Abstract
Celiac disease is a common disease with a prevalence of ∼1%. A recent genome-wide association study (GWAS) and follow-up study identified eight loci significantly associated with celiac disease risk. We genotyped the top 1020 non-HLA single nucleotide polymorphisms (SNPs) from the GWAS study that were genotyped in the previous follow-up study. After quality control assessments, 975 SNPs were analyzed for association with 906 celiac disease cases and 3819 controls, using logistic regression. Additional genotype data were generated by imputation and analyzed across the regions showing the strongest statistical evidence for association. Twenty SNPs were associated with celiac disease with P < 0.01 in the current study as well as in the previous follow-up study, of which 16 had P < 0.001 and 11 had P < 1 × 10−11. Five of eight regions identified in the follow-up study were strongly associated with celiac disease, including regions on 1q31, 3q25, 3q28, 4q27 and 12q24. The strongest associations were at 4q27, the region most strongly associated in the GWAS and follow-up study and containing IL2 and IL21, and at 3q28 harboring LPP. In addition, we provide new evidence for an association, not previously reported, on 2q31 harboring a strong candidate gene, ITGA4. In conclusion, in this first follow-up study of celiac cases from the USA, we provide additional evidence that five of eight previously identified regions harbor risk alleles for celiac disease, and new evidence for an association on 2q31. The underlying functional mutations responsible for these replicated associations need to be identified.
INTRODUCTION
Celiac disease (gluten-sensitive enteropathy, celiac sprue) is a common disease caused by sensitivity to the dietary protein gluten. The prevalence of the disease has been estimated to be near 1% of the US and European populations (1). Celiac disease is a familial, immune-mediated disease, with significant morbidity if untreated. It is one of the most significant causes of chronic malabsorption in children. Complications of celiac disease include lymphoma, osteoporosis, anemia, miscarriages, seizures and vitamin deficiencies. Co-occurrence of celiac disease with other autoimmune disease has been noted and recent genetic studies have shown evidence for shared genetic risk factors, e.g. at 4q27 (2). The only current effective treatment for the disease is a gluten-free diet; minor dietary indiscretions can lead to recurrence of symptoms and complications.
The HLA class II DQ2 genotype, composed of the DQA1*05 and DQB1*02 alleles, has a strong association with celiac disease. The genotype is found in greater than 90% of Northern European celiac disease cases and in ∼12–26% of individuals without the disease (3–5). The HLA association is estimated to explain less than half the sibling risk (6–8), indicating the presence of additional susceptibility loci for celiac disease or autoimmunity in general. Several genome-wide linkage studies of celiac disease have been conducted. Other than a common locus at HLA, there has been little replication of results. In 2007, the results of a genome-wide association study (GWAS) of 778 celiac disease cases and 1422 population controls from the UK using 310 605 single nucleotide polymorphisms (SNPs) were reported (9). The SNP rs13119723 on chromosome 4q27 was the only SNP outside of the HLA region to show genome-wide statistical significance. This SNP was located in an ∼500 kb linkage disequilibrium (LD) block that contained the IL2 and IL21 genes, both strong candidates for celiac disease. The association to the chromosome 4q27 region was replicated in Dutch and Irish case–control collections and it was estimated that the association in the region explains less than 1% of the familial risk of celiac disease. In order to identify additional celiac disease loci and account for a greater proportion of the familial risk, Hunt et al. (10) studied the 1020 most significantly associated, non-HLA region SNPs from the GWAS in an independent set of 1643 European celiac cases and 3406 controls from the UK, the Netherlands and Ireland. The 4q27 region, with the strongest association in the GWAS, as well as seven regions that did not meet genome-wide statistical significance in the GWAS were found to be strongly associated (P-value<5 × 10−7) in the follow-up replication study. The eight associated chromosome regions, excluding HLA, harbored strong candidate genes for celiac disease including RGS1 at 1q31, IL1RL1, IL118R1, IL18RAP and SLC9A4 at 2q11–q12, CCR1 and CCR3 at 3q21, IL12A and SCHIP1 at 3q25–q26, LPP at 3q28, IL2 and IL21 at 4q27, TAGAP at 6q25 and SH2B3 and ATXN2 at 12q24. The eight loci combined were estimated to account for 3–4% of the familial risk of celiac disease, suggesting that additional disease loci remain unknown. In order to identify new disease loci and provide additional replication for the eight known loci, we analyzed 975 of 1020 SNPs genotyped in the follow-up study (10), in a sample of 906 celiac cases of self-reported European descent from the USA and 3819 ethnicity-matched controls.
RESULTS
Nine hundred and seventy-five SNPs, of the top 1020 non-HLA SNPs from the GWAS study subsequently analyzed in the follow-up study by Hunt et al. (10), passed the SNP quality control analyses and were included in the current association analysis. The full sample included 928 celiac disease cases and 3905 European controls from the Illumina iControlDB. After excluding 17 case and 38 control samples that failed the quality control assessment, the analyzed sample was 911 cases and 3867 controls. Given that the genotype data for the 3867 European controls were downloaded from a repository with data submissions from multiple sources, there was a potential for measurable genetic substructure to exist in the data (11). Principal components analysis of the identity-by-state (IBS) distance matrix computed from the 975 SNPs in 4778 case and control individuals identified three clusters (denoted Clusters 1, 2 and 3) (12). Clusters 1 and 2 constituted 99% of the samples, did not show a clear boundary and were largely attributed to clustering of the control samples. Cluster 1 included 783 cases (86%) and 2455 controls (63%), and Cluster 2 included 123 cases (13%) and 1364 controls (35%). Cluster 3 was clearly separated and included only 5 cases and 48 controls, which were excluded from subsequent analyses, leaving 906 cases and 3819 controls for the association analysis. The clustering could not be attributed to specific ancestral sources and was most likely due to experimental variation. Each SNP was analyzed for association with a case–control sample made up of individuals from Clusters 1 and 2 (subsequently referred to as the full sample), as well as the sample of cases and controls that occurred only in Cluster 1. For the former association analysis, a dichotomous cluster variable was included in the logistic regression model. The case and control samples in the full sample were 72 and 61% females, respectively, and were 72 and 63% females in Cluster 1.
Twenty SNPs with P-values for association of less than 0.01 in the full sample, and also with P-values less than 0.01 in the previous follow-up study, are shown in Table 1. All but one of these twenty SNPs also had P-values for association of less than 0.01 in the Cluster 1 sample only (rs13010713, cluster 1 P-value = 0.026). Odds ratios (OR) were computed for each of the twenty SNPs shown in Table 1 to determine if the associations were due to an excess of the major allele (OR < 1.0) or an excess of the minor allele (OR > 1.0) in the cases when compared with the controls. As shown in Table 1, the OR estimates for 18 of 20 SNPs were in the same direction as in the follow-up study (10). For the two associated SNPs on chromosome 12q24 (rs3184504 and rs653178), the OR estimates were 0.81 in our analysis, similar to the GWAS, but were greater than 1.0 in the Hunt follow-up study. This current study provides additional evidence for a celiac disease susceptibility region on chromosome 2q31. SNPs rs6433894 and 13010713 on chromosome 2q31 showed P-values of 0.00066 and 0.0023 in the current study and 0.00079 and 0.0052, respectively, in the previous follow-up study (10). The OR estimates showed that the same alleles were associated and with similar effect on risk. An additional sixteen SNPs, distributed across 15 distinct genomic regions had P-values less than 0.01 but failed to meet this significance threshold in the Hunt et al. study (Supplementary Material, Table), so they were not considered further. One of the sixteen SNPs on chromosome 9q23 (rs4008031) had a P-value less than 0.001 (P-value = 0.00036 for Clusters 1 and 2 combined). The P-values reported in Table 1 do not take into account the number of independent tests carried out in the study nor the analysis of the Cluster 1 subsample. The 975 SNPs analyzed in the study were not independent, due to LD between SNPs in specific genomic regions. However, with a conservative Bonferroni correction for 1000 tests, SNPs on chromosomes 3q28 and 4q27 maintained high statistical significance, whereas none of the other seven SNPs reported in Table 1 exceeded nominal significance.
Table 1.
Association of SNPs with P < 0.01 in both this study and the previous follow-up study
| Cytogenetic location | Position | SNP | Full sample |
Cluster 1 |
Hunt et al. (10) | Combined P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | P-value | OR | P-value | OR | ||||
| 1q31 | 190803436 | rs2816316 | 0.00038 | 0.76 | 0.002 | 0.77 | 5.11 × 10−9 | 0.71 | 4.09 × 10−10 |
| 2q31 | 181685472 | rs6433894 | 0.00066 | 1.20 | 0.0084 | 1.17 | 0.00079 | 1.16 | 1.32 × 10−5 |
| 181704290 | rs13010713 | 0.0023 | 1.18 | 0.026 | 1.14 | 0.0052 | 1.13 | 0.00022 | |
| 3q25 | 161147744 | rs17810546 | 0.00046 | 1.32 | 0.0055 | 1.26 | 7.77 × 10−7 | 1.34 | 7.36 × 10−9 |
| 161179692 | rs9811792 | 0.0014 | 1.19 | 0.0077 | 1.17 | 5.42 × 10−6 | 1.21 | 6.87 × 10−8 | |
| 3q28 | 189555207 | rs9865818 | 8.74 × 10−10 | 0.71 | 4.94 × 10−10 | 0.69 | 0.00052 | 0.86 | 1.06 × 10−9 |
| 189570322 | rs9851967 | 6.64 × 10−8 | 0.75 | 2.27 × 10−8 | 0.72 | 7.13 × 10−6 | 0.82 | 2.17 × 10−11 | |
| 189571948 | rs13076312 | 2.42 × 10−9 | 1.38 | 8.60 × 10−10 | 1.43 | 1.21 × 10−5 | 1.21 | 7.55 × 10−12 | |
| 189595248 | rs1464510 | 1.61 × 10−9 | 1.38 | 8.92 × 10−10 | 1.43 | 1.21 × 10−5 | 1.21 | 7.80 × 10−12 | |
| 189607048 | rs1559810 | 5.86 × 10−7 | 1.31 | 2.43 × 10−7 | 1.36 | 3.28 × 10−5 | 1.19 | 5.66 × 10−10 | |
| 4q27 | 123292459 | rs11938795 | 1.42 × 10−10 | 0.66 | 2.76 × 10−9 | 0.65 | 1.07 × 10−5 | 0.80 | 1.20 × 10−11 |
| 123334952 | rs13151961 | 4.24 × 10−10 | 0.60 | 2.79 × 10−9 | 0.60 | 4.32 × 10−8 | 0.72 | 1.19 × 10−14 | |
| 123437763 | rs13119723 | 8.29 × 10−5 | 0.74 | 0.0003 | 0.72 | 1.23 × 10−7 | 0.73 | 9.67 × 10−11 | |
| 123447563 | rs11734090 | 6.42 × 10−10 | 0.67 | 1.08 × 10−8 | 0.66 | 2.95 × 10−5 | 0.81 | 9.14 × 10−11 | |
| 123560609 | rs7684187 | 1.44 × 10−11 | 0.67 | 1.70 × 10−9 | 0.65 | 3.11 × 10−5 | 0.82 | 3.76 × 10−11 | |
| 123727951 | rs12642902 | 2.58 × 10−11 | 0.70 | 1.64 × 10−8 | 0.67 | 7.35 × 10−6 | 0.82 | 1.88 × 10−11 | |
| 123728871 | rs6822844 | 1.69 × 10−11 | 0.58 | 4.95 × 10−10 | 0.57 | 9.84 × 10−9 | 0.71 | 6.92 × 10−16 | |
| 123774157 | rs6840978 | 4.95 × 10−12 | 0.62 | 9.69 × 10−10 | 0.60 | 2.32 × 10−7 | 0.75 | 4.74 × 10−14 | |
| 12q24 | 110368991 | rs3184504 | 0.0017 | 0.84 | 0.0015 | 0.83 | 3.22 × 10−5 | 1.19 | 0.015 |
| 110492139 | rs653178 | 0.0022 | 0.85 | 0.0017 | 0.83 | 2.98 × 10−5 | 1.19 | 0.013 | |
Combined P-value includes the Cluster 1 P-value and Hunt et al.
The genomic control parameter, lambda (13) was estimated from the χ2 association statistics computed for all 975 SNPs with the Cluster 1 sample to be 2.129 (SE = 0.031), indicating a considerable deviation from the null expectation of 1.0. When all of the twenty SNPs shown in Table 1 as well as all other SNPs in the five regions (and HLA) were excluded, lambda was estimated at 1.068 (SE = 0.0016), indicating a slight deviation from the null which could be attributed to other SNP associations that did not meet the defined statistical significance criteria.
The combined P-values from the meta-analysis of the Cluster 1 sample and the previous follow-up study (10) are shown in Table 1. The differences in the ratio of cases to controls and the sample sizes of the two studies as well as the direction of the effect on the disease were taken into account in the meta-analysis. For all regions except chromosome 12q24, the meta-analysis resulted in a substantial increase in the significance of the association. The significance of the chromosome 12q24 SNP associations was reduced in the meta-analysis because in the current study, the minor allele decreased disease risk while in the previous follow-up study (10), the effect was the opposite. Our results were consistent with the GWAS (9) results that reported odd ratios less than 1.0 for the two SNPs on chromosome 12q24.
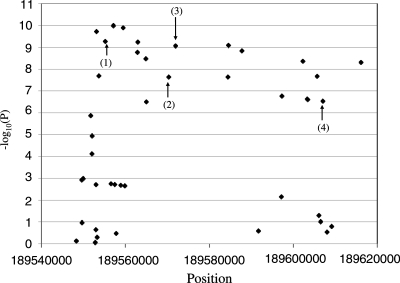
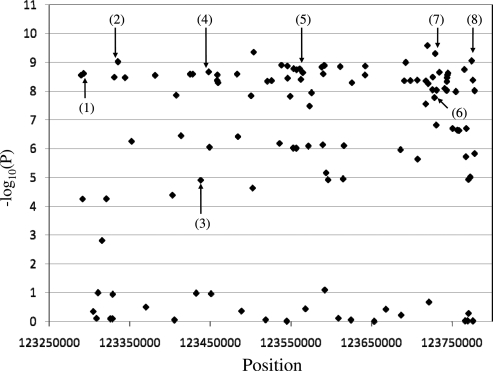
The regions on chromosomes 3q28 and 4q27 showed the strongest statistical evidence for association (Table 1). The associations on chromosome 3q28 had slightly lower statistical significance in the full sample when compared with the Cluster 1 sample, whereas the opposite relationship was observed for the chromosome 4q27 SNPs. Five SNPs spanning 51 841 bp of chromosome 3q28 were associated with P-values ranging from 2.43 × 10−7 (rs1559810) to 4.94 × 10−10 (rs9865818) with the 783 cases and 2115 controls in Cluster 1. All five of the 3q28 SNPs map within the LPP gene. Eight SNPs spanning 481 698 bp of chromosome 4q27 showed P-values for association with celiac disease ranging from 3.00 × 10−4 (rs7684187) to 4.95 × 10−10 (rs6822844) with the Cluster 1 sample. The nearly 482 kb region contains three known genes, ADAD1, IL2 and IL21. Within both regions, all of the SNPs showed high levels of LD (D′ ≥ 0.80 and R2 ≥ 0.5) Using the HapMap database, the LD across the chromosome 3q28 and 4q27 associated regions was analyzed to identify the boundaries of continuous strong LD, i.e. LD blocks. Genotype imputation was used to generate data from unmeasured SNPs within the regions of strong LD using haplotype information from the HapMap database. The association analysis results for four of five SNPs on chromosome 3q28 and an additional 46 SNPs that were generated through imputation are shown in Figure 1. The SNP rs1464510 was not included in this analysis shown in Figure 1, because HapMap genotype data were unavailable. The strongest evidence for association was found near the genotyped SNP rs9865818. Eighteen of the imputed SNPs showed P-values less than 0.00001 and were distributed across the entire region of high LD. The association analysis results for the eight SNPs genotyped on chromosome 4q27 and an additional 138 SNPs generated through imputation are shown in Figure 2. Seventy-six of the imputed SNPs were associated with P-values less than 1 × 10−5. For both regions, the imputed SNPs did not provide any additional localization information.
Figure 1.
Association results for 47 SNPs generated by imputation and four SNPs with measured genotypes across chromosome 3q28. The region spans ∼72 kb between positions 189 545 207 and 189 617 048. Measured SNPs are indicated in the figure as: (1) rs9865818, (2) rs9851968, (3) rs13076312 and (4) rs1559810.
Figure 2.
Association results for 137 SNPs generated by imputation and eight SNPs with measured genotypes across chromosome 4q27. The region spans ∼492 kb between positions 123 287 459 and 123 779 157. Measured SNPs are indicated in the figure as: (1) rs11938795, (2) rs13151961, (3) rs13119723, (4) rs11734090, (5) rs7684187, (6) rs12642902, (7) rs6822844 and (8) rs6840978.
A stepwise regression analysis was used to determine whether the associations observed across the chromosome 3q28 and 4q27 regions could be attributed to multiple independent genetic effects. LD analysis of the five chromosome 3q28 SNPs shown in Table 1 identified two sets of SNPs defined by having high pairwise correlations (r2 > 0.80). Set 1 included rs9865818 and rs9851967, and Set 2 included rs13076312, rs1464510 and rs1559810. The SNPs showing the strongest associations within each of the two sets were tested in the stepwise analysis. Regression analysis indicated the two SNPs have independent effects (rs9865818 P-value=0.0099 and rs13076312 P-value=0.025) with no significant statistical interaction (rs9865818 × rs13076312 P-value=0.82). All of the imputed SNPs were then tested in a logistic regression model that included rs9865818 and rs13076312 and none were significant at P-value less than 0.05. LD analysis of the eight genotyped 4q27 SNPs shown in Table 1 identified four sets of highly correlated SNPs. Set 1 included rs11938795, rs11734090 and rs7684187. Set 2 included rs13151961, rs13118723 and rs6822844. Sets 3 and 4 included the individual SNPs rs12642902 and rs6840978, respectively. In stepwise regression analysis of rs7684187, rs6822844, rs12642902 and 6840978, the association was accounted for by rs6882284 alone with no independent effects from the other three SNPs. All imputed SNPs were then tested in a logistic regression model that included rs6882284. A set of 25 highly correlated SNPs (r2 > 0.90) showed P-values for association ranging from 0.0047 to 0.0098, with the most significant association being with the imputed SNP rs2086346. Twenty-five imputed SNPs all showed poor quality assessments from the MACH1 program (mean quality score = 0.60 and mean R2 = 0.19), suggesting that their reliability was low and diminishing the evidence for a second independent effect at the chromosome 4q27 locus. There was no statistical evidence for interaction.
DISCUSSION
For the current study, the same 1020 SNPs as in the previous follow-up study were genotyped in a sample of 911 North American celiac cases and 3867 controls of self-reported European descent. After quality control assessments, genotype data for 975 SNPs in 906 cases and 3819 controls were analyzed for association. All 21 SNPs showing strong association in the previous replication study of Hunt et al. were among the SNPs analyzed in this study. Five of the eight previously identified regions (1q31, 3q25, 3q28, 4q27 and 12q24) showed evidence for association in this study defined by at least one SNP having a P-value less than 0.01. Seventeen of the 21 SNPs reported by Hunt et al. (10) had P-values for association that were less than 0.01 in the current study, 14 had P-values less than 0.001 and a further 11 had P-values less than 1 × 10−7. The strongest associations were to the regions 3q28 (rs9865818 P-value = 4.94 × 10−10) and 4q27 (rs6840978 P-value = 4.95 × 10−12). Evidence for two independent genetic associations was observed at both of the regions; however, the evidence for multiple disease alleles at chromosome 4q27 was based on imputed genotype data with low reliability. The findings of multiple genetic effects require replication at both regions. Associations to the other four regions (1q31, 2q31, 3q25 and 12q24) did not meet nominal statistical significance after applying highly conservative Bonferroni multiple corrections that assumed all 975 SNPs were independent. A meta-analysis that combined the current results with those reported by Hunt et al. (10) resulted in the P-values for the two SNPs on chromosome 12q24 increasing to greater than 0.01. The decrease in statistical significance was due to the minor alleles at the chromosome 12q24 SNPs being protective in the current study and deleterious in the previous follow-up study. The lack of consistency in the direction of the effects suggests that the association to chromosome 12q24 is less likely to be real. Interestingly, of the five regions we replicated, three regions harbor genes controlling immune response. The 3q25–q26 region spans the IL12A gene. The RGS1 and SH2B3 genes are located in the 1q31 and 12q24 regions, respectively. The 4q27 region spans both IL2 and IL2a, two genes controlling t-cell activation. LPP is located in the 3q28 region and is a relatively uncharacterized gene which is highly expressed in the small intestine and may be important in maintaining cell shape and motility at adhesion sites.
Nine SNPs that tagged the eight celiac disease associated regions were tested for association in an Italian population sample (14). The most significant associations were to chromosome 3q28 (P = 0.0004), followed by 12q24 (P = 0.005), with a moderately significant association to 4q27 (P = 0.0313). In a Scandanavian replication study of the 4q27 (IL2/IL21) association, rs13119723 and rs6822844 were significantly associated with celiac disease, showing haplotype P-value of 0.0002 (15). These two regions 3q28 and 4q27 have now been replicated in multiple populations and require additional study to identify the causal variants. We found no evidence for association at chromosome 2q11–q12 (rs13015714 and rs917997), 3p21 (rs6441961) and 6q25 (rs1738074). These three regions all showed strong associations (P < 1 × 10−5) in the previous follow-up study and contain immune response genes. Similar to our results, the Italian study found no association to 3p21 (CCR3) and 2q11–q12 (IL18RAP) (14). They reported a moderate association at 6q25 (TAGAP) with a P-value of 0.049. In a study in Hungarian, Finnish, and Italian populations, two SNPs in IL18RAP on 2q11–q12 were tested for association with celiac disease with only the Hungarians showing significant evidence (P = 0.0001 for haplotype) (16). The failure to replicate is unlikely to be due to allelic heterogeneity, given that the associated SNP alleles showing inconsistent replication were common in all of the populations tested. It is also possible that the follow-up studies lacked adequate power to detect the weak individual effects on celiac disease risk attributed to each of the loci and greater sample sizes will be required for consistent replication. Finally, the statistically significant associations to these regions observed in the GWAS and initial follow-up could be false-positives. Determining whether these unconfirmed celiac disease loci are true will require additional association studies with large case–control samples.
The SNPs rs6433894 and rs13010713 on chromosome 2q31 showed P-values of 0.00066 and 0.0023 in the current study and 0.00079 and 0.0052, respectively, in the previous follow-up study (10), with similar effects on risk. The region was not reported in the previous follow-up study (10), because it did not meet the statistical significance threshold of P-value less than 1 × 10−5 that they defined for a multiple test corrected replication. Although the 2q31 association would not meet strict Bonferroni multiple test correction in each individual study, the combined evidence (P-value = 1.32 × 10−5) warrants further study of the locus. The associated SNPs on 2q31 are ∼325 kb from the integrin alpha-4 (ITGA4) gene. Integrins are cell surface glycoproteins involved in the adhesion (both extracellular matrix and cell–cell), migration, and activation of immune cells. Alpha-4 integrins are heterodimeric receptors consisting of an a4 subunit and either a b1 or b7 subunit, expressed on the surface of lymphocytes and monocytes. Specifically, a4b1 integrin binds to vascular-cell adhesion molecule-1 which is up-regulated on the vascular endothelium at many sites of chronic inflammation including the intestine (17,18). Tissue transglutaminase (tTG), a marker of celiac disease, mediates the interaction with a4b1 integrins with fibronectins, to promote cell adhesion and spreading (19). The a4b7 dimer interacts with mucosal addressin-cell adhesion molecule (MAdCAM-1) which specifically mediates homing of lymphocytes to the gut (20,21). MAdCAM-1 is primarily detected in inflamed intestinal tissue and has been shown to be increased at sites of inflammation in the intestinal tract of patients with inflammatory bowel disease (22,23). Thus, α4 integrin-dependent adhesion pathways appear to play a major role in celiac disease. While the SNPs rs6433894 and rs13010713 on 2q31 are not in strong LD with SNPs in ITGA4, and 40 additional SNPs spanning the candidate SNPs and the gene did not show evidence for association in the GWAS, it is possible that the SNPs on 2q31 are associated with functional variants in a regulatory region of ITGA4. Additional follow-up studies and combined data analysis of the 2q31 region are required.
In conclusion, we replicated five of eight regions identified as significantly associated with celiac disease from the previous GWAS follow-up study in a sample of US celiac disease cases and ethnically matched controls. The most significant associations were at 4q27 harboring IL2 and IL21 and at 3q28 harboring LPP. Given the statistical significance of the associations to these two regions and that they span genes that appear to play important roles in celiac disease, there is strong evidence that these loci contain alleles that affect the risk of celiac disease. DNA resequencing and functional studies of the 4q27 and 3q28 regions need to be performed so the associations can be specifically related to the disease process. In addition, we report a new association to chromosome 2q31, near the candidate gene ITGA4. All of the associations reported here and in the previous follow-up study account for less than 10% of the individual risk for celiac disease, indicating that a substantial non-HLA genetic risk for the disease is unaccounted for. Additional GWAS and follow-up studies likely will continue to identify low-risk common alleles for the disease.
MATERIALS AND METHODS
Study subjects
Celiac disease cases
The 928 celiac disease cases were enrolled in previous studies, with DNA and diagnoses data available for this study. All participants were Caucasian and had signed informed consent with approval by the respective Institutional Review Boards. Five hundred and thirty of the celiac disease cases from the Mayo Clinic were subjects previously recruited from the Rochester Epidemiology Project with a potential diagnosis of celiac disease between 1 January 1950 and 31 December 2007, from attendees at the celiac disease clinic and the pediatric gastroenterology clinics at the Mayo Clinic, and from the local celiac disease support group and records of the Mayo Clinic Department of Pathology and Laboratory Medicine. Medical records of each candidate case were reviewed, as were the biopsy slides of the small intestinal samples taken at the time of the original diagnosis. To be defined as celiac disease, each case was required to have: (i) a proximal small intestinal biopsy compatible with celiac disease and (ii) either clinical and/or histological improvement with a gluten-free diet fulfilling the accepted criteria for celiac disease or had to have a positive celiac-specific autoantibody (IgA EMA or IgA tTG antibodies). The majority of patients fulfilled all three criteria of characteristic histological change, positive celiac-specific serology and an objective response to a gluten-free diet. A small proportion of patients, predominantly those diagnosed before modern serology came into use, did not have celiac-specific serology, and another small minority of patients had positive IgA EMA and IgA tTG, but had not yet had their biopsies. An additional 398 celiac disease cases were from a family based study (268 cases with one per family) and a singleton study (132 cases) of celiac disease conducted by Dr Neuhausen at the University of California Irvine. Of the 398 cases, 242 were diagnosed with positive serology and biopsy, 125 with biopsy only and 31 with positive serology (EMA and tTG). For both sites, individuals who were self-declared celiacs because of a self administered gluten-free diet and without a biopsy, or individuals whose biopsy demonstrated only minor changes such as increased intraepithelial lymphocytes and/or crypt hyperplasia, were not included as cases.
Controls
We used existing genotype data from the Illumina iControlDB controls database, matching by age, sex and ethnicity (all Caucasian). Given the prevalence of celiac disease and a published analysis, we did not expect a significant reduction in power to result from using the US population controls from iControlDB (24).
Genotyping and quality control
For genotyping, we utilized the pooled oligo set (OPA) from the Hunt et al. replication study composed of 1025 SNPs. Genotyping was performed using the GoldenGate assay (Illumina) on a BeadStudio (Illumina). Quality control tests were carried out using the GenABEL library (25) of the R-statistical package (http://www.r-project.org/). SNPs with less than 95% complete data across all samples were excluded. SNPs were filtered from subsequent analysis if they had P-values for Hardy–Weinberg equilibrium (HWE) less than 0.00001. All of the SNPs showing strong evidence for association were in HWE in the controls and combined samples. Samples with less than 95% complete data were excluded from the association analysis. The IBS was assessed for each sample pair to identify related individuals or duplicate samples. Duplicate samples that were included in the genotyping process to assess intra-sample variation showed greater than 99% concordance and no unplanned duplicate samples were identified.
Genotype statistical analysis
The 975 SNPs that met the quality control standards were used to test for genetic substructure among the case and control samples. The procedure implemented in GenABEL closely resembles the principal components approach first described by Price et al. (12). First, a matrix of average IBS sharing for all pairs of individuals is computed using the autosomal markers. The IBS matrix is transformed to a distance matrix to which classical multidimensional scaling is applied. The first two principal components were plotted and the number of clusters was inferred. The analysis identified two major clusters and a third cluster made up of several ethnic outliers. The outliers were excluded from the association analysis. The dichotomous celiac disease variable was analyzed for association with the SNP genotypes using a logistic regression model that included sex as a covariate. Genotypes were coded as 0, 1 or 2, indicating the number of copies of the minor allele in the genotype. The effect of the additive genotype parameter was estimated assuming that the variable had a continuous distribution. The ORs reflect the effect from the addition of one copy of the minor allele to the genotype. Missing genotypes and genotypes from unmeasured SNPs across the two regions were imputed using the program MACH1 (26). Phased haplotypes were downloaded from the HapMap database (http://www.hapmap.org) as input for the imputation. The estimated allele dose for each imputed SNP was analyzed as described for the measured SNPs. The allele dose was the product of the computed posterior probability of each genotype given the measured genotype data and the HapMap phased haplotype data and the allele dose for the genotype (0, 1 or 2 reflecting the number of minor alleles in the genotype), summed over the three possible genotypes. For the meta-analysis, P-values were combined using methods described by Rosenthal (27) for adding weighted Z statistics, where the weights in the current study were the sample sizes. The proportion of cases to controls was uneven and different between the two studies so a standardized sample size for 1:1 matching was first computed for each study based on a specific power and genetic model. P-values were converted to standard normal deviates (Zs) with the sign of the deviate reflecting the direction of the genetic effect, i.e. ORs less than 1 were given negative Zs and positive otherwise. A Zs for the combined studies was calculated as the sum of the products of the weights (standardized sample sizes) and the observed Zs. The sum was divided by the square root of the sum of the squared weights. The calculated Zs from the two studies were then converted to P-values.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health DK50678 (to S.L.N.), DK57892 (to J.A.M.) and DK80490 (to S.L.N. and J.A.M.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Linda Steele for providing us with the sample lists and accompanying data.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fasano A., Berti I., Gerarduzzi T., Not T., Colletti R.B., Drago S., Elitsur Y., Green P.H., Guandalini S., Hill I.D., et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch. Intern. Med. 2003;163:286–292. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- 2.Zhernakova A., Alizadeh B.Z., Bevova M., van Leeuwen M.A., Coenen M.J., Franke B., Franke L., Posthumus M.D., van Heel D.A., van der Steege G., et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am. J. Hum. Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balas A., Vicario J.L., Zambrano A., Acuna D., Garcia-Novo D. Absolute linkage of celiac disease and dermatitis herpetiformis to HLA-DQ. Tissue Antigens. 1997;50:52–56. doi: 10.1111/j.1399-0039.1997.tb02834.x. [DOI] [PubMed] [Google Scholar]
- 4.Sollid L.M., Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910–922. doi: 10.1016/0016-5085(93)90912-v. [DOI] [PubMed] [Google Scholar]
- 5.Tighe M.R., Ciclitira P.J. The implications of recent advances in coeliac disease. Acta Paediatr. 1993;82:805–810. doi: 10.1111/j.1651-2227.1993.tb17614.x. [DOI] [PubMed] [Google Scholar]
- 6.Bevan S., Popat S., Braegger C.P., Busch A., O'Donoghue D., Falth-Magnusson K., Ferguson A., Godkin A., Hogberg L., Holmes G., et al. Contribution of the MHC region to the familial risk of coeliac disease. J. Med. Genet. 1999;36:687–690. [PMC free article] [PubMed] [Google Scholar]
- 7.Petronzelli F., Bonamico M., Ferrante P., Grillo R., Mora B., Mariani P., Apollonio I., Gemme G., Mazzilli M.C. Genetic contribution of the HLA region to the familial clustering of coeliac disease. Ann. Hum. Genet. 1997;61:307–317. doi: 10.1046/j.1469-1809.1997.6140307.x. [DOI] [PubMed] [Google Scholar]
- 8.Risch N. Assessing the role of HLA-linked and unlinked determinants of disease. Am. J. Hum. Genet. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 9.van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton D.G., Walker N.M., Smyth D.J., Pask R., Cooper J.D., Maier L.M., Smink L.J., Lam A.C., Ovington N.R., Stevens H.E., et al. Population structure, differential bias and genomic control in a large-scale, case–control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 12.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 14.Romanos J., Barisani D., Trynka G., Zhernakova A., Bardella M.T., Wijmenga C. Six new celiac disease loci replicated in an Italian population confirm association to celiac disease. J. Med. Genet. 2009;61:60–63. doi: 10.1136/jmg.2008.061457. [DOI] [PubMed] [Google Scholar]
- 15.Adamovic S., Amundsen S.S., Lie B.A., Gudjonsdottir A.H., Ascher H., Ek J., van Heel D.A., Nilsson S., Sollid L.M., Torinsson Naluai A. Association study of IL2/IL21 and FcgRIIa: significant association with the IL2/IL21 region in Scandinavian coeliac disease families. Genes Immun. 2008;9:364–367. doi: 10.1038/gene.2008.27. [DOI] [PubMed] [Google Scholar]
- 16.Einarsdottir E., Koskinen L.L., Dukes E., Kainu K., Suomela S., Lappalainen M., Ziberna F., Korponay-Szabo I.R., Kurppa K., Kaukinen K., et al. IL23R in the Swedish, Finnish, Hungarian and Italian populations: association with IBD and psoriasis, and linkage to celiac disease. BMC Med. Genet. 2009;10:8. doi: 10.1186/1471-2350-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobb R.R., Hemler M.E. The pathophysiologic role of alpha 4 integrins in vivo. J. Clin. Invest. 1994;94:1722–1728. doi: 10.1172/JCI117519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bevilacqua M.P. Endothelial-leukocyte adhesion molecules. Ann. Rev. Immun. 1993;11:767–804. doi: 10.1146/annurev.iy.11.040193.004003. [DOI] [PubMed] [Google Scholar]
- 19.Akimov S.S., Krylov D., Fleischman L.F., Belkin A.M. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postigo A.A., Teixido J., Sanchez-Madrid F. The alpha 4 beta 1/VCAM-1 adhesion pathway in physiology and disease. Res. Immun. 1993;144:723–735. doi: 10.1016/s0923-2494(93)80057-6. discussion 754–762. [DOI] [PubMed] [Google Scholar]
- 21.Farstad I.N., Halstensen T.S., Kvale D., Fausa O., Brandtzaeg P. Topographic distribution of homing receptors on B and T cells in human gut-associated lymphoid tissue: relation of L-selectin and integrin alpha 4 beta 7 to naive and memory phenotypes. Am. J. Pathol. 1997;150:187–199. [PMC free article] [PubMed] [Google Scholar]
- 22.Briskin M., Winsor-Hines D., Shyjan A., Cochran N., Bloom S., Wilson J., McEvoy L.M., Butcher E.C., Kassam N., Mackay C.R., et al. Human mucosal addressin cell adhesion molecule-1 is preferentially expressed in intestinal tract and associated lymphoid tissue. Am. J. Pathol. 1997;151:97–110. [PMC free article] [PubMed] [Google Scholar]
- 23.Issekutz T.B. Inhibition of in vivo lymphocyte migration to inflammation and homing to lymphoid tissues by the TA-2 monoclonal antibody. A likely role for VLA-4 in vivo. J. Immunol. 1991;147:4178–4184. [PubMed] [Google Scholar]
- 24.Garner C. The use of random controls in genetic association studies. Hum. Hered. 2006;61:22–26. doi: 10.1159/000091833. [DOI] [PubMed] [Google Scholar]
- 25.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 26.Li Y., Abecasis G.R. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 27.Rosenthal R. Combining results from independent studies. Psychol. Bull. 1978;85:185–193. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.