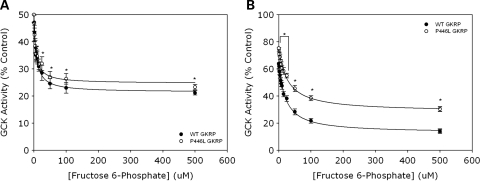
GCK activity was observed over 0–500 µm F6P in the presence of either equal activities or equivalent concentrations of both GKRPs (Fig. 3A and B, respectively). It is well established that F6P-mediated regulation of GKRP leads to enhanced GCK inhibition, and hence a corresponding reduction in observed enzyme activity (7,9,18). This was seen with both WT and P446L-GKRP when using the same activity of regulatory protein, although the reduction in GCK activity was significantly less over the physiologically relevant 25–500 µm F6P concentration range with the variant protein (31.9 ± 2.6 to 23.3 ± 1.0% GCK activity versus 28.5 ± 2.8 to 21.2 ± 0.8% GCK activity for WT; means ± SEM; n = 18; P = 0.04, 0.0001, 0.006 and 0.03 for 25, 50, 100 and 500 µm F6P, respectively) (Fig. 3A).
Figure 3.
(A and B) F6P-mediated regulation of P446L-GKRP is significantly diminished compared with WT regulatory protein. Inhibition of 10 m U/ml GCK by (A) one unit GKRP, 5 mm glucose (n = 18) and (B) 100 nM GKRP, 10 mm glucose (n = 3) was observed over 0–500 µm F6P (black circles=WT, white circles=P446L). GCK activity was plotted as a percentage of that obtained in the absence of regulatory protein. Statistical analysis showed there to be a significant difference between the response of WT and P446L-GKRP (A) over the physiologically relevant range 25–500 µm F6P (31.9 ± 2.6 to 23.3 ± 1.0% GCK activity for P446L versus 28.5 ± 2.8 to 21.2 ± 0.8% GCK activity for WT; means ± SEM; P = 0.04, 0.0001, 0.006 and 0.03 for 25, 50, 100 and 500 µm F6P, respectively) and (B) over the entire 0–500 µm F6P range (P < 0.001). Dixon plots of [F6P] versus 1/GCK activity showed Kiapparent was also significantly different between WT and variant regulatory protein in both (A) the comparable activity (31.0 ± 4.4 versus 21.1 ± 2.7 µm, respectively; P = 0.05) and (B) equimolar concentration experiments (16.6 ± 1.7 versus 9.2 ± 1.0 µm, respectively; P < 0.001).
These results were confirmed in experiments using the same concentration of both regulatory proteins, as there was a significant difference between the response of WT and P446L-GKRP over the entire F6P concentration range (63.7 ± 2.6 to 14.1 ± 1.2% versus 71.9 ± 1.4 to 33.8 ± 3.1%, respectively; n = 9; P < 0.001) (Fig. 3B).
Increasing F6P concentration has been shown in this study to enhance GKRP-mediated inhibition of GCK activity, and thus it would be expected that Kiapparent would be decreased as a result of reduced dissociation of the inhibitor GKRP from GCK (i.e. ‘tighter’ enzyme:regulatory protein binding). This was reflected in our data, as Dixon plots of [F6P] versus 1/GCK activity showed a decrease in Kiapparent upon addition of F6P. Kiapparent was also significantly different between WT and P446L-GKRP for both the equal activity (31.0 ± 4.4 µm versus 21.1 ± 2.7 µm, respectively; P = 0.5) and equimolar concentration experiments (16.6 ± 1.7 µm versus 9.2 ± 1.0 µm, respectively; P < 0.001). The IC50 (concentration of F6P needed to inhibit enzyme activity by 50%) was also significantly different between WT and P446L-GKRP; 23.8 versus 35.0 µm, respectively, for matched activity experiments (P = 0.01), and 25.0 versus 56.3 µm, respectively, for equal concentration experiments (P < 0.001), reflecting a decreased affinity of the variant regulatory protein for F6P in both experiments.
GCKR is highly expressed relative to GCK in human liver, with very low levels in human pancreatic islets
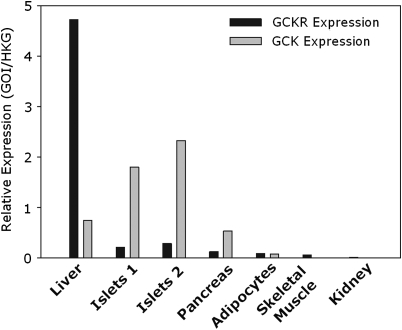
Owing to the association of P446L-GKRP with fpg levels, we investigated whether the observed modulation in the P446L-GKRP response to physiologically relevant F6P concentrations could have consequent metabolic affects in human pancreatic islets as well as human liver. To address this question, we quantified GCKR and GCK mRNA levels in appropriate human tissues. Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analysis of liver (n = 1), islets (n = 2), whole pancreas (n = 1) and adipocytes (n = 1) demonstrated that GCKR mRNA is expressed in molar excess in human liver compared with GCK, thus supporting a functional role for the regulatory protein in the hepatocyte (19). Conversely, although GCKR is expressed at very low levels in human islets, whole pancreas and adipocytes (only a tenth of that seen in liver), the much higher relative expression of GCK suggests regulation of this enzyme in these tissues is largely not GKRP mediated (Fig. 4).
Figure 4.
GCKR and GCK expression in human liver, pancreas, isolated islets, adipocytes, skeletal muscle and kidney. One microgram RNA was converted to cDNA via RT–PCR and expression of the human GCKR and GCK genes observed via Taqman gene expression analysis. Liver (n = 1), islets (n = 2), pancreas (n=1) and adipocytes (n = 1) were studied (as well as appropriate negative controls). These data show GCKR to be highly expressed in molar excess in the liver compared with GCK. However, in the pancreas and islets, GCK levels are much higher than that of the regulatory protein. In adipocytes, skeletal muscle and kidney both genes are only present at negligible levels.
DISCUSSION
In this study, we have performed the first functional characterization of the human P446L-GCKR variant, which has been reproducibly associated with an average 0.15 mmol/l increase in triglyceride and a 0.06 mmol/l decrease in fpg levels in the general population (1–4). Our data demonstrate both human WT and variant regulatory protein to be functional and able to inhibit GCK activity in a dose-dependent manner. Both GKRPs were also equally sensitive to F1P-mediated regulation, which is consistent with that shown previously using rodent proteins (16). However, our analyses have revealed a previously unappreciated change in F6P-mediated modulation of the P446L regulatory protein at physiologically relevant concentrations of this phosphate ester. These results differ from those reported in earlier rodent studies which showed no differences between the two proteins and confirm the important differences in the properties of human GKRP compared with rodent homologues (7).
The differing responses of the P446L-variant regulatory protein to F1P and F6P may be explained by looking at the binding site of these molecules. The crystal structure of GKRP has not been solved; however, it does have homology with other proteins of known crystal structure, such as the isomerase domain of bacterial glucosamine 6-phosphate synthase (GlmS), which catalyses the conversion of F6P into either glucose 6-phosphate or glucosamine 6-phosphate (9,20–22). This isomerase domain consists of two sugar isomerase sub-domains (23), and analysis of the GlmS crystal structure (21) and functional studies on rat GKRP mutants (9) suggests a single substrate binding site lies between the interface of these two sub-domains. It has also been suggested that GKRP adopts different structural conformations in order to accommodate either F1P or F6P at this single binding site, and only one of these (that favoring F6P-binding) can complex with GCK (9,24). Residue 446 lies between two sequence motifs thought to be involved directly in binding of phosphate esters. As previously reported, mutating certain residues outside of these motifs can still affect binding (and hence regulation of GKRP activity) (9). Also, residue 446 is a conserved proline across human, rat and Xenopus laevis regulatory protein, and all bar the latter are responsive to both F1P and F6P (7,9). Therefore, we propose that residue 446 may not play a direct role in the binding of phosphate esters, but may be important structurally within GKRP. This amino acid substitution could then either reduce the ability of the regulatory protein to adopt the F6P-binding conformation or hinder the F6P–GKRP complex from binding to and inhibiting GCK.
Our data show F6P-mediated regulation of P446L-GKRP to be significantly attenuated compared with WT over a physiological range of F6P concentrations, which ultimately leads to a reduction in GCK-inhibition by the variant regulatory protein. This is predicted to increase glycolytic flux and hence glucose uptake by the liver. This enhanced rate of glycolysis (as is seen with GCK over-expression in rats) may raise levels of other liver metabolites such as malonyl-CoA, increasing triglyceride levels via two mechanisms; first by acting as a substrate for de novo lipogenesis and secondly by inhibiting carnitine-palmitoyl transferase-1 (thus blocking fatty acid oxidation) (13). This perturbation of hepatic metabolism could account for the reduced glucose and raised triglycerides seen in L446-GKRP individuals. However, the very low GCKR mRNA levels detected in human islets (compared with human liver) suggest that GKRP has only a very minor (if any) functional role in the regulation of GCK in the pancreas. Consequently, pancreatic GCK is unlikely to be affected in individuals with this variant regulatory protein confirming that the phenotype observed is driven by a hepatic effect.
Broader implications of these findings regard the alteration of GCK activity to treat diabetes. Aberrant GCK function has been implicated in both monogenic forms of diabetes and in the variation of fpg levels within the general population, and as such it is a well-established therapeutic target (25,26). The initial genetic association data concerning the P446L-GCKR variant (1–4) supported by our functional studies confirm that changing the regulation of GCK activity solely in the liver could alter other hepatic metabolic pathways, thus resulting in an unfavorable lipid profile. This has already been shown in one study in rodents, where over-expressing hepatic gck lowers plasma glucose as well as raising triglyceride levels (13). However, a number of other studies have shown that hepatic over-expression of gck does not always result in an increase in triglycerides, suggesting that the degree of increased gck activity is critical (27,28). As such, potential new therapies targeting GKRP in the liver should be approached with caution.
In conclusion, our functional studies demonstrate that altered F6P-mediated GKRP regulation in carriers of the L446 allele explains the association between the common P446L-GCKR variant and higher triglyceride and lower fpg levels in several human populations. This work demonstrates that with suitable functional studies it is possible to establish mutational mechanisms for common variants of modest effect size identified by GWA studies.
MATERIALS AND METHODS
Site-directed mutagenesis
p-FLAG CTC vector (Sigma Aldrich Company Ltd, Gillingham, UK) containing human GKRP coding sequence was kindly supplied by Katy Brocklehurst (7). The P446L mutation was introduced via PCR-based site-directed mutagenesis using Pfu-polymerase (Promega, Southampton, UK) and primers shown in Supplementary Material, Table S1 (Operon Biotechnologies GmbH, Cologne, Germany). Mutant and WT plasmids were then used to transfect BL21 competent cells (Stratagene Europe, Amsterdam, The Netherlands), and DNA obtained using QIAprep Spin Miniprep Kit (Qiagen Ltd, Crawley, UK). Mutant and WT sequences were confirmed via direct sequencing using primers in Supplementary Material, Table S2.
Protein extraction
Glutathione S-transferase-tagged human pancreatic GCK was prepared as described previously (29). The pancreatic isoform was used as it has the same affinity for GKRP as liver GCK (30). The extracted enzyme was pure and at a concentration of 1.2 mg/ml, as determined by the Agilent 230 Protein kit (Agilent Technologies UK Ltd, Stockport, UK) and Bio-Rad Bradford reagent assay (Bio-Rad Laboratories Ltd, Hemel Hempstead, UK), respectively. Three preparations each of FLAG-tagged human WT and P446L-GKRP were extracted based on a protocol previously described but without the use of a DEAE-sepharose ion-exchange resin (7). The extracted regulatory proteins were confirmed as being greater than 95% pure via Agilent analysis, and concentrations quantified as before (average 0.19 mg/ml for WT and 0.12 mg/ml for P446L-GKRP; n = 3). Storage buffer [0.7 m glycerol, 0.2 m KCl, 0.06 m Tris–HCl (pH 8), 5 mm dithiothreitol (DTT), 0.06 m glucose (GCK preparation only); all Sigma Aldrich Company Ltd] was added to recombinant GCK and GKRP in 1:1 volume ratio prior to snap-freezing and storage at −80°C. For initial regulatory protein preparations (n = 1), storage buffer including glucose was added to samples. As this affected competitive inhibition assays, storage buffer was changed using Vivaspin20 columns containing diafiltration cups (Sartorius Mechatronics, Epsom, UK) to one in which glucose was absent.
GKRP assays
GKRP inhibition of GCK activity was determined spectrophotometrically using glucose 6-phosphate dehydrogenase (G6PDH)-linked assays (Sigma Aldrich Ltd). Experiments using both comparable activity/mg (amount of WT and P446L-GKRP used in each experiment inhibited 10 m U/ml GCK by the same extent at 5 mm glucose in the absence of the phosphate esters) (7) and equimolar concentrations (either 0, 50, 100 or 150 mm of both regulatory proteins used per assay) (9) were conducted. Assays were based on previously described protocols (7,29,30) but with the following modifications. Competitive inhibition assays were at 37°C, pH 7.1, and contained 2 mm MgCl2, 3.8 µm bovine serum albumin, 25 mm KCl, 25 mm HEPES, 0.5 mm NADP+, 1 mm ATP, 1 mm DTT, 0–100 mm glucose, 4 U/ml G6PDH, 10 m U/ml GCK and 1 U GKRP. All assay reagents (except GCK and GKRP which were generated in-house) were obtained from Sigma Aldrich Ltd. One m-unit of GCK was defined as that which converts 1 nmol substrate per minute, whereas one GKRP unit was defined as that which inhibits 10 m U/ml GCK by 50% under the standard assay conditions defined above (7). On average at 5 mm glucose, one GKRP unit resulted from 15.87 µg/ml of WT and 14.05 µg/ml of P446L-GKRP (n = 3 independent protein preparations). One GKRP unit of both WT and variant P446L regulatory protein were subsequently used per equimolar activity assay.
F1P and F6P assays were performed using the same assay conditions as for competitive inhibition experiments, but only 5 mm glucose (7). Both phosphate esters were purchased from Sigma Aldrich Ltd.
Ki (inhibitor dissociation constants) were determined from Dixon plots of 1/GCK activity versus [GKRP] as previously described (7) for the competitive inhibition assays. Plots of 1/GCK activity versus [F6P] or [F1P] were also used to determine apparent inhibitor dissociation constants, ‘Kiapparent’, from these experiments. As the phosphate esters are secondary regulators of GCK activity (act via the inhibitor GKRP), Dixon plots provided an insight into dissociation of the phosphate esters, GKRP and GCK, but could not be used to distinguish specifically between the individual components of these interactions.
Gene expression analysis
GCKR and GCK expressions were observed via qRT–PCR in human liver (n = 1), human islets (n = 2), human pancreas (n = 1) and human adipocytes (n = 1). All samples were part of a commercially available human RNA tissue panel (Clontech-Takara Bio Europe, Saint-Germain-en-Laye, France), except for the human islets which were collected as part of a previous study at Oxford University which had been conducted with full ethical consent. RNA samples were initially treated with DNase I (Applied Biosystems, Warrington, UK) to remove all genomic contamination. This was followed by random primed first-strand cDNA synthesis using 1 µg of each RNA sample and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Reactions in which the enzyme was absent were also carried out to produce negative controls for the final qRT–PCR. cDNA was diluted 1/100 and 4 µl used in a total reaction volume of 10 µl [5.5 µl of Taqman gene expression master mix and 0.5 µl of the appropriate Taqman gene expression assay (both Applied Biosystems)] and each sample run in triplicate. GCKR and GCK expressions were made relative to three house-keeping genes (HPRT, B2M and ACTB), and normalized to the 1/100 dilution of the standard curve (for each gene-specific assay).
Statistical analysis
Paired two-tail t-tests were used for statistical analysis of all assay data, with a cut-off value for statistical significance of 0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in Oxford by the Medical Research Council [Grant number 81696 to A.L.G.]. C.R. is a recipient of a European Foundation for the Study of Diabetes (EFSD) Albert Renold Travel Fellowship. Funding to pay the Open Access publication charges for this article was provided by MRC Grant Reference 81696.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Katy Brocklehurst for providing the human GKRP plasmid and are also grateful to both Katy Brocklehurst and Rick Davies for their technical advice, and Geoff Gibbons and Sandy Humphries for their invaluable input into the assay analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 2.Orho-Melander M., Melander O., Guiducci C., Perez-Martinez P., Corella D., Roos C., Tewhey R., Rieder M.J., Hall J., Abecasis G., et al. A common missense variant in the glucokinase regulatory protein gene (GCKR) is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:10. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sparso T., Andersen G., Nielsen T., Burgdorf K.S., Gjesing A.P., Nielsen A.L., Albrechtsen A., Rasmussen S.S., Jorgensen T., Borch-Johnsen K., et al. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 4.Vaxillaire M., Cavalcanti-Proenca C., Dechaume A., Tichet J., Marre M., Balkau B., Froguel P. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57:2253–2257. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matschinsky F.M. Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes. 1990;39:647–652. doi: 10.2337/diab.39.6.647. [DOI] [PubMed] [Google Scholar]
- 6.Matschinsky F.M. Regulation of pancreatic beta-cell glucokinase: from basics to therapeutics. Diabetes. 2002;51(Suppl. 3):S394–S404. doi: 10.2337/diabetes.51.2007.s394. [DOI] [PubMed] [Google Scholar]
- 7.Brocklehurst K.J., Davies R.A., Agius L. Differences in regulatory properties between human and rat glucokinase regulatory protein. Biochem. J. 2004;378:693–697. doi: 10.1042/BJ20031414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandercammen A., Detheux M., Van Schaftingen E. Binding of sorbitol 6-phosphate and of fructose 1-phosphate to the regulatory protein of liver glucokinase. Biochem. J. 1992;286:253–256. doi: 10.1042/bj2860253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veiga-da-Cunha M., Van Schaftingen E. Identification of fructose 6-phosphate- and fructose 1-phosphate-binding residues in the regulatory protein of glucokinase. J. Biol. Chem. 2002;277:8466–8473. doi: 10.1074/jbc.M105984200. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez E., Roncero I., Chowen J.A., Vazquez P., Blazquez E. Evidence that glucokinase regulatory protein is expressed and interacts with glucokinase in rat brain. J. Neurochem. 2002;80:45–53. doi: 10.1046/j.0022-3042.2001.00677.x. [DOI] [PubMed] [Google Scholar]
- 11.Grimsby J., Coffey J.W., Dvorozniak M.T., Magram J., Li G., Matschinsky F.M., Shiota C., Kaur S., Magnuson M.A., Grippo J.F. Characterization of glucokinase regulatory protein-deficient mice. J. Biol. Chem. 2000;275:7826–7831. doi: 10.1074/jbc.275.11.7826. [DOI] [PubMed] [Google Scholar]
- 12.Zawalich W.S., Rognstad R., Pagliara A.S., Matschinsky F.M. A comparison of the utilization rates and hormone-releasing actions of glucose, mannose, and fructose in isolated pancreatic islets. J. Biol. Chem. 1977;252:8519–8523. [PubMed] [Google Scholar]
- 13.O'Doherty R.M., Lehman D.L., Telemaque-Potts S., Newgard C.B. Metabolic impact of glucokinase overexpression in liver: lowering of blood glucose in fed rats is accompanied by hyperlipidemia. Diabetes. 1999;48:2022–2027. doi: 10.2337/diabetes.48.10.2022. [DOI] [PubMed] [Google Scholar]
- 14.Slosberg E.D., Desai U.J., Fanelli B., St Denny I., Connelly S., Kaleko M., Boettcher B.R., Caplan S.L. Treatment of type 2 diabetes by adenoviral-mediated overexpression of the glucokinase regulatory protein. Diabetes. 2001;50:1813–1820. doi: 10.2337/diabetes.50.8.1813. [DOI] [PubMed] [Google Scholar]
- 15.Mookhtiar K.A., Kalinowski S.S., Brown K.S., Tsay Y.H., Smith-Monroy C., Robinson G.W. Heterologous expression and characterization of rat liver glucokinase regulatory protein. Diabetes. 1996;45:1670–1677. doi: 10.2337/diab.45.12.1670. [DOI] [PubMed] [Google Scholar]
- 16.Veiga-da-Cunha M., Delplanque J., Gillain A., Bonthron D.T., Boutin P., Van Schaftingen E., Froguel P. Mutations in the glucokinase regulatory protein gene in 2p23 in obese French caucasians. Diabetologia. 2003;46:704–711. doi: 10.1007/s00125-003-1083-y. [DOI] [PubMed] [Google Scholar]
- 17.Warner J.P., Leek J.P., Intody S., Markham A.F., Bonthron D.T. Human glucokinase regulatory protein (GCKR): cDNA and genomic cloning, complete primary structure, and chromosomal localization. Mamm. Genome. 1995;6:532–536. doi: 10.1007/BF00356171. [DOI] [PubMed] [Google Scholar]
- 18.Vandercammen A., Van Schaftingen E. Species and tissue distribution of the regulatory protein of glucokinase. Biochem. J. 1993;294:551–556. doi: 10.1042/bj2940551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Scahftingen E., Veiga-da-Cunha M. In: Glucokinase and Glycemic Disease: From Basics to Novel Therapeutics. FM M., MA M., editors. Vol. 16. Basel: Karger; 2004. pp. 193–207. [Google Scholar]
- 20.Teplyakov A., Obmolova G., Badet-Denisot M.A., Badet B. The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase. Protein Sci. 1999;8:596–602. doi: 10.1110/ps.8.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teplyakov A., Obmolova G., Badet-Denisot M.A., Badet B., Polikarpov I. Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 A crystal structure of the isomerase domain. Structure. 1998;6:1047–1055. doi: 10.1016/s0969-2126(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 22.van Schaftingen E., Veiga-da-Cunha M., Niculescu L. The regulatory protein of glucokinase. Biochem. Soc. Trans. 1997;25:136–140. doi: 10.1042/bst0250136. [DOI] [PubMed] [Google Scholar]
- 23.Bateman A. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 1999;24:94–95. doi: 10.1016/s0968-0004(99)01357-2. [DOI] [PubMed] [Google Scholar]
- 24.Detheux M., Vandercammen A., Van Schaftingen E. Effectors of the regulatory protein acting on liver glucokinase: a kinetic investigation. Eur. J. Biochem. 1991;200:553–561. doi: 10.1111/j.1432-1033.1991.tb16218.x. [DOI] [PubMed] [Google Scholar]
- 25.Gloyn A.L., Noordam K., Willemsen M.A., Ellard S., Lam W.W., Campbell I.W., Midgley P., Shiota C., Buettger C., Magnuson M.A., et al. Insights into the biochemical and genetic basis of glucokinase activation from naturally occurring hypoglycemia mutations. Diabetes. 2003;52:2433–2440. doi: 10.2337/diabetes.52.9.2433. [DOI] [PubMed] [Google Scholar]
- 26.Grimsby J., Sarabu R., Corbett W.L., Haynes N.E., Bizzarro F.T., Coffey J.W., Guertin K.R., Hilliard D.W., Kester R.F., Mahaney P.E., et al. Allosteric activators of glucokinase: potential role in diabetes therapy. Science. 2003;301:370–373. doi: 10.1126/science.1084073. [DOI] [PubMed] [Google Scholar]
- 27.Desai U.J., Slosberg E.D., Boettcher B.R., Caplan S.L., Fanelli B., Stephan Z., Gunther V.J., Kaleko M., Connelly S. Phenotypic correction of diabetic mice by adenovirus-mediated glucokinase expression. Diabetes. 2001;50:2287–2295. doi: 10.2337/diabetes.50.10.2287. [DOI] [PubMed] [Google Scholar]
- 28.Ferre T., Pujol A., Riu E., Bosch F., Valera A. Correction of diabetic alterations by glucokinase. Proc. Natl Acad. Sci. USA. 1996;93:7225–7230. doi: 10.1073/pnas.93.14.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y., Kesavan P., Wang L.Q., Niswender K., Tanizawa Y., Permutt M.A., Magnuson M.A., Matschinsky F.M. Variable effects of maturity-onset-diabetes-of-youth (MODY)-associated glucokinase mutations on substrate interactions and stability of the enzyme. Biochem. J. 1995;309:167–173. doi: 10.1042/bj3090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veiga-da-Cunha M., Xu L.Z., Lee Y.H., Marotta D., Pilkis S.J., Van Schaftingen E. Effect of mutations on the sensitivity of human beta-cell glucokinase to liver regulatory protein. Diabetologia. 1996;39:1173–1179. doi: 10.1007/BF02658503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.