Abstract
Fluorescein-labeled oligodeoxynucleotides (oligos) were introduced into cultured rat myoblasts, and their molecular movements inside the nucleus were studied by fluorescence correlation spectroscopy (FCS) and fluorescence recovery after photobleaching (FRAP). FCS revealed that a large fraction of both intranuclear oligo(dT) (43%) and oligo(dA) (77%) moves rapidly with a diffusion coefficient of 4 × 10−7 cm2/s. Interestingly, this rate of intranuclear oligo movement is similar to their diffusion rates measured in aqueous solution. In addition, we detected a large fraction (45%) of the intranuclear oligo(dT), but not oligo(dA), diffusing at slower rates (≤1 × 10−7 cm2/s). The amount of this slower-moving oligo(dT) was greatly reduced if the oligo(dT) was prehybridized in solution with (unlabeled) oligo(dA) prior to introduction to cells, presumably because the oligo(dT) was then unavailable for subsequent hybridization to endogenous poly(A) RNA. The FCS-measured diffusion rate for much of the slower oligo(dT) population approximated the diffusion rate in aqueous solution of oligo(dT) hybridized to a large polyadenylated RNA (1.0 × 10−7 cm2/s). Moreover, this intranuclear movement rate falls within the range of calculated diffusion rates for an average-sized heterogeneous nuclear ribonucleoprotein particle in aqueous solution. A subfraction of oligo(dT) (15%) moved over 10-fold more slowly, suggesting it was bound to very large macromolecular complexes. Average diffusion coefficients obtained from FRAP experiments were in agreement with the FCS data. These results demonstrate that oligos can move about within the nucleus at rates comparable to those in aqueous solution and further suggest that this is true for large ribonucleoprotein complexes as well.
An understanding of the physical environment inside the cell nucleus is central to a coherent view of gene expression. It is important to know how the viscosity and molecular diffusion rates in the nucleus of a living cell compare with experimental conditions in vitro, where interactions between nucleic acids and proteins are studied at high dilution in aqueous solution. For example, it is not clear whether ribonucleoprotein (RNP) complexes can diffuse freely about the nucleus, impeded only by locally high concentrations of macromolecules or, alternatively, are bound or compartmentalized in such a way as to constrain their motion. Some observations suggest that pre-mRNA transcripts are tethered to elements of the transcriptional, splicing, and/or polyadenylation machinery (1–3), and it has been proposed that processed mRNAs make their way out of the nucleus by molecular diffusion (4, 5). However, mediated processes have not been ruled out, and the functional relationships between RNA export and nuclear structure remain unclear (6).
Recently, Politz et al. (7) characterized nucleic acid uptake and hybridization in living cells by in situ reverse transcription and found that fluorescently labeled oligo(dT) can be taken up by living cells and form hybrids with intranuclear poly(A) RNA. These hybrids are stable for at least 24 h in the living cell and cause no obvious cytotoxic effects. The development of this system suggested the use of fluorescently labeled oligo(dT) as a probe to measure both the diffusion rate of a small molecule and, as well, of poly(A) RNA [by virtue of hybridized oligo(dT)], in the nucleus of living cells by using biophysical techniques.
Fluorescence recovery after photobleaching (FRAP) has traditionally been used to measure rates of diffusion in membranes and in cells (8). A small volume of a living cell containing fluorescently labeled molecules is photobleached, and the rate at which fluorescent molecules move back into the bleached volume is monitored. A diffusion coefficient can be calculated from the rate of fluorescence recovery. However, technical limitations in FRAP studies have typically precluded the identification of more than one diffusing component (9), so that detection of different molecular populations moving at different rates concurrently, as can be anticipated in many intracellular situations, is not practical.
Fluorescence correlation spectroscopy (FCS), a type of concentration correlation analysis, has been widely applied to study molecular diffusion both in solution and in model membranes (10, 11). FCS measures fluctuations in fluorescence intensity that result from diffusion of fluorescent molecules in and out of a small open volume and then employs autocorrelation analysis to determine the correlation between intensity deviations at some point in time with intensity deviations at later points in time. The diffusion coefficient is related to the rate of decay of correlation; the more slowly a species diffuses, the longer correlation persists. Recently, refined algorithms that allow the modeling of up to three components diffusing at different rates have been coupled with avalanche photodiode detection and confocal optics systems to increase the sensitivity of FCS and potentiate the study of molecular movement in living cells (12–14). These more sensitive systems have been used to monitor hybridization of nucleic acids in solution (15–17) but not yet in living cells.
In the present investigation, FCS was used to measure the molecular motion of both oligo(dT) and oligo(dA) inside the nucleus of living cells. FRAP measurements were also carried out for comparison. We found that the majority (77%) of oligo(dA) diffused at similar rates in both the nucleus and in aqueous solution. However, about 45% of intranuclear oligo(dT), but not oligo(dA), moved more slowly, consistent with its hybridization to intranuclear poly(A) RNA. A substantial fraction of this slower-moving population moved at rates within the range predicted for the diffusion rate of a pre-mRNA:heterogeneous nuclear RNP particle (hnRNP particle) in aqueous solution.
MATERIALS AND METHODS
Oligo Synthesis, Purification, and Labeling.
Oligo(dT) and oligo(dA) 43 mers containing a 5′-aminohexyl-modified thymidine (Glen Research, Sterling, VA) at nucleotide positions 2, 12, 22, 32, and 42 were synthesized by using standard phosphoramidite chemistry and coupled with fluorescein isothiocyanate as described (18). Oligos were labeled with an average of 2–4 fluorochromes/molecule (calculated from A488/A260 by using extinction coefficients for the fluorochrome and the homopolymers).
Cell Culture and Oligo Uptake.
L6 rat myoblasts were cultured in DMEM with 10% heat-inactivated fetal calf serum (hereafter referred to as “serum”). For fluorescence correlation spectroscopy, about 12,500 cells in 500 μl DMEM/10% serum were plated per well in small eight-chambered glass-bottomed dishes (Nunc) and allowed to grow for ≈24 h at 37°C in 5% CO2. For FRAP, 25,000 cells were plated onto 12-mm round glass coverslips and grown overnight as above. All data reported were obtained with cells actively growing at subconfluence.
For FCS studies, cells were incubated with 2 μM oligo in medium (with serum). For FRAP measurements, 1.5 μl of cationic lipid (Pfx no. 4, Invitrogen, San Diego) was premixed with 0.45 pmol of oligo in 300 μl DMEM and then added to cells. After a 2-h incubation, cells were rinsed three times with DMEM/10% serum and incubated at least 1 h at 37°C in 5% CO2 before analysis. These oligo concentrations and incubation times were found to give maximal levels of oligo(dT) hybridization (detected as in ref. 7) with no obvious effects on cell morphology or growth rate (again see ref. 7). In one experiment, cells were incubated for 90 min with 20 mM NaN3 (19) in DMEM/10% serum and subjected to FCS (see below) in Leibovitz L-15 (GIBCO/BRL) medium containing 20 mM NaN3.
FRAP.
FRAP measurements were performed by using a previously described workstation (8). All FRAP measurements were made at room temperature with a Zeiss ×63 oil 1.4 NA plan Apochromat objective. Solution measurements were performed by using 0.05-mm square capillary tubes (Vitrodynamics, Rockaway, NJ) containing 20 nM fluorescein-labeled oligo(dT) or oligo(dA) in Leibovitz L-15 medium. FRAP measurements on nuclei were performed on cells growing in Leibovitz L-15 (with serum), on coverslips mounted on slides. Photobleaching was at ≈1.3 mW for 10 ms with an exp(−2) beam radius of 1 μm (≈40 kW/cm2). Monitoring intensity was ≈0.13 μW. Measured diffusion coefficients were normalized by using the published diffusion coefficient for BSA (see ref. 20) to correct for changes in beam radius along the optical axis.
FCS Measurements.
FCS measurements were made by using a Zeiss-Evotec (Jena, Dusseldorf, Germany and Thornwood, NY) ConfoCor spectrofluorimeter (see ref. 14). This system is similar to a conventional confocal microscope in that the laser beam is focused to an exp(−2) radius of ω ≈0.2 μm in the x-y plane perpendicular to the optic axis and an exp(−2) radius of δ along the optic axis of ≈0.6 μm. The illumination source is an Ar laser (488 nm and 514 nm) coupled with a fiber optic to a Zeiss Axiovert microscope. This microscope has a electronic x-y-z stage and an electronically adjustable and positionable aperature at the image plane. The detector is a fast quenching avalanche photodiode with a dead time of 33 ns. The system also has a charge-couple device (CCD) camera that is used for adjustment, calibration, and location of the appropriate region of the sample. Data are processed by a 288-channel logarithmic autocorrelator with adjustable sampling times from 200 ns to 3438.8 s.
In FCS, one measures fluctuations with time, t, in fluorescence intensity, ΔI(t), about the mean intensity, 〈I〉, which result from diffusion of fluorescent molecules in and out of a confocal volume element, as well as from other phenomena that can alter fluorescence (21, 22). We calculate the correlation between one point in time and later points in time by dividing the data stream into a series of counting intervals of duration Δt. Then each experimental point in time can be expressed as the product of an integer (either m or n) and Δt, where m and n are time bin numbers. To calculate the correlation between the fluctuation in intensity at any arbitrary point in time, nΔt, and later points in time, nΔt + mΔt, the autocorrelation function G(m) is determined
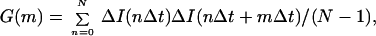 |
1 |
where ΔI(t) represents the fluctuation in intensity at a given time, I(t) − 〈I〉. In integral form this becomes
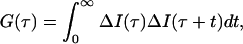 |
2 |
where we have replaced mΔt in the limit of infinitesimal Δt with τ.
In the case of multiple component diffusion in three dimensions, the autocorrelation function takes the form
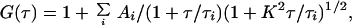 |
3 |
where Ai and τi represent the fraction and diffusion time for a given component i, and where the so-called structural parameter K = ω/δ. These diffusion times may in turn be related to their respective diffusion coefficients, D, by
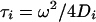 |
4 |
The structure parameter and ω are determined by fitting the diffusion of free fluorescein, assuming D = 3.6 × 10−6 cm2/s, to Eq. 3 with one component.
For solution measurements, 200 μl of 20 nM oligo in water (or in Leibovitz L-15 medium) were placed in one well of an eight-chambered dish and the laser beam was focused 150 μm above the dish bottom in a known confocal volume of ≈1 × 10−15 liter. For live cell FCS, cells were incubated in Leibovitz L-15 medium (without serum or phenol red). This medium supported continued cell viability on the microscope stage at room temperature without CO2 and also contributed only low background autofluorescence signal to FCS readings. The laser beam was then focused (again in the same known confocal volume) inside the nucleus of a treated (or untreated) cell by adjusting both the stage position and focal plane. Neutral density filters were used to attenuate laser intensity and reduce detection of cellular autofluorescence. The fluorescence intensity was noted and the z plane was adjusted up or down 1–2 μm to obtain the maximal intensity reading. Care was taken to focus in the interior of the nucleus, which in these myoblasts is typically 5–8 μm thick, and measurements were not taken inside or touching nucleoli. A series of 10 measurements of the fluorescence intensity fluctuation (each over 20 s) was then captured and stored. Cells were exposed to ≈5 μW of excitation light which is equivalent to ≈100 μJ over the 0.23-μm2 average cross section of the confocal volume. Cells remained viable for at least 24 h after treatment when returned to the CO2 incubator at 37°C. Photoexposure in this general range has also been used to study Drosophila embryos with no detectable side effects (23).
Solution Hybridization.
A 7.5-kb poly(A) RNA that was transcribed in vitro from a chimeric plasmid was purchased from GIBCO/BRL. The manufacturer estimates about 40% of the RNA molecules contain poly(A) tails with an average length of 150 nt. Aliquots (1 μl) of a 0.8 μM solution containing this RNA were added sequentially to 200 μl of 20 nM fluorescein-labeled oligo(dT) in PBS/10 mM MgCl2. Before and after each addition, FCS was performed as described above for solution studies.
Data Analyses.
The Evotec software integrated with the ConfoCor instrument fits data by nonlinear least squares analysis to one, two, or three diffusion components as well as quantifying a fraction undergoing intersystem conversion from the fluorescein singlet to triplet state. The optimal fit was chosen by comparing the following parameters among possible fits: shape of residual curve, value of relative least square, and standard deviations associated with computed fractions and diffusion times. In some data sets, the optimal fit required postulating a very rapidly moving fraction with a transit time (through the confocal volume) of less than 0.1 ms. This fraction typically represented about 20% of the total signal in an FCS measurement, both in solution and in cells. Diffusion rates for various medium components and free fluorescein fall into this range, and other investigators have observed this ultrafast fraction in FCS experiments as well (16). We do not know what this fraction represents, but we did not exclude it from the fits when it appeared.
To combine and compare data among different experiments in cells, the best fits from all the measurements from each experiment were sorted into five distinct ranges of diffusion times. The average fraction of oligo and its average diffusion time within each of the five ranges was then calculated after weighting each value to the number of fluorescent particles present in each range.
RESULTS
Diffusion Times of Oligos in Solution Measured By Using FCS and FRAP.
Initial experiments were carried out to measure diffusion times of oligos in aqueous medium (Leibovitz L-15) by using both FCS and FRAP. Oligos were 43 nt long and labeled with fluorescein as described. In FCS measurements, solutions containing either oligo most often fit a one-component model with an average diffusion time (transit time through the confocal volume) of about 0.3 ms (Table 1, Fig. 1A). In some cases, the presence of a second, very rapidly moving component was also observed (see Materials and Methods). Oligo diffusion coefficients were also measured by using conventional FRAP techniques and found to be in the same range as diffusion coefficients calculated from FCS diffusion times (5–7 × 10−7 cm2/s, Table 1).
Table 1.
Diffusion coefficients in aqueous solution
| BSA | oligo(dA) | oligo(dT) | |
|---|---|---|---|
| FCS t (ms) | 0.25 ± 0.01 | 0.34 ± 0.01 | 0.38 ± 0.01 |
| FCS D (10−7 cm2/s) | 7.6 ± 0.1 | 5.7 ± 0.2 | 5.0 ± 0.2 |
| FRAP D (10−7 cm2/s) | 6.5 ± 0.6 | 7.0 ± 0.6 | 4.9 ± 0.6 |
t, Diffusion time; D, diffusion coefficient. Errors are standard errors of the mean.
Figure 1.
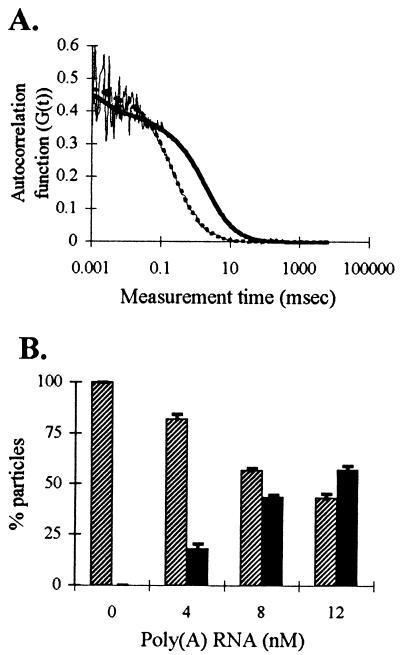
Hybridization of oligo(dT) to poly(A) RNA in solution measured by FCS. Aliquots containing 0.4 pmol unlabeled poly(A) RNA (7.5 kb) were added to 20 nM fluorescein-labeled oligo(dT) and, before and after each addition, ten 20-s FCS samplings were performed. (A) Autocorrelation curves (thin lines) obtained from oligo(dT) alone in solution and after last addition of poly(A) RNA. The dashed thick [oligo(dT) alone] and solid thick [hybridized oligo(dT)] lines are the best fit to each autocorrelation curve by using nonlinear least-squares analysis. (B) Histogram showing the extent of hybridization with increasing concentrations of poly(A) RNA. Hatched bars represent the fraction of oligo(dT) moving at 0.23 ms (unhybridized); solid bars represent the fraction of oligo(dT) moving at 1.9 ms (hybridized).
Solution Hybridization of Oligo(dT) to Poly(A) RNA.
We next used FCS to monitor hybridization of oligo(dT) to poly(A) RNA and to measure the mobility of the oligo(dT)/poly(A) RNA hybrid in aqueous solution. When a 7.5-kb polyadenylated RNA was added to fluorescein-labeled oligo(dT) in solution, the autocorrelation curve shifted to the right, indicating the presence of a slower-moving component (Fig. 1A). This curve best fit a two-component model, with one species fixed at the free oligo diffusion time (0.23 ms) and a slower component moving at a diffusion time of 1.9 ms (D = 1.0 × 10−7 cm2/s). This diffusion coefficient is close to the value calculated for a globular molecule the size of this poly(A) RNA (D = 1.1 × 10−7 cm2/s). As more RNA was added, an increasingly greater percentage of oligo(dT) moved at a slower diffusion time, indicating an increase in the amount of oligo(dT) hybridized (Fig. 1B). Oligo(dA) diffusion time was not altered upon addition of poly(A) RNA (not shown), confirming that the observed binding of oligo(dT) to poly(A) RNA represented specific hybridization.
Diffusion Rates of Oligos in Nuclei of L6 Myoblasts.
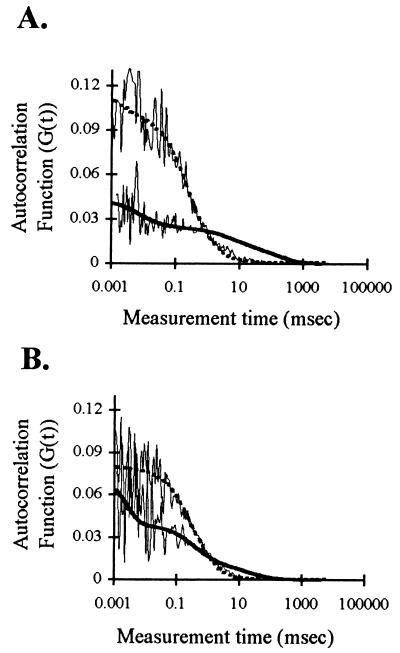
Cells were grown in medium containing fluorescein-labeled oligo(dT) or oligo(dA) for 2 h followed by incubation for at least 1 h in oligo-free medium to allow efflux of excess unbound oligo. Earlier work has shown that under these conditions, fluorescein-labeled oligo(dT) is taken up by cultured L6 myoblasts and can hybridize to intranuclear poly(A) RNA targets (7). The diffusion times of the oligos within the nucleus were then measured by FCS. As shown in Fig. 2A, the shapes of the autocorrelation functions were different for oligo(dT) vs. oligo(dA). A substantial amount of oligo(dT) moved at a slower rate than oligo(dA) as evidenced by the tail of the correlation curve for oligo(dT) extending out to slower diffusion times. To confirm that this fraction represented oligo(dT) molecules hybridized to poly(A) RNA in the nucleus, fluorescein-labeled oligo(dT) was prehybridized in vitro with a 1.5 molar excess of unlabeled oligo(dA) before incubation with cells. This prehybridization step changed the shape of the autocorrelation curve to show a reduction in the amount of oligo(dT) moving at the slower rate in the nucleus, as would be expected if the oligo(dT) already bound to oligo(dA) were unable to hybridize to the intranuclear poly(A) RNA targets (Fig. 2B).
Figure 2.
FCS autocorrelation curves of oligo(dT) and oligo(dA) in nuclei. L6 myoblasts were incubated with fluorescein-labeled oligo(dT) or oligo(dA) and the movements of the oligos in nuclei were measured by FCS as described. (A) Autocorrelation curves (thin lines) and best fit curves (obtained by using nonlinear least-squares analysis) for nuclei containing oligo(dT) (thick solid line) or oligo(dA) (thick dashed line). (B) Fluorescein-labeled oligo(dT) was prehybridized in vitro with unlabeled oligo(dA) and then incubated with cells as usual before FCS analysis. The thick dashed line is the best fit curve for the prehybridized oligo(dT) autocorrelation curve (thin line); the solid thick line is the best fit curve for an autocorrelation curve of oligo(dT) (not prehybridized) from the same experiment.
The y-intercept of the autocorrelation curves is the inverse of the number of fluorescent particles in the confocal volume. It was interesting to note that the controls, oligo(dA) and prehybridized oligo(dT), had higher y-intercepts, indicating fewer particles were present in the confocal volume (20 ± 1 and 15 ± 2, respectively), than with oligo(dT) alone (36 ± 1). However, these particle numbers do not neccessarily predict oligo concentration because more than one fluorescent molecule may be present in a particle.
To determine diffusion times, autocorrelation functions were fit to a diffusion model by using nonlinear least-squares analysis, as above. In contrast to solution measurements, the cell experiments with oligo(dT), and to a lesser extent oligo(dA), showed variation between FCS measurements, both in different areas of the same nucleus and in different nuclei. This result was evidenced by the fact that, although autocorrelation functions derived from cells containing oligo(dT) most often fit a model which predicted the presence of two components diffusing at different rates, in some cases the fit was significantly improved with a one- or three-component model (see Materials and Methods). Data for intranuclear oligo(dA) most often best fit a one-component model. We attribute this variation to the presence of different microenvironments in the nucleus and not experimental error, because (i) the variation was more pronounced in the situation [i.e., oligo(dT)] where we expected to see more than one category of molecules moving at different rates, and (ii) we did not see this type of variation when using FCS to measure oligo diffusion in solution. We therefore selected the best fit for each measurement within an experiment.
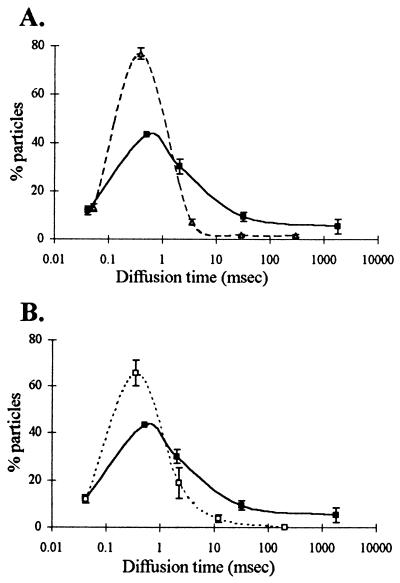
This finding required us to subsequently sort the data sets from several experiments into bins or ranges based on mean translational diffusion times. An average of 77% intranuclear oligo(dA) molecules and 43% intranuclear oligo(dT) molecules fell into a range representing diffusion times between 0.1 and 1 ms (Table 2). The diffusion times of these oligos in aqueous solution also fall in this range (see Table 1). This correlation indicates that a significant fraction of oligodeoxynucleotides diffuse in the cell nucleus at a rate similar to that observed in aqueous solution.
Table 2.
Oligonucleotide mobility in cell nuclei
| 0–0.1 ms | 0.1–1 ms | 1–10 ms | 10–100 ms | >100 ms | |
|---|---|---|---|---|---|
| Oligo(dT) (n = 228) | |||||
| % Particles | 12.0 ± 1.4 | 43.2 ± 2.9 | 30.1 ± 2.9 | 9.4 ± 1.1 | 5.3 ± 0.9 |
| t (ms) | 0.042 ± 0.002 | 0.517 ± 0.015 | 2.07 ± 0.07 | 32.45 ± 1.00 | 1,838 ± 211 |
| D (10−8 cm2/s) | 452.4 ± 21.5 | 36.75 ± 1.07 | 9.07 ± 0.29 | 0.59 ± 0.02 | 0.010 ± 0.001 |
| Oligo(dA) (n = 217) | |||||
| % Particles | 13.0 ± 1.6 | 76.7 ± 2.3 | 7.1 ± 1.2 | 1.7 ± 0.4 | 1.5 ± 0.5 |
| t (ms) | 0.054 ± 0.003 | 0.387 ± 0.014 | 3.55 ± 0.22 | 29.2 ± 1.06 | 301.8 ± 14.6 |
| D (10−8 cm2/s) | 351.9 ± 19.6 | 49.1 ± 1.8 | 5.36 ± 0.33 | 0.65 ± 0.02 | 0.062 ± 0.003 |
| Prehybridized oligo(dT) (n = 73) | |||||
| % Particles | 11.9 ± 1.7 | 65.5 ± 5.6 | 18.8 ± 6.5 | 3.8 ± 1.6 | 0.01 ± 0.01 |
| t (ms) | 0.043 ± 0.004 | 0.347 ± 0.017 | 2.27 ± 0.09 | 12.13 ± 0.13 | 203.4 ± 0.1 |
| D (10−8 cm2/s) | 441.9 ± 30.8 | 54.8 ± 2.68 | 8.37 ± 0.31 | 1.57 ± 0.02 | 0.092 ± 0.000 |
Errors are standard error of the mean.
The slow-moving molecules present in oligo(dT) treated cells, with diffusion times of 1 ms or more (diffusion coefficients between 1 × 10−10 and 9 × 10−8 cm2/s), represented an average of 45% of the total oligo(dT) molecules detectable in the nucleus by FCS (Table 2, columns 3–5; Fig. 3). As explained above, this fraction likely contained oligo(dT) hybridized to poly(A) RNA because we saw much less of this fraction in cells treated with oligo(dA) (10%) or in cells treated with oligo(dT) that had been prehybridized in vitro to oligo(dA) (22%) (Table 2, Fig. 3 A and B, respectively). The diffusion times of this slow-moving population were very heterogeneous (1 ms to more than 1,000 ms), which suggested that poly(A) RNA might be moving at a variety of rates within the nucleus. The proportion of oligo(dT) present in this slow moving fraction did not change when cells were treated with sodium azide under conditions that deplete cells of ATP, suggesting that movement at this rate did not require energy (results not shown).
Figure 3.
Summary of FCS-determined oligo diffusion rates in L6 nuclei. Multiple FCS readings were performed in nuclei of cells treated with oligo(dT), oligo(dA) or prehybridized oligo(dT) (see Fig. 2), and the data obtained from the best fit for each measurement were sorted into five categories representing various diffusion times. The average fraction of oligo and its average diffusion time within each of the five categories were calculated after weighting each value by using the number of fluorescent particles present in each category (Table 2) and then plotted. (A) Plot showing distribution of oligo(dT) (solid line) compared with oligo(dA) (dashed line). (B) Distribution of oligo(dT) (solid line) compared with prehybridized oligo(dT) (dashed line, see Fig. 2). The vertical bars represent standard error of the mean.
An additional oligo(dT) component appeared in some cells that we were unable to study by FCS. This component was very bright and bleached rapidly during the first few measurements made at some sites, indicating that some oligo(dT)-bound molecules did not move from the confocal volume throughout the sampling time (20 s).
FRAP Analysis of Oligo Diffusion in Nuclei.
Because our FRAP instrument has a beam radius approximately four times larger than the FCS instrument, we had to increase the amount of fluorescent oligo signal in cells to carry out FRAP measurements. To do this, L6 myoblasts were incubated with oligo in the presence of a cationic lipid; this procedure increases the amount of internalized oligo enough to allow intranuclear detection by using standard fluorescence microscopy (unpublished results). When intranuclear oligo movement was measured by FRAP, oligo(dT) populations were found to move with a diffusion coefficient of 1.2 × 10−7 cm2/s (Table 3), a rate intermediate between the fast and slower rates observed for oligo(dT) by using FCS (see Table 2). This finding is consistent with the fact that our FRAP instrument calculates one overall average rate of diffusion back into the bleached area. In fact, the FRAP-determined diffusion coefficient for oligo(dT) corresponded well to the average FCS diffusion coefficient of 8.7 × 10−8 cm2/s calculated from FCS data treated as a one-component model (Table 3). When FRAP was used to measure diffusion in oligo(dA) treated cells, an average diffusion coefficient of 2.6 × 10−7 cm2/s was obtained (Table 3), reflecting the higher percentage of oligo(dA) as free oligo. The average FCS measured diffusion coefficient for oligo(dA) in cells calculated from data treated as a one-component model (5.05 × 10−7 cm2/s) was also larger than that for oligo(dT), thus indicating the presence of free oligo(dA). Therefore, under the conditions used in this study, FCS-measured diffusion rates in the nucleus of living cells are well within the range of values obtained with FRAP.
Table 3.
Diffusion coefficients of oligos in cell nuclei: FRAP and FCS
| FRAP* D, 10−7 cm2/s | FCS D, 10−7 cm2/s | |
|---|---|---|
| Oligo(dT) | 1.2 ± 0.6 | 0.87 ± 0.02 |
| Oligo(dA) | 2.6 ± 0.6 | 5.05 ± 0.52 |
FRAP recovery rates were 80 ± 3%. Errors are standard errors of the mean.
DISCUSSION
We have shown that fluorescence correlation spectroscopy can be used to measure diffusion rates of oligodeoxynucleotides in the nucleus of living cells. Diffusion coefficients obtained by using FCS correlate well with FRAP measurements, under the conditions we report, where both techniques can be applied. We find that oligos can move with diffusion coefficients as fast as 4 × 10−7 cm2/s in the cell nucleus. This rapid rate is similar to the rate at which the oligos move in aqueous solution and indicates that diffusion can occur in the nucleus at rates approximating those measured in solution. Although the viscosity of the cytoplasm has been investigated (24–27), only a few measurements of molecular diffusion rates in the nucleus have been reported, primarily in isolated nuclei (20, 28, 29). Some of these studies report that nuclear viscosity is greater than water, and others suggest that molecular movement in the nucleus occurs at aqueous solution rates. A recent study of the movement of dextrans in nuclei of intact cells by using FRAP reported intranuclear diffusion rates close to rates observed in aqueous solution (27), as we observe for oligos in the present investigation with both FCS and FRAP.
In the present measurements, the fraction of oligo diffusing at this rapid “aqueous” rate varied among nuclei much more than the narrow range of variation observed for the same oligos diffusing in solution. Because the intracellular volume analyzed is very small, less than a femtoliter, it is possible that these variations reflect microenvironment differences among the various nuclear regions measured. Although we routinely selected areas outside the nucleoli, all other areas of nuclear structure (homogeneous in bright field microscopy) were randomly sampled. Similar spatial variation has been reported in the study of membrane protein diffusion and has been suggested to represent the presence of diffusional microdomains (30, 31). Such microdomains are thought to arise from variations in effective viscosity and/or “corralling” effects due the probe’s vicinity to cellular structure. It is seems likely, therefore, that we have sampled an array of nuclear domains that permit, in the case of oligo(dA), diffusional mobility more often than not. Hence, although we do not have information on the size, number, or interconnectivity of the aqueous intranuclear domains we detect, our results suggest that these domains can be found virtually anywhere throughout the nucleus.
Even though the oligo(dA) measurements showed some variability, we nonetheless observed a significant difference in the behavior of oligo(dT) compared with oligo(dA) in the nucleus of living cells. Although the majority of oligo(dA) always moved at the rapid “aqueous” rate, almost 45% of oligo(dT) molecules moved much slower, with diffusion coefficients between 1 × 10−10 and 9 × 10−8 cm2/s. We consider it likely that the majority of this latter fraction represents oligo(dT) hybridized to poly(A) RNA in the nucleus for three reasons. (i) Earlier in situ reverse transcription experiments have shown that the synthetic oligo(dT) used in these experiments hybridizes to intranuclear poly(A) targets in the living cell (7). (ii) Only a small fraction of synthetic oligo(dA) was observed to move at these slower rates in the nucleus. (iii) When oligo(dT) was prehybridized with unlabeled oligo(dA) [making the oligo(dT) unavailable for hybridization] before incubation with cells, only a small amount of the slow-moving oligo(dT) component was observed.
These results therefore suggest that endogenous poly(A) RNA [tagged with the hybridized oligo(dT)] moves in the nucleus at rates between 1 × 10−10 and 9 × 10−8 cm2/s. As with the oligo(dA) measurements, there was considerable variation between different nuclear sites in the amount of oligo(dT) diffusing at any given rate, and we believe this variation suggests the presence of different nuclear microenvironments in which the oligo(dT)/poly(A) RNA hybrids move at different rates. One would not expect to see this variation among readings if all areas of the nucleus contained equal concentrations of freely diffusing oligo(dT)/poly(A) RNA hybrids (see below).
About one-third of this slower oligo(dT) population [15% of total oligo(dT), see Table 2, columns 4 and 5] moved at average rates of 5.9 × 10−9 cm2/s or slower. If our interpretation is correct that the slower oligo(dT) components represent hybridization to nuclear poly(A) RNA, this suggests that a sizable fraction of nuclear poly(A) RNA may be tethered to large macromolecular complexes that move very slowly. Other results have also indicated that poly(A) RNA may be associated with large nuclear structures. For example, in situ hybridization studies have shown poly(A) RNA is retained in detergent-insoluble nuclear structures (32) and ultrastructural studies have shown that nuclear poly(A) RNAs are associated with perichromatin fibrils and granules and interchromatin granule clusters (33, 34). Moreover, recent biochemical studies strongly suggest that pre-mRNA molecules are tethered to large complexes containing transcription, polyadenylation, and splicing machinery (1–3), and it is anticipated that complexes of this size would diffuse very slowly.
The other two-thirds of the oligo(dT) population that we interpret as being hybridized to intranuclear poly(A) RNA [30% of total oligo(dT), see Table 2, column 3] was moving more rapidly (average diffusion coefficient of 9 × 10−8 cm2/s), in the same range as might be expected for free diffusion of a full length poly(A) RNA molecule in solution. The 7.5-kb RNA we monitored with FCS (molecular mass about 2.5 × 106 Da) moved at 1 × 10−7 cm2/s in solution. Even if a poly(A) RNA molecule of this size were complexed with four times its weight in protein, as would be expected for a typical hnRNP particle (35), its diffusion coefficient would still fall in this range. (For an average mammalian hnRNP particle (20 nm × 100 nm (35), molecular mass of 1.25 × 107 Da), the calculated diffusion coefficients range (depending on shape assumptions) from 2.2 × 10−8 to 9.2 × 10−8 cm2/s.) Therefore, one interpretation of our data is that a rather large number of poly(A) RNA molecules are able to diffuse freely in the nucleus. In agreement with this interpretation, this population is observed even in cells depleted of ATP, suggesting that movement of these molecules is not powered by an ATP-requiring system. This interpretation is also consistent with our finding that the oligos used in our experiments can themselves diffuse freely in the nucleus, and is consistent with another study (4) that reports that some pre-mRNAs move in the nucleus at rates consistent with free diffusion. However, because oligo(dT) will hybridize to all available poly(A) RNA in the nucleus, including nonmessenger polyadenylated RNA which remains in the nucleus (33, 34), the results presented here do not prove that intranuclear pre-messenger RNAs necessarily move at this rapid rate (or at the slower rate discussed above). This information awaits future FCS studies in which diffusion rates of specific mRNA molecules in the nucleus are measured.
Acknowledgments
We are grateful to Carl Zeiss, Inc. and Evotec for the opportunity to use their new FCS ConfoCor, and to Reinhard Janka of Carl Zeiss and Birgit Hecks of Evotec for their help in getting the instrument operational. We thank Christine Thompson for her expert assistance in the FRAP measurements, Robert Singer (Albert Einstein College of Medicine) for the kind gift of oligos synthesized in his laboratory, and Elizabeth Ryder (Worcester Polytechnic Institute) for helpful statistical advice. This investigation was supported by National Institutes of Health National Research Service Award Postdoctoral Fellowship AR-08361 to J.C.P., National Institutes of Health Postdoctoral Training Grant HD-07439 to E.S.B., National Institutes of Health Grant NS-28760 to D.E.W., and National Institutes of Health Grant GM-21595-21 to T.P.
ABBREVIATIONS
- oligo
oligonucleotide
- FCS
fluorescence correlation spectroscopy
- FRAP
fluorescence recovery after photobleaching
- hnRNP
heterogeneous nuclear ribonucleoprotein
References
- 1.Du L, Warren S L. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:257–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 3.Zeng C, Kim E, Warren S L, Berget S M. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zachar Z, Kramer J, Mims I P, Bingham P M. J Cell Biol. 1993;121:729–742. doi: 10.1083/jcb.121.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zirbel R M, Mathieu U R, Kurz A, Cremer T, Lichter P. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]
- 6.Pederson T. J Mol Biol. 1998;277:147–159. doi: 10.1006/jmbi.1997.1618. [DOI] [PubMed] [Google Scholar]
- 7.Politz J C, Taneja K, Singer R H. Nucleic Acids Res. 1995;23:4946–4953. doi: 10.1093/nar/23.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf D E. Methods Cell Biol. 1989;30:271–306. doi: 10.1016/s0091-679x(08)60983-8. [DOI] [PubMed] [Google Scholar]
- 9.Gordon G W, Chazotte B, Wang X F, Herman B. Biophys J. 1995;68:766–778. doi: 10.1016/S0006-3495(95)80251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berland K M. Biophys J. 1997;72:1487–1488. doi: 10.1016/S0006-3495(97)78796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigler R. J Biotechnol. 1995;41:177–186. doi: 10.1016/0168-1656(95)00054-t. [DOI] [PubMed] [Google Scholar]
- 12.Berland L M, So P T C, Gratton E. Biophys J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widengren J. Ph.D. thesis. Stockholm: Karolina Institutet; 1996. [Google Scholar]
- 14.Maiti S, Haupts U, Webb W W. Proc Natl Acad Sci USA. 1997;94:11753–11757. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinjo M, Rigler R. Nucleic Acids Res. 1995;23:1795–1799. doi: 10.1093/nar/23.10.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwille P, Oehlenschlager F, Walter N G. Biochemistry. 1996;35:10182–10193. doi: 10.1021/bi960517g. [DOI] [PubMed] [Google Scholar]
- 17.Walter N G, Schwille P, Eigen M. Proc Natl Acad Sci USA. 1996;93:12805–12810. doi: 10.1073/pnas.93.23.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kislauskis E J, Li, Zhifang, Singer R H, Taneja K L. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat R, Weaver J A, Wagner C, Bodwell J E, Bresnick E. J Biol Chem. 1996;271:32551–32556. doi: 10.1074/jbc.271.51.32551. [DOI] [PubMed] [Google Scholar]
- 20.Lang I, Scholz M, Peters R. J Cell Biol. 1986;102:1183–1190. doi: 10.1083/jcb.102.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aragon S R, Pecora R. J Chem Phys. 1976;64:1791–1803. [Google Scholar]
- 22.Magde D, Webb W W, Elson E L. Biopolymers. 1978;17:361–376. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 23.Swedlow J R, Sedat J W, Agard D A. Cell. 1993;73:97–108. doi: 10.1016/0092-8674(93)90163-k. [DOI] [PubMed] [Google Scholar]
- 24.Fushimi K, Verkman A S. J Cell Biol. 1991;112:719–725. doi: 10.1083/jcb.112.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arrio-Dupont M, Cribier S, Foucault G, Devaus P F, d’Albis A. Biophys J. 1996;70:2327–2332. doi: 10.1016/S0006-3495(96)79798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janson L W, Ragsdale K, Luby-Phelps K. Biophys J. 1996;71:1228–1234. doi: 10.1016/S0006-3495(96)79367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seksek O, Biwersi J, Verkman A S. J Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorscher S M, Bartholomew J C, Klein M P. Biochim Biophy Acta. 1980;610:28–46. doi: 10.1016/0005-2787(80)90053-2. [DOI] [PubMed] [Google Scholar]
- 29.Burgoyne L A, Skinner J D, Marshall A. J Cell Sci. 1978;31:1–11. doi: 10.1242/jcs.31.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Saxton M J. Biophys J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxton M J. Biophys J. 1997;72:1744–1753. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter K C, Taneja K L, Lawrence J B. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puvion E, Puvion-Dutilleul F. Exp Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, Deerinck T J, Ellisman M K, Spector D L. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kish V M, Pederson T. Methods Cell Biol. 1978;17:377–399. doi: 10.1016/s0091-679x(08)61155-3. [DOI] [PubMed] [Google Scholar]