SUMMARY
LIM-homeodomain (HD) and POU-HD transcription factors play critical roles in neurogenesis. However, it remains largely unknown how they cooperate in this process and what downstream target genes they regulate. Here we show that ISL1, a LIM-HD protein, is co-expressed with BRN3B, a POU-HD factor, in nascent, post-mitotic retinal ganglion cells (RGCs). Similar to the Brn3b-null retinas, retina-specific deletion of Isl1 results in the apoptosis of a majority of RGCs and in RGC axon guidance defects. The Isl1 and Brn3b double null mice display more severe retinal abnormalities with a near complete loss of RGCs, indicating the synergistic functions of these two factors. Furthermore, we show that both Isl1 and Brn3b function downstream of Math5 to regulate the expression of a common set of RGC-specific genes. Whole retina chromatin immunoprecipitation and in vitro transactivation assays reveal that ISL1 and BRN3B concurrently bind to and synergistically regulate the expression of a common set of RGC-specific genes. Thus, our results uncover a novel regulatory mechanism of BRN3B and ISL1 in RGC differentiation.
Keywords: LIM-homeodomain, POU-domain, Math5, Atoh7, Pou4f2, RGC, retinal development, transcription factor
INTRODUCTION
Due to its easy accessibility and well-organized laminar structure, the retina has long served as an ideal experimental model for studying the central nervous system (CNS). Neurogenesis in vertebrate retinas is stereotyped in an ordered fashion that RGCs are always born first, immediately followed by horizontal, amacrine, and cone photoreceptor cells, and lastly by bipolar, rod photoreceptor, and Müller cells (Cepko et al., 1996; Young, 1985). The basic helix-loop-helix (bHLH) transcription factors (TFs) are believed to play critical roles in specifying retinal cell fates (Hatakeyama and Kageyama, 2004; Yan et al., 2005). During RGC development, the bHLH factor MATH5 (ATOH7-Mouse Genome Informatics) is expressed in post-mitotic retinal precursors and provides them with the competency to become RGCs (Yang et al., 2003). MATH5 functions to promote the expression of early RGC-specific factors such as the POU-homeodomain (POU-HD) factor BRN3B (POU4F2-Mouse Genome Informatics) and the LIM-homeodomain (LIM-HD) factor ISL1(Wang et al., 2001; Yang et al., 2003). Furthermore, MATH5 suppresses the expression of NGN2, NEUROD, MATH3 (NEUROG2, NEUROD1, and NEUROD4, respectively – all Mouse Genome Informatics), and BHLHB5, which are the bHLH factors required for the specification of non-RGC fates (Feng et al., 2006; Hatakeyama and Kageyama, 2004; Inoue et al., 2002; Liu et al., 2001; Mu et al., 2005a). In Math5-deficient mice, RGC genesis is disrupted, resulting in less than 5% of RGCs being detected in adult retinas. Concurrently, the number of amacrine cells and cone photoreceptors are increased (Brown et al., 2001; Wang et al., 2001). In contrast to the early function of MATH5 in RGC genesis, BRN3B expression starts in nascent RGCs and is required for their terminal differentiation, including axon outgrowth and pathfinding, and cell survival (Erkman et al., 2000; Gan et al., 1999; Pan et al., 2005; Xiang, 1998). In Brn3b–null mice, RGCs are initially generated but 80% of them undergo apoptosis before birth (Erkman et al., 1996; Gan et al., 1996). Moreover, the compound mutation of Brn3b and Brn3c (Pou4f3-Mouse Genome Informatics) results in the further reduction of RGCs in adult mice (Wang et al., 2002a). While previous studies have shown clearly the importance of BRN3 factors in RGC development, it still remains poorly understood how RGC differentiation is regulated and whether other TFs control this process in parallel to BRN3 factors.
Studies of CNS development in both vertebrates and invertebrates have shown that TFs, often acting in distinct combinatorial manner, play important roles in neuronal development (Bang and Goulding, 1996; Castro et al., 2006; Lee and Pfaff, 2001). The POU-HD and LIM-HD TFs appear to function primarily at the later stages of neurogenesis. For example, in the spinal cord, the unique combinatorial expression of LIM-HD TFs confers motor neuron subtypes with specific axon targeting pathways (Appel et al., 1995; Kania et al., 2000; Sharma et al., 2000; Sharma et al., 1998; Tsuchida et al., 1994). BRN3B is required in the development of RGCs (discussed above). In Drosophila, Acj6 and Drifter, both POU-HD TFs, are required for the dendritic targeting in olfactory system (Komiyama et al., 2003).
LIM-HD TFs consist of two LIM-domains mediating the protein-protein interaction and one homeodomain required for DNA-binding (Hobert and Westphal, 2000). LIM-HD factors can bind to other TFs directly or through the mediation of LIM domain binding (LDB) family cofactors (Jurata et al., 1998; Lichtsteiner and Tjian, 1995). LIM-HD TFs have the potential to assemble homomeric or heteromeric complexes in a cellular context specific manner and to regulate a wide range of developmental events (Lee and Pfaff, 2003; Shirasaki and Pfaff, 2002; Tsuchida et al., 1994). Moreover, LIM-HD and POU-HD TFs have been shown to cooperate in regulating neuronal differentiation. One such example is the regulation of touch receptor differentiation in C. elegans by POU-HD factor UNC-86 and LIM-HD factor MEC-3. UNC-86 dimerizes with MEC-3 on the mec-3 promoter, which is required for the maintenance of mec-3 expression and touch cell differentiation (Lichtsteiner and Tjian, 1995; Xue et al., 1993). UNC-86 and MEC-3 also synergistically activate touch cell-specific genes, mec-4 and mec-7 (Duggan et al., 1998).
ISL1 is one of the founding members of LIM–HD TFs and belongs to the Islet subgroup. The function of ISL1 in the spinal cord has been intensively studied. ISL1 is expressed in all motor neurons immediately after they exit cell cycle (Pfaff et al., 1996). In Isl1-null mice, motor neurons fail to develop (Pfaff et al., 1996). In contrast, Drosophila Islet (a homolog of mammalian ISL1) is not required for the genesis of motor neurons, but rather is involved in axonal trajectory selection and neurotransmitter expression (Thor and Thomas, 1997). Here, we demonstrate the co-expression of BRN3B and ISL1 in post-mitotic, differentiating RGCs. To investigate the role of ISL1 in RGC development, we generate Isl1-lacZ knock-in and Isl1 conditional knockout mice. We reveal that in Isl1-null retinas, RGCs appear to be generated normally but 67% RGCs subsequently undergo apoptosis and RGC axon growth is defective, a phenotype strongly resembling that of Brn3b mutants (Erkman et al., 2000; Gan et al., 1999). In Isl1 and Brn3b double null retinas, greater than 95% nascent RGCs die of apoptosis, suggesting their cooperative relationship in RGC development. Furthermore, in vivo chromatin immunoprecipitation (ChIP) and in vitro transactivation assays demonstrate that both factors bind to and regulate the expression of RGC-specific genes. Our data strongly argue for the involvement of both parallel and cooperative functions of ISL1 and BRN3B in RGC development.
MATERIALS AND METHODS
Animals
Brn3blacZ, Brn3bAP, and Six3-cre mice have been previously described (Furuta et al., 2000; Gan et al., 1999). Isl2lacZ knock-in mice (L. Gan, unpublished) were generated by replacing all the Isl2 coding exons and the flanking introns with a lacZ-SV40 polyA-Neo cassette. The Isl1lacZ targeting construct was generated by inserting the Isl1 2.6 kb 5′- and 4.2 kb 3′-flanking sequences into the 5′ and 3′ multiple cloning sites of PKII-lacZ vector (L. Gan, unpublished), where a cassette with lacZ-SV40 polyA-Neo replaced Isl1 coding regions of Exon 1–2 and the adjacent intron sequences. To make Isl1CKO targeting construct, we inserted a neomycin resistance gene cassette along with a loxP site at the 5′ of Exon 2 and another loxP site at the 3′ of Exon 2. The targeting vectors were electroporated into W4 mouse embryonic stem cells (Auerbach et al., 2000) to generate Isl1lacZ/+ and Isl1CKO/+ mice using standard techniques. Isl1loxP/+ mice were obtained by crossing Isl1CKO/+ mice with the ROSA26-FLPe mice (The Jackson Laboratory, Stock Number: 003946) to remove FRT-flanked neomycin resistance cassette.
The PCR genotyping of these animals was performed by using the following primer sets: 5′-AGGGCCGCAAGAAAACTATCC and 5′-ACTTCGGCACCTTACGCTTCTTCT to detect a 404 bp product of Isl1lacZ allele, 5′-GGTGCTTAGCGGTGATTTCCTC and 5′-GCACTTTGGGATGGTAATTGGAG to detect a 452 bp product of wild type Isl1 allele and a 512 bp product of Isl1loxP allele, and 5′-GTGGAATCGCTGAATCTTGAC and 5′-GCCCAAATGTTGCTGGATAGT to detect Six3-cre allele. The mouse strains were maintained in the C57BL/6J and 129S6 mixed background. Embryonic day 0.5 (E0.5) was defined as the day when the vaginal plug appeared. All animal procedures were approved by University Committee of Animal Resources at the University of Rochester.
Hematoxylin and Eosin (H&E) staining, immunohistochemistry, X-Gal staining and in situ hybridization
Embryos were harvested from E11.5 to E18.5, decapitated and fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) for several hours and were processed for paraffin sections or cryosections. Horizontal retina sections across optic disc were collected from controls and mutant littermates were mounted side by side for comparisons. BrdU labeling, H&E and X-Gal staining were carried out as previously reported (Pan et al., 2005). The primary antibodies used in immunohistochemistry were: mouse anti-BRN3A (POU4F1–Mouse Genome Informatics) (Santa Cruz, 1:200), goat anti-BRN3B (Santa Cruz, 1:200), mouse anti-ISL1 (Developmental Studies Hybridoma Bank, 1:200), mouse anti-bromodeoxyuridine (BrdU) (Becton Dickson, 1:400), rabbit anti-phosphorylated Histone3 (Santa Cruz, 1:400), mouse anti-SMI32 (Sternberger Monoclonals, 1:1000) and rabbit anti-activated caspase3 (R&D system, 1:100). The Alexa-conjugated secondary antibodies (Molecular Probes) were used at 1:1000 dilution. Non-radioactive in situ hybridization was performed using digoxigenin-UTP labeled riboprobes (Radde-Gallwitz et al., 2004). The specific cDNA sequences used to generate riboprobes were: Brn3a (3′UTR); Olf1 (L12147, nts865–1180); Irx4 (NM_018885, nts1556–2265); Ablim1 (AF316037, nts12–31); L1cam (NM_008478, nts3083–3748). Isl1 and Isl2 probes were described previously (Yang et al., 2003). The Shh probe was a generous gift from Dr. Valerie A. Wallace (Jensen and Wallace, 1997). Confocal images were acquired on an Olympus microscope (BX50WI) with Fluoview 4.3 laser scanning. Other pictures were taken with a Nikon Eclipse TE2000-U inverted microscope with a Nikon DXM1200F digital camera.
Cell counts and statistical analysis
For apoptosis analysis, five pairs of matched retina sections of Isl1-null and littermate controls were collected at regularly spaced intervals to completely survey each retina. After anti-activated caspase-3 immunolabeling, images were taken and the immunoreactive cells were counted with Image J program (NIH). Results from five sections were averaged to obtain the apoptotic cell number for each eye. For analyses of BRN3A+ or BRN3B+ cells, whole-mount retinas were used. In these cases, three pictures were obtained from the central region of the retinas. The immunoreactive cells were counted and averaged. To compare the optic nerve size, three pairs of matched cross sections of null and control optic nerves were collected and processed for H&E staining. The boundary of each optic nerve was outlined using Adobe Photoshop. The size of optic nerve was determined by measuring the number of pixels contained in the outlined area.
Lipophilic dye tracing
For anterograde labeling of the optic pathway, mouse heads at E13.5 and E15.5 were fixed overnight in 4% paraformaldehyde in PBS. After enucleation of the right eye, DiI crystals (Molecular Probes) were implanted unilaterally in the optic disc. After incubation in PBS containing 0.1% sodium azide at 37°C for 1–2 weeks, the brains were dissected to expose the optic chiasm and visualized under a Nikon SMZ1500 fluorescent stereomicroscope.
Chromatin immunoprecipitation (ChIP)
Chromatin from retinas at indicated stages was collected according to the protocol supplied with the ChIP assay kit (Upstate Biotechnologies). Mouse anti-ISL1 and goat anti-BRN3B were used for immunoprecipitation. Promoter regions with Brn3-binding consensus sequences were detected in the precipitated material by PCR using the primer sets listed in Fig. S4A (supplementary material). Brn3b ORF was used as a negative control.
Luciferase activity assay
CV1 epithelial cells were cultured in 24 well plates in DMEM with 10% FBS. Transfections were carried out with Lipofectamine 2000 (Invitrogen) when cells reached 70% confluence. Brn3b expression plasmid and Brn3a-luciferase reporter construct were generous gifts of Dr. Eric Turner (Trieu et al., 2003). Isl1 expression plasmid was generated by inserting Isl1 cDNA into pcDNA expression vector (Invitrogen). For each transfection, 100 ng of Isl1 and/or Brn3b expression plasmid, 200 ng of Brn3a-luciferase reporter construct and 5 ng of PRL (Promega) Renilla luciferase control plasmid were used. The total amount of DNA was balanced by adding empty pcDNA vector. Cells were harvested 36 hours after transfection and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase activity was normalized by renilla luciferase activity.
RESULTS
Co-localization of ISL1 and BRN3B in post-mitotic RGCs
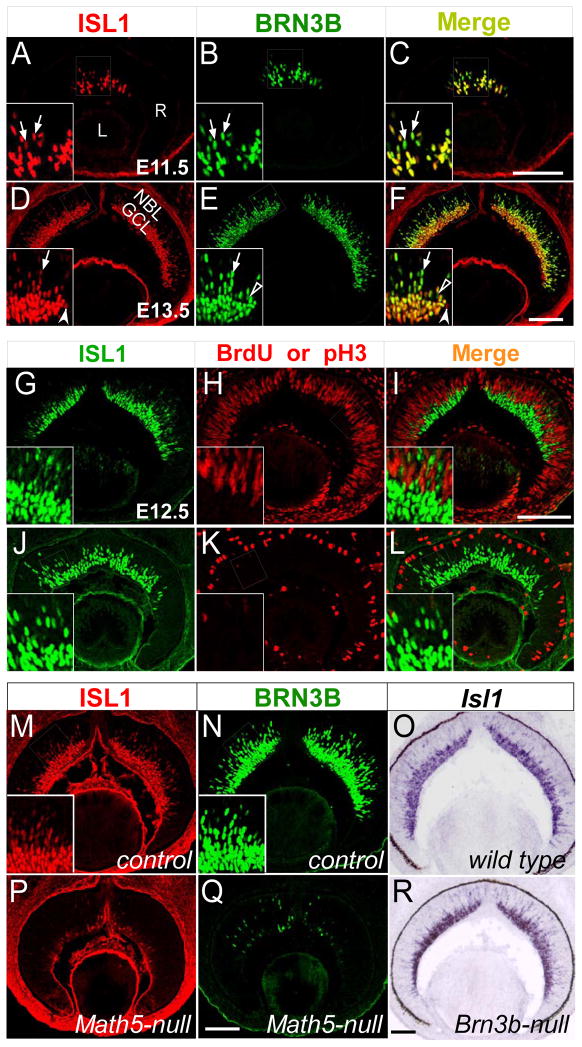
To address the functions of ISL1 in RGC differentiation, we first studied its spatiotemporal expression pattern in developing retinas and how its expression pattern correlated with BRN3B, one of the earliest RGC markers. At E11.5, nascent RGCs expressed BRN3B and were first found in the central retina (Fig. 1B). At this age, BRN3B+ cells also expressed ISL1 (Fig. 1A–C, arrows). At E13.5, as retinogenesis proceeded in a central-to-peripheral wave, more RGCs were co-labeled with ISL1 and BRN3B (Fig. 1D–F). Although the overall expression pattern was almost identical for these two factors, there was a slight difference in the signal intensity in the labeled cells. While BRN3B was expressed uniformly in both migrating RGCs in the neuroblast layer (NBL) and the post-migrated RGCs in the ganglion cell layer (GCL), ISL1 expression level appeared to be higher in the GCL and lower in the NBL. The expression of ISL1 was sustained in RGCs in adult mice and was also found in cholinergic amacrine and ON-bipolar cells (Elshatory et al., 2007a; Elshatory et al., 2007b; Galli-Resta et al., 1997). Anti-ISL1 antibody is raised against the C-terminal of ISL1 homeodomain and reacts to ISL1 specifically but also recognizes ISL2 with a lower affinity (Tanabe et al., 1998). In mouse, weak ISL2 expression is first detected in a very few RGCs in the central GCL at E13.5 and in about 30% RGCs postnatally (see Fig. S1A-D in the supplementary material) (Brown et al., 2000; Pak et al., 2004). Targeted deletion of Isl2 reveals that Isl2 is not necessary for the generation and survival of RGCs (Pak et al., 2004). To confirm that the above anti-ISL1 labeling reveals the true Isl1 expression pattern, we compared the ISL1-immunostaining in wild type and Isl2-null retinas at E13.5 to E15.5 and found no difference in the labeling patterns (Fig. 1D–F, data not shown and see Fig. S1E-J in the supplementary material). Thus, at E11.5 to E15.5, all cells labeled with anti-ISL1 antibody in the retina represent ISL1-expressing cells.
Figure 1.
Expression of ISL1 in developing mouse retina. (A–F) Horizontal sections of retinas at E11.5 (A–C) and E13.5 (D–F) were immunolabeled with anti-ISL1 (red) and anti-BRN3B (green). The expression of ISL1 is detected from E11.5 and is largely co-localized with that of BRN3B in both migrating RGCs in the NBL and post-migrated RGCs in the GCL (arrows). Open and filled arrowheads indicate the occasional RGCs expressing BRN3B or ISL1 alone. (G–L) Co-localization of ISL1 (green) with S- and M-phase markers (red), anti-BrdU (G–I) and anti-pH3 (J–L), reveals the post-mitotic expression of ISL1 in the developing retina at E12.5. (M–R) Expression of Isl1 is regulated by Math5 but not by Brn3b. Immunolabeling of E13.5 retinal sections shows that the expression of ISL1 (red) and BRN3B (green) is dramatically decreased in Math5-null mice (P,Q) compared with that in heterozygous controls (M,N). (O,R) Section in situ hybridization demonstrates the comparable Isl1 expression in wild type (O) and Brn3b-null (R) retinas at E14. Inserts show the enlarged views of the corresponding boxed regions. Abbreviations for this and other figures: R, retina; L, lens; NBL, neuroblast layer; GCL, ganglion cell layer; Scale bar: 100 μm.
The detection of ISL1 in the NBL suggested that it is expressed in retinal progenitors. To test this possibility, we performed immunostaining with anti-ISL1 antibody in conjunction with several cell cycle markers, BrdU labeling to mark S-phase cells and anti-phosphorylated histone 3 (pH3) staining to identify M-phase cells. At E12.5, when both proliferating progenitors and post-mitotic RGCs were readily detectable, ISL1 expression was detected mostly in cells negative for BrdU and pH3 (Fig. 1G–L). Thus, ISL1 is predominantly expressed in post-mitotic cells during early retinogenesis.
The early expression of ISL1 in nascent RGCs, its co-localization with BRN3B, and the known cooperativity of these two types of TFs suggested that ISL1 could function in parallel to BRN3B or immediately upstream or downstream of BRN3B during RGC development. To distinguish these possibilities, we analyzed the expression of ISL1 in Math5-null and Brn3b-null mice. In Math5-null retinas at E13.5, the expression of ISL1 and BRN3B was dramatically decreased (Fig. 1M,N,P,Q), consistent with our previous finding that Brn3b and Isl1 are downstream genes of Math5 (Wang et al., 2001; Yang et al., 2003). In contrast, we observed no discernible changes in Isl1 expression in Brn3b-null retinas at E14.5 (Fig. 1O,R), a stage before the onset of RGC death caused by the absence of Brn3b (Gan et al., 1999). Therefore, Isl1 likely functions upstream of or in parallel to Brn3b during RGC development.
Targeted disruption of Isl1 in retina
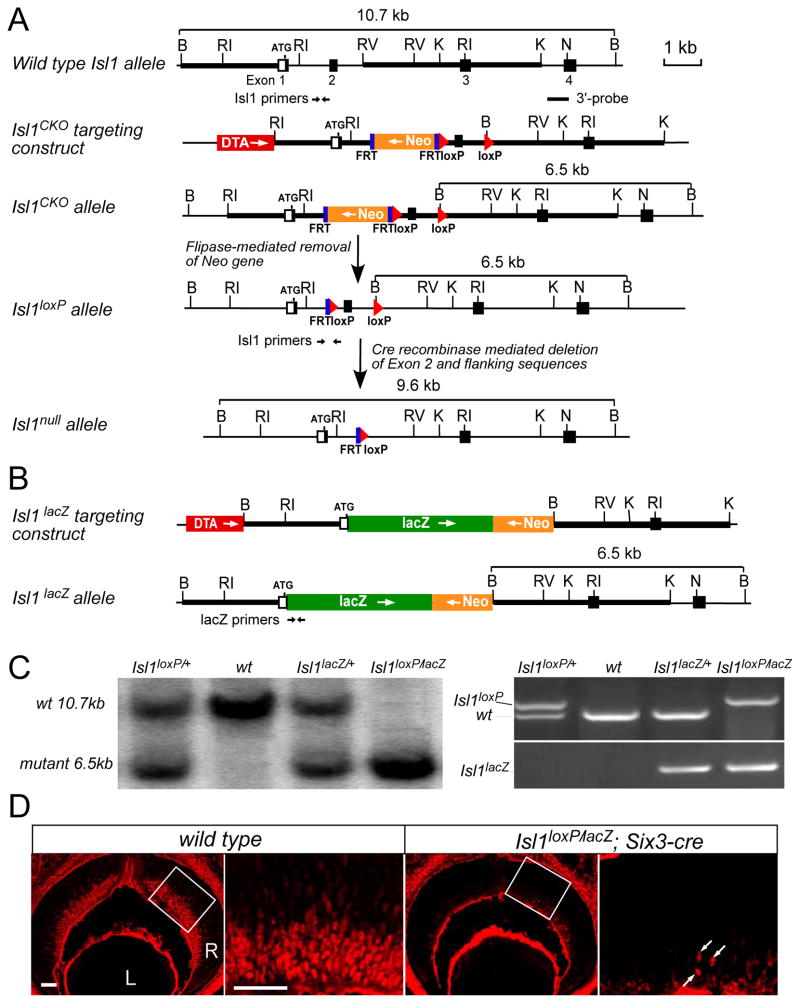
Conventional Isl1 knockout mice do not survive beyond E11.5 probably due to the failure in vascular development (Pfaff et al., 1996). To assess the role of ISL1 in RGC development that occurs during mid- to late gestation stages, we generated an Isl1 conditional knockout allele (Isl1loxP) by flanking Exon 2, which encodes the first LIM domain, with loxP sequences (Fig. 2A). Cre recombinase-mediated deletion of the loxP-flanked Exon 2 resulted in a null mutation via a reading frame shift. Additionally, a lacZ knock-in allele, Isl1lacZ, was created by replacing the Exon 2 with a nuclear lacZ reporter gene (Fig. 2B). The genotypes of Isl1 mutant mice were confirmed by Southern blotting and PCR (Fig. 2C). The expression of β–galactosidase in Isl1lacZ/+ mice faithfully recapitulated the pattern of endogenous Isl1 as shown by in situ hybridization and immunostaining (see Fig. S2 in the supplementary material) and thus, served as an excellent marker of Isl1-expressing cells.
Figure 2.
Generation of Isl1 conditional knockout and Isl1-lacZ knock-in alleles. (A) Generation of Isl1 conditional allele. Isl1 genomic structure and restriction enzyme map is shown at the top. Open boxes are the non-coding exon sequences and filled boxes the coding sequences. Thick bars are the sequences used to generate the homologous arms in the targeting vector. The Isl1CKO targeting vector is made by inserting the FRT-flanked neomycin gene and the 5′ loxP sequence into Intron 1. The 3′ loxP sequence is inserted at the EcoRV site at Intron 2 and the EcoRV site is changed to BamHI site. ROSA26-FLPe mice are used to remove the neomycin resistance gene and generate Isl1 conditional knockout allele (Isl1lox). Tissue-specific deletion of Isl1 in the retina (Isl1null) is achieved by crossing Isl1lox mice to Six3-cre deleter mice. (B) Generation of Isl1lacZ knock-in allele. The DNA fragment containing reporter lacZ and neomycin resistance genes is used to replace the coding region of Exon 1, Exon2 and the flanking intron sequences in the targeting vector. Abbreviations: Neo, PGK-neomycin resistance gene; DTA, diphtheria toxin gene for negative selection of embryonic stem cells; lacZ, β-galactosidase reporter gene; FRT, flipase recognition sequence; loxP, Cre recombinase recognition sequence. Restriction enzymes: B, BamHI; RI, EcoRI; RV, EcoRV; K, KpnI; N, NotI. (C) Left panel, Southern genotyping confirmation of Isl1lacZ and Isl1lox mice using 3′ probe and BamHI digestion. Right panel, PCR genotyping using primers indicated in A&B can distinguish Isl1lox, Isl1lacZ, and wild type allele. (D) Anti-ISL1 immumolabeling of E13.5 retina sections confirms the deletion of ISL1 in the retina of Isl1loxP/lacZ; Six3-Cre mice. The enlarged views of the corresponding boxed regions are showed on the right. Deletion efficiency is greater than 90%. Arrows indicate residual ISL1 expression. Scale bar: 50 μm.
Retina-specific removal of Isl1 was achieved by breeding heterozygous Isl1lacZ/+ or Isl1loxP/+ with Six3-cre mice and subsequently crossing with Isl1loxP/loxP mice. Six3-cre mice express Cre recombinase in the eye field and the ventral forebrain from E9 to E9.5 (Furuta et al., 2000), and have been used successfully as an effective retina-specific deleter (Mu et al., 2005b). In our experiments, we observed consistently a greater than 90% deletion of ISL1 in Isl1loxP/lacZ; Six3-cre and Isl1loxP/loxP; Six3-cre retinas at E13.5 (Fig. 2D and data not shown). In the following experiments, Isl1loxP/lacZ; Six3-cre and Isl1loxP/loxP; Six3-cre mice were used interchangeably as Isl1-nulls. Isl1loxP/+, Isl1loxP/loxP and the wild type mice were phenotypically indistinguishable and were designated as controls hereafter. The Isl1-nulls were born at Mendelian frequencies (26%, n=96) with no overt morphological defects, but exhibited moderate growth retardation postnatally. Their body weights were about 91% of their littermate controls at P10 (n=7 pairs) and about 80% at 6 weeks (n=5 pairs).
Major loss of RGCs in the absence of Isl1
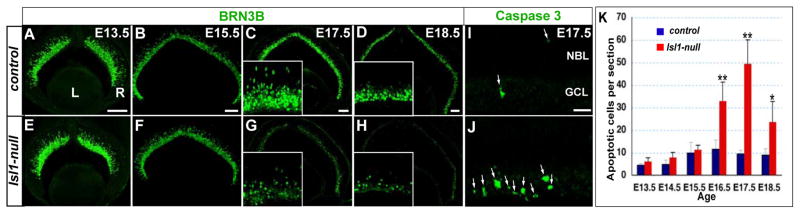
To assess the importance of ISL1 during RGC development, we analyzed BRN3B expression in Isl1-null retinas at different embryonic stages (Fig. 3A–H). At E13.5 and E15.5, the peak period of RGC generation, there was no substantial difference in the number and distribution of BRN3B+ RGCs in Isl1-null retinas compared with controls (Fig. 3A,B,E,F), indicating that Isl1 is dispensable for the generation and migration of RGCs as well as for the onset of BRN3B expression. However, BRN3B+ RGCs were drastically reduced in the absence of Isl1 at E17.5, (Fig. 3C,G). This reduction progressed in a central to peripheral wave and was most evident at E18.5 (Fig. 3D,H).
Figure 3.
Targeted disruption of Isl1 results in the developmental loss of RGCs. (A–H) Immunostaining of retina sections with anti-BRN3B shows that in Isl1-null retina, BRN3B+ RGCs are generated and positioned properly at E13.5 (A,E) and E15.5 (B,F). However, a significant loss of BRN3B+ RGCs is detected at E17.5 (C,G) and E18.5 (D,H). Inserts show the enlarged views of the central retina. (I,J) Anti-activated caspase3 immunostaining shows an increase in apoptotic cells (arrows) in the GCL of Isl1-null retina at E17.5. (K) Quantification of apoptotic cells in developing retina by averaging activated caspase3 positive cells in 5 sections per retina. The difference of apoptotic cell numbers in wild type and Isl1-null retina is insignificant at E13.5 to E15.5. However, in Isl1-nulls, the apoptotic cells are significantly increased from E16.5 and are peaked at E17.5. Each histogram represents the mean ± s.d. n=3–4 for each genotype per stage. (**t-test P<0.01; *P<0.05;). Scale bar: 100 μm in A–D; 25 μm in I.
To determine whether the progressive loss of BRN3B+ RGCs resulted from apoptosis, we examined the number of apoptotic cells in control and Isl1-null retinas. The developmental RGC death normally takes place in the first postnatal week to eliminate false axon connections (Farah and Easter, 2005; Perry et al., 1983). Consistently, we detected few apoptotic cells in wild type retinas throughout embryogenesis. In Isl1-null retinas, while there was no significant change in the number of apoptotic cells during the peak of RGC genesis at E13.5 to E15.5, a significant increase in the number of apoptotic cells was detected from E16.5 (2.8-fold of the wild type level at E16.5, t-test P<0.01; 5.3-fold at E17.5, P<0.01; and 2.6 fold at E18.5, P<0.05, Fig. 3K)). The apoptotic cells were primarily detected in the GCL (Fig. 3I,J) and the timing of increased apoptosis corresponded to the massive loss of BRN3B+ RGCs.
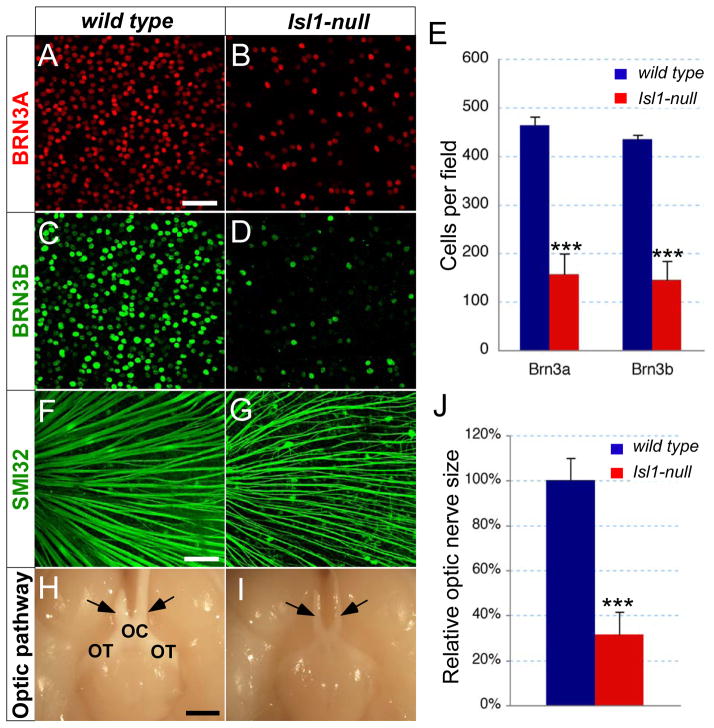
To quantify RGC loss as a result of Isl1-null mutation, we performed whole-mount immunostaining of adult retinas using anti-BRN3A and BRN3B antibodies (Fig. 4A–E). In adult retinas, BRN3A and BRN3B are each expressed in approximate 70% RGCs in a partially overlapping pattern and their combined expression reveals nearly the entire RGC population (Xiang et al., 1995). In Isl1-null retinas, both BRN3A+ and BRN3B+ cells were reduced to about 33% of those in wild type littermates. To rule out the possibility that this reduction was due to the down-regulation of BRN3A and BRN3B rather than the loss of RGCs, we also analyzed RGC axons as another parameter of RGC number. SMI32 antibody recognizes a non-phosphorylated epitope on the neurofilament H-chain and specifically labels large RGCs and their nerve fibers (Nixon et al., 1989). In Isl1-null retinas, the number of SMI32+ axon bundles was significantly reduced and the remaining ones appeared to be less fasciculated (Fig. 4F,G). Consistently, the ventral view of mouse brains revealed that Isl1-nulls have much thinner optic nerves, optic chiasms, and optic tracts (Fig. 4H,I). Quantification analysis showed a 69% reduction in the cross-section area of optic nerves in Isl1-nulls. These data indicate that Isl1 and Brn3b function, downstream of Math5, in parallel or corporative pathways to regulate the terminal differentiation and survival of RGCs and that ISL1 is required for RGC survival but not for the generation and migration of RGCs or for the initiation of BRN3B expression in retinas.
Figure 4.
Loss of RGCs in adult Isl1-null retina. (A–G) Immunostaining of whole-mount retina with anti-BRN3A (A,B), BRN3B (C,D) and SMI32 (F,G) antibodies reveals the reduction of RGCs in Isl1-null retina. (E) Quantification of BRN3A+ and BRN3B+ cells in the central retinal region of the whole-mounts (n=4 for each genotype) reveals a loss of 66% RGCs in Isl1-null retina. (H,I) Ventral views of brain show the thinner optic nerves (arrows) in Isl1-nulls. (J) Quantification of optic nerve size by measuring the cross area of H&E stained optic nerve transverse sections at the level indicated by arrows. Mean size of optic nerve in Isl1-null mice (red, n=6) is reduced to 31% of that in wild type (Blue, n=6). Each histogram represents the mean ± s.d. (***t-test P<0.001). OC, optic chiasm; OT, optic tract. Scale bar: 50 μm in A; 100 μm in F; 1 mm in H.
ISL1 and BRN3B regulate common downstream target genes
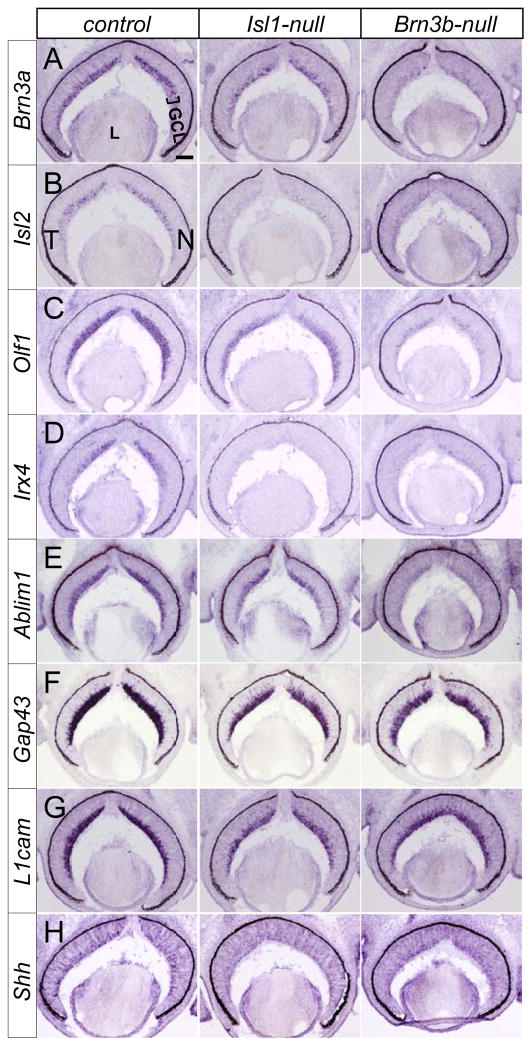
The co-expression of ISL1 and BRN3B raises the possibility that they regulate a common set of downstream target genes in differentiating RGCs. To test this possibility, we examined retinas at E14.5, a time point that neither the reduction of BRN3B expression nor the progressive RGC apoptosis has occurred. BRN3A, ISL2, IRX4, and OLF1 (EBF1–Mouse Genome Informatics), are TFs whose expression immediately follows BRN3B during retinal development. BRN3A is expressed in the post-migrated RGCs starting from E12.5 and is thought to play a redundant role with BRN3B (Pan et al., 2005). ISL2 is selectively expressed in one-third of contralaterally projecting RGCs and represses the ipsilateral targeting program (Pak et al., 2004). IRX4 is involved in intra-retina pathfinding of RGCs (Jin et al., 2003). OLF1, with an early onset of retinal expression from E12.5 (Xiang, 1998), has no identified function in RGC differentiation. ABLIM1, GAP43 and L1CAM all play crucial roles in RGC axon growth and pathfinding by mediating cytoskeleton changes or cell-cell interactions (Demyanenko and Maness, 2003; Erkman et al., 2000; Suh et al., 2004; Zhang et al., 2000). SHH is secreted by differentiated RGCs and negatively regulates the differentiation of retinal progenitors into RGCs (Wang et al., 2005; Wang et al., 2002b; Zhang and Yang, 2001). When compared with the controls, the expression of the above genes in the GCL was reduced in mice lacking either Isl1 or Brn3b (Fig. 5 and Table S1). Interestingly, we also observed that the reduction in the expression of Brn3a, Olf1 and Ablim1 was less severe in Isl1-null than in Brn3b-null retinas (Fig. 5A,C,E). In contrast, a more significant decrease in Isl2 expression was seen in Isl1-null retinas (Fig. 5B). The expression analysis presented here not only supports a direct role of ISL1 in RGC differentiation but also suggests that ISL1 and BRN3B likely function cooperatively through regulating the common downstream target genes.
Figure 5.
ISL1 and BRN3B regulate the expression of a common set of RGC-specific genes. (A–H) Compared with controls (left panels), in situ hybridization shows that at E14.5, the expression of RGC specific genes Brn3a, Isl2, Olf1, Irx4, Ablim1, Gap43, L1cam, and Shh decreases in the GCL (bracket) of both Isl1-null (middle panels) and Brn3b–null retinas (right panels). GCL, ganglion cell layer; L, lens; T, temporal; N, nasal. Scale bar: 100 μm.
Isl1-null mutants are defective in RGC axon growth and pathfinding
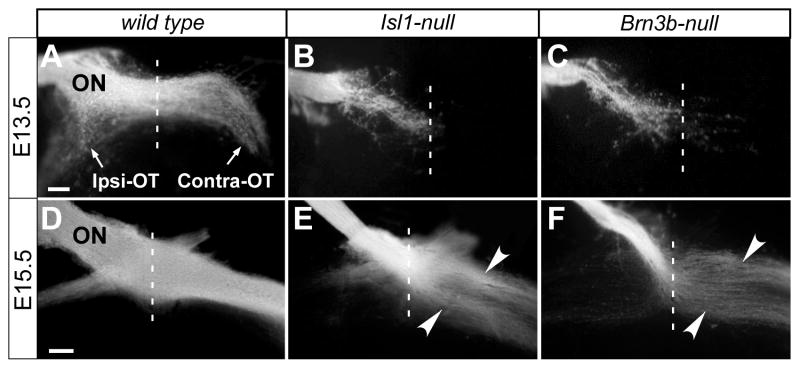
Since several Isl1downstream genes are involved in axon growth and/or pathfinding, we suspected that ISL1 might regulate axon behavior and that axon targeting defects might be the trigger of RGC apoptosis in Isl1-nulls. We anterogradely traced RGC axons by placing DiI crystals at the optic nerve-heads of fixed embryos. In mice, the pioneer RGC axons reach the optic chiasm at E12.5 (Marcus and Mason, 1995). Consistently, our analysis revealed that in the wild type embryo at E13.5, a large portion of RGC axons already passed the midline of the optic chiasm and extended in the contralateral optic tract. At the same time, a sizeable portion of axons turned away from the midline and proceeded in the ipsilateral optic tract (Fig. 6A). However, in Isl1-null and Brn3b-null mice (Fig. 6B,C), most axons failed to reach the midline at E13.5. At E15.5, the optic chiasm structure in wild type mice was well defined and resembled its adult-like morphology (Fig. 6D). In both Isl1-null and Brn3b-null mutants, though RGC axons projected into the contra- and ipsi-lateral optic tracts, several pathfinding defects were apparent (Fig. 6E,F). First, the optic nerves were smaller, indicating that a substantial fraction of RGCs failed to send out axons or that the axons did not exit the optic disc. Second, the axons in the optic tracts were less fasciculated and appeared to be grouped into two bundles (Fig. 6E,F, arrows). These observations are consistent with the recognized function of BRN3B in RGC axon growth and pathfinding (Erkman et al., 2000; Gan et al., 1999), and the similar roles of ISL1 in the projections of spinal motor neuron and sensory neuron axons (Kania and Jessell, 2003; Segawa et al., 2001; Thor and Thomas, 1997). Taken together, our data suggest that the axon growth defects occur prior to the onset of RGC apoptosis and may contribute to RGC death in both mutants.
Figure 6.
Axon growth defects in mice deficient for Isl1 or Brn3b. (A–F) After DiI was placed at the right optic nerve head, brains were dissected to expose the optic pathways at the ventral diencephalons. RGC axons of wild type pass the midline (dot line), project both contra- and ipsi-laterally, and form the X-shaped optic chiasm at E13.5 (A). At E15.5 (D), the wild type chiasm resembles its mature morphology. In Isl1-nulls and Brn3b-nulls, a majority of RGC axons fail to reach the midline at E13.5 (B,C). At E15.5, although the chiasms are formed in the null mice (E,F), the optic nerves are noticeably thinner and axons are de-fasciculated in the optic tract (arrowheads). ON, optic nerve; Ipsi-OT, ipsilateral optic tract; Contra-OT, contralateral optic tract. Scale bar: 100 μm.
More severe RGC loss in Isl1 and Brn3b compound null mice
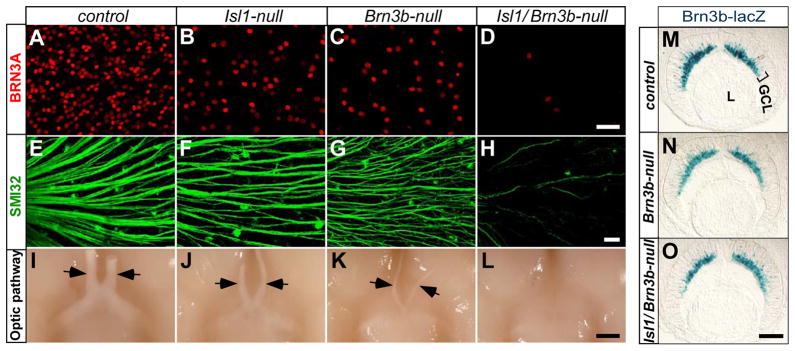
Based on the similarities in their expression patterns and roles during RGC development, we reasoned that BRN3B and ISL1 could function synergistically in RGCs and expected to observe more severe phenotypes in Isl1 and Brn3b compound null (Isl1/Brn3b-null) mutants. We therefore compared the number of BRN3A+ RGCs in whole-mount adult retinas of control, Isl1-null, Brn3b-null, and Isl1/Brn3b-null mice. We observed a reduction in the number of BRN3A+ cells in Isl1-null and Brn3b-null retinas compared with controls (Fig. 7A–C). Strikingly, only a very few BRN3A+ cells were identified in Isl1/Brn3b-null retinas (Fig. 7D). Similarly, compared with the high density of SMI32+ axon bundles in controls (Fig. 7E) or even the reduced axon bundles in Isl1-null or Brn3b-null (Fig. 7F,G) retinas, only a very limited number of SMI32+ axon bundles were detected in Isl1/Brn3b-nulls (Fig. 7H). Consistent with the reduction of axon bundles in the retinas, the optic nerves of Isl1/Brn3b-nulls were barely detectable and those of Isl1-nulls and Brn3b-nulls were reduced to about 31% and 20% of the controls, respectively (Fig. 7I–L, n=3 each genotype). The striking RGC loss in the compound mutants resembled that in Math5-null mice where only about 5% of RGCs are generated (Brown et al., 2001; Lin et al., 2004; Wang et al., 2001). However, unlike Math5-null mutation, loss of both Isl1 and Brn3b did not affect the genesis of RGCs. X-Gal staining of E13.5 retina sections revealed the comparable number of Brn3b-lacZ-labeled nascent RGCs in the NBL and GCL of control, Brn3b-null, and Isl1/Brn3b-null mice (Fig. 7M-O). Taken together, these data imply that Isl1 and Brn3b function together downstream of Math5 to control the terminal differentiation but not the genesis of almost all RGCs.
Figure 7.
More severe RGC loss in Isl1 and Brn3b compound null mice. (A–H) Immunostaining of adult whole-mount retinas with anti-BRN3A (A–D) and SMI32 (E–H). Compared with control (A,E), Isl1-null (B,F), and Brn3b-null (C,G), a more severe loss of BRN3A+ RGCs (red) and SMI32+ axon bundles (green) is observed in Isl1/Brn3b-null retina (D,H). (I–L) Ventral views of brains reveal optic nerves (arrows). Compared with the control (I), optic nerves are reduced in Isl1-nulls (J) and Brn3b-nulls (K). The optic nerves are barely detectable in Isl1/Brn3b-null mice (L). (M–O) X-Gal staining of Brn3b-lacZ-expressing RGCs in E13.5 retina sections shows that compared with the control (M), the genesis and migration of RGCs are undisturbed in Brn3b-null (N) and Isl1/Brn3b-null (O) retinas. Scale bar: 50 μm in D; 200 μm in H; 800 μm in L and 100 μm in O.
Simultaneous binding of ISL1 and BRN3B to RGC-specific promoters
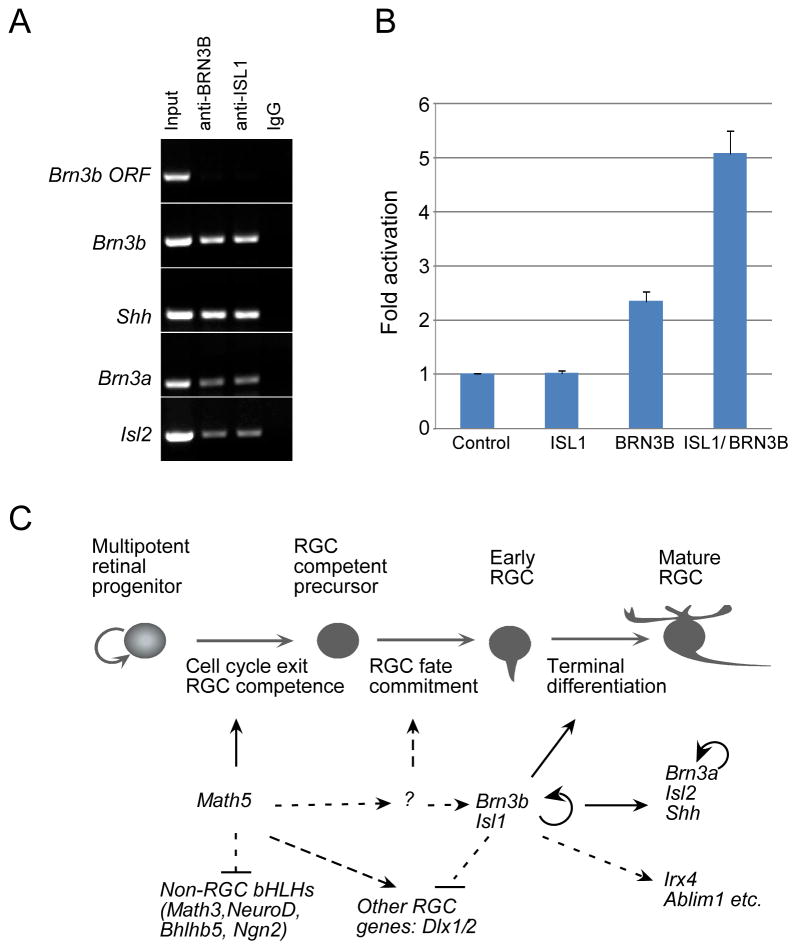
To investigate whether the cooperative function of ISL1 and BRN3B during RGC development was mediated by their direct regulation of RGC-specific genes, we explored the co-occupancy of these two factors on RGC-specific promoters in vivo with ChIP assays. The BRN3-binding site, ATNA(A/T)T(T/A)AT (Gruber et al., 1997; Trieu et al., 2003), was found in many genes expressed in RGCs. We examined four of them, Brn3b, Shh, Brn3a and Isl2 (see Fig. S4 in the supplementary material), to determine whether they are directly regulated by both ISL1 and BRN3B. Besides Brn3b, these genes were chosen because their expression in RGCs immediately follows that of ISL1 and BRN3B (Quina et al., 2005; Wang et al., 2005; Xiang, 1998) and depends on ISL1 and BRN3B (Fig. 5). We immuno-precipitated the chromatin from wild type retinas at E13.5-E14.5 and PCR-amplified the promoter regions containing Brn3-binding consensus sequence (see Fig. S4 in the supplementary material). Both anti-ISL1 and anti-BRN3B antibodies co-precipitated with the promoters of Brn3b, Shh, Brn3a and Isl2 (Fig. 8A and Fig. S3 in the supplementary material). Neither of these antibodies precipitated with Brn3b coding sequences in the controls (Fig. 8A). To further sustain the specificity of our assays, we also incorporate several negative controls, including IgG-precipitation, anti-BRN3B precipitation of chromatin derived from Brn3b-null retinas and anti-ISL1 precipitation with cerebellum tissues where ISL1 was not expressed. (Fig. 8A, Fig. S3). Moreover, anti-ISL1 was not able to co-precipitate with these promoters in Brn3b-null retinas, implying that the binding of ISL1 to these promoters depends on BRN3B.
Figure 8.
Functional mechanism of ISL1 and BRN3B in the development of RGCs. (A) Concurrent binding of ISL1 and BRN3B to RGC-specific promoters. Anti-BRN3B and anti-ISL1 antibodies co-precipitate with the promoters of Brn3b, Shh, Brn3a and Isl2. Both antibodies do not precipitate with control Brn3b ORF. Moreover, control IgG precipitation does not show any non-specific binding to these promoters. (B) Functional synergy of ISL1 and BRN3B on Brn3a luciferase reporter gene expression. CV1 cells are co-transfected with Brn3a luciferase reporter construct, different expression plasmids and PRL encoding CMV-renilla luciferase. Cells transfected with reporter, PRL and empty pcDNA expression vector are used as controls. Luciferase activity is determined by normalizing firefly activity with renilla activity. Then fold activation is calculated by dividing the luciferase activity of experimental group with that of control. Each histogram represents the mean + s.d. (n=4) (C) The Math5-Brn3b/Isl1 pathway in the development of RGCs. MATH5 expression endows the post-mitotic precursors with RGC competence by activating BRN3B and ISL1 expression and inhibiting the expression of non-RGC bHLH TFs. In this report, we show that ISL1 cooperates with BRN3B to regulate RGC terminal differentiation by maintaining the expression of Brn3b and activating a RGC-specific gene cascade, including Brn3a, Isl2, Shh, and Irx4. Solid lines indicate the direct regulation identified.
We further explored whether ISL1 and BRN3B synergistically regulate the expression of their targets using in vitro transactivation assay. We used the established Brn3a-luciferase transactivation assays in CV1 epithelial cells (Trieu et al., 2003) and co-transfected CV1 cells with Brn3a-luciferase reporter and Isl1 and/or Brn3b expression plasmids. While BRN3B alone activated Brn3a-luciferase expression to 2.34-fold of the control level (t-test P<0.001) and ISL1 had no effect on Brn3a-luciferase expression (1.01-fold, P>0.05), co-expression of ISL1 and BRN3B activated Brn3a-luciferase expression by 5.07-fold (P<0.001, Fig. 8B). Thus, ISL1 and BRN3B simultaneously bind to and synergistically regulate the expression of their target genes.
DISCUSSION
TF networks have been shown to regulate the sequential events of neurogenesis including cell fate specification, differentiation, and neuronal subtype determination (Castro et al., 2006; Lee and Pfaff, 2003; Shirasaki and Pfaff, 2002). Among these TFs, the POU-HD and LIM-HD TFs frequently appear together in the same differentiating neurons and play essential roles during late stage of the neuronal differentiation process in both invertebrates and vertebrates. In this study, we use neural retina to explore the functional mechanism of LIM-HD and POU-HD TFs during neurogenesis. Using retina-specific knockout of Isl1 and gene expression assays, we show that similar to those in Brn3b-null retinas, a majority of RGCs in Isl1-null retinas are defective in axon growth and pathfinding, and die prenatally. Moreover, a loss of greater than 95% RGCs in Isl1/Brn3b-null retinas, the necessity of both ISL1 and BRN3B for the expression of common downstream genes, and their simultaneous binding to and synergistic regulation of RGC-specific genes demonstrate the parallel and cooperative nature of ISL1 and BRN3B function in RGC development.
Expression and function of ISL1 in RGC development
The onset of ISL1 expression in post-mitotic cells at E11.5 and its complete co-localization with BRN3B in nascent RGCs suggest a role in RGC development. We found that targeted disruption of Isl1 does not affect the genesis of RGCs. Rather, it results in the apoptosis of a significant number of RGCs (Fig. 3). The observed RGC defect in Isl1-null retinas is consistent with ISL1’s spatiotemporal expression in nascent RGCs and supports a role for it in the late stages of RGC development. In addition to RGCs, ISL1 is also expressed in developing cholinergic amacrine and ON-bipolar cells in mouse retina (Elshatory et al., 2007a). In a separate study, we have examined the effect of the Isl1-null mutation on these other two types of retinal neurons and found the postnatal loss of nearly all these cells in the absence of Isl1 (Elshatory et al., 2007b), further supporting its essential role in the late stages of retinal neurogenesis.
In normal retina development, about half of RGCs die during the first postnatal week due to the deprivation of target-derived neurotrophins as the result of axon mis-projection (Farah and Easter, 2005; Perry et al., 1983). It is possible that the massive RGC death in Isl1-null or Brn3b-null retinas could also result from a similar mechanism based on their recognized roles in axon guidance. In Drosophila, Islet is required for proper axon trajectory of spinal motor neurons (Thor and Thomas, 1997). Mis-expression of Isl1 in chicken also causes axon targeting errors of the spinal motor neurons (Kania and Jessell, 2003). Moreover, mis-guidance of RGC axons at multiple decision-making points has been reported in Brn3b–null mutants (Erkman et al., 2000). In this study, we show that the defect in RGC axons arises prior to RGC apoptosis in Isl1-null or Brn3b-null mutants. It is possible that the defect in axon outgrowth and/or targeting contribute to the excessive apoptosis of developing RGCs. At the molecular level, in mice null for Isl1 or Brn3b, there is a significant down-regulation of genes implicated in axon growth (Gap43), fasciculation (L1cam) and guidance (Isl2, Ablim1) (Fig. 5). Furthermore, DiI anterograde tracing experiments reveal that at E13.5, a significant amount of pioneer RGC axons fail to reach the midline in mice null for Isl1 or Brn3b (Fig. 6). At E15.5, although the optic chiasm does form in these null mutants, it is always associated with axon growth and fasciculation defects.
Since loss of Brn3b results in a similar phenotype of RGC apoptosis, it is possible that ISL1 could regulate the terminal differentiation and survival of RGCs by maintaining Brn3b expression. Using anti-BRN3B immunostaining, we show that starting from E16.5, both the number of BRN3B+ RGCs and its expression intensity per cell decline in Isl1-null retinas (Fig. 3). ChIP assays show that both ISL1 and BRN3B bind to Brn3b promoter in vivo. Previously, it has been shown that after the initial expression, BRN3B maintains its own expression by positive autofeedback regulation (Liu et al., 2001). Taken together, these data suggest that ISL1 and BRN3B act jointly in retina to maintain Brn3b expression by directly binding to and activating Brn3b promoter. The continuous expression of Brn3b then maintains the survival of developing RGCs.
Alternatively, ISL1 could directly control the terminal differentiation and survival of RGCs by regulating the expression of genes essential for these processes. Supporting this hypothesis are our findings that genes with expression in differentiating RGCs are significantly down regulated in Isl1-null retinas before the reduction of BRN3B expression and the initiation of RGC apoptosis (Fig. 5). The expression of these genes is also diminished in Brn3b-nulls, suggesting that these two TFs regulate a common set of downstream target genes. Interestingly, we have also noticed that the extent of down-regulation of certain RGC genes differs in Isl1-nulls than in Brn3b-nulls. For example, the expression of Brn3a, Olf1 and Ablim1 is reduced more severely in Brn3b-nulls while Isl2 expression is more significantly down regulated in Isl1-nulls (Fig. 5). There are two possible explanations to this. First, since the Six3-cre-mediated deletion of Isl1 in retinas is incomplete (10% ISL1+ cells remain), the expression of certain downstream target genes could be maintained in cells with residual ISL1 expression. On the other hand, the expression of these genes could have different dependency on ISL1 than on BRN3B. For example, BRN3B alone may be sufficient to maintain the expression of Brn3a, Olf1 and Ablim1 in certain RGCs, whereas ISL1 is essential for the expression of Isl2. The resolution of this different dependency awaits the future transcriptional regulation analysis of these genes.
ISL1 and BRN3B co-regulate the RGC differentiation program
Our data indicate that ISL1 and BRN3B simultaneously bind to the promoter regions of RGC-specific genes and synergistically regulate their expression during RGC differentiation. This finding is consistent with prior studies of the cooperative function of POU-HD and LIM-HD factors. The POU-HD factor PIT1 (POU1F1–Mouse Genome Informatics) and the LIM-HD factor P-LIM (LHX3–Mouse Genome Informatics) are co-expressed during pituitary development. PIT1 and P-LIM interact with each other and both bind to promoter sequences containing PIT1 binding sites (Bach et al., 1995). The LIM-domains of P-LIM are not required for DNA binding but are critical for the synergistic interaction with PIT1 on distal target genes, including Pit1. In C. elegans, UNC-86 dimerizes with MEC-3 to play an essential role in regulating the terminal differentiation of touch sensory cells (Duggan et al., 1998; Lichtsteiner and Tjian, 1995; Xue et al., 1992; Xue et al., 1993). However, in contrast to the interaction between PIT1 and P-LIM, the coupling of UNC-86 and MEC-3 does not require the LIM-domains and MEC-3 alone binds poorly to the promoters. We found similar results with ChIP experiments that anti-ISL1 does not co-precipitate with promoters tested in the absence of BRN3B (Fig. S3A), suggesting the binding of ISL1 to these promoters depends on BRN3B. We also found that ISL1 alone is not sufficient to activate Brn3a-luciferase reporter, but is essential for the synergetic activation of Brn3a when co-expressed with BRN3B (Fig. 8B).
The consensus sequence for ISL1-binding has not been clearly identified. ISL1 is thought to bind to A/T rich sequences like the other homeodomain TFs (Dodou et al., 2004; Lee and Pfaff, 2003). Thus, it is likely that in RGCs, ISL1 dimerizes with BRN3B and binds to BRN3-binding sites with an AT extension. Previous studies have shown by gel-shift assays that BRN3B binds to the BRN3-binding site (SBRN3) in the first intron of Shh and activates the expression of a reporter gene containing SBRN3 in cultured HEK293 cells (Mu et al., 2004). Using ChIP assay, we demonstrate that BRN3B does bind to this SBRN3-containing region in vivo in the developing retinas. Additionally, we reveal the simultaneously binding of ISL1 to the same region in the first intron of Shh. Intriguingly, ISL1 has also been reported as an upstream regulator of Shh during cardiac morphogenesis (Lin et al., 2006). It would be interesting to test whether ISL1 controls Shh expression through its first intron in developing heart.
Taken together, our expression and targeted deletion analysis suggests a Math5- Isl1/Brn3b pathway of RGC development (Fig. 8C). The transient expression of MATH5 endows the post-mitotic precursors with a period of RGC competence. During this competence period, MATH5 and other factor(s) yet to be identified activate the expression of Isl1 and Brn3b. This activation initiates the RGC differentiation program and suppresses the non-RGC differentiation pathways by negatively regulating the non-RGC specifying factors such as NEUROD, MATH3, NGN2, and BHLHB5 (Feng et al., 2006; Mu et al., 2005a). Since the loss of either Isl1 or Brn3b does not affect the initial expression of the other, their expression is regulated in parallel by upstream TFs such as MATH5. The joint action of ISL1 and BRN3B leads to the expression of RGC-specific genes including Brn3a, Shh, Olf1 and Ablim1 as well as the autofeedback regulation of Brn3b. Although our data imply that the majority of RGCs require both factors to activate their terminal differentiation, the more severe RGC loss observed in Isl1 and Brn3b compound null mutants suggests the existence of RGCs that rely on either BRN3B or ISL1 alone. Published studies show that other TFs, such as DLX1 and DLX2, also participate in regulating the terminal differentiation and survival of 33% RGCs (de Melo et al., 2005). Interestingly, the expression of Dlx1 and Dlx2 is up-regulated in Brn3b-null retinas, suggesting BRN3B normally represses Dlx1/2 expression (Mu et al., 2004; Pan et al., 2005). Thus, it is possible that BRN3B and DLX1/2 are required for the development of complementary sets of RGCs (de Melo et al., 2005). Though the remaining RGCs in Brn3b-null retinas represent most or all RGC subtypes judging by their dendritic structures (Lin et al., 2004), it remains to be tested whether any specific RGC subtype is selectively lost in Isl1-null or Dlx1/2-null mice.
In addition to retina, both ISL1 and BRN3 TFs are known to be expressed in the developing dorsal root ganglia, trigeminal ganglia, and the spiral and vestibular ganglia of the inner ear (Artinger et al., 1998; Avivi and Goldstein, 1999; Huang et al., 2001; Radde-Gallwitz et al., 2004; Sohal et al., 1996). Our findings that BRN3B and ISL1 cooperate in RGC differentiation strongly argue for a common co-regulatory mechanism of these two classes of TFs in neurogenesis in these other areas of the developing nervous systems. It remains unknown whether these two factors directly couple with each other in transcriptional complexes. Interactions of LIM-HD proteins are generally mediated by LDB family cofactors with the direct interaction between UNC-86 and MEC-3 as the only exception (Hobert and Westphal, 2000; Lichtsteiner and Tjian, 1995). In combination with immunoprecipitation, future experiments, such as in vitro pull down, in vitro DNA-binding, and yeast two-hybrid assays, can be used to resolve the biochemistry nature of this interaction.
Supplementary Material
Acknowledgments
We thank Dr. Alexandra Joyner for the W4 mouse ES cells, Drs., Richard Libby, Eric Turner, Jason Lanier, and the members of the Gan Laboratory for or many helpful discussions and technical assistance. This work was supported by NIH grants (EY013426 and EY015551), Kilian and Caroline F. Schmitt Program on Integrative Brain Research to L.G., and the Research to Prevent Blindness challenge grant to the Department of Ophthalmology at the University of Rochester.
References
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–25. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Artinger KB, Fedtsova N, Rhee JM, Bronner-Fraser M, Turner E. Placodal origin of Brn-3-expressing cranial sensory neurons. J Neurobiol. 1998;36:572–85. doi: 10.1002/(sici)1097-4695(19980915)36:4<572::aid-neu10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Auerbach W, Dunmore JH, Fairchild-Huntress V, Fang Q, Auerbach AB, Huszar D, Joyner AL. Establishment and chimera analysis of 129/SvEv- and C57BL/6-derived mouse embryonic stem cell lines. Biotechniques. 2000;29:1024–8. 1030–1032. doi: 10.2144/00295st04. [DOI] [PubMed] [Google Scholar]
- Avivi C, Goldstein RS. Differential expression of Islet-1 in neural crest-derived ganglia: Islet-1 + dorsal root ganglion cells are post-mitotic and Islet-1 + sympathetic ganglion cells are still cycling. Brain Res Dev Brain Res. 1999;115:89–92. doi: 10.1016/s0165-3806(99)00054-1. [DOI] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse RV, 2nd, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci U S A. 1995;92:2720–4. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang AG, Goulding MD. Regulation of vertebrate neural cell fate by transcription factors. Curr Opin Neurobiol. 1996;6:25–32. doi: 10.1016/s0959-4388(96)80005-5. [DOI] [PubMed] [Google Scholar]
- Brown A, Yates PA, Burrola P, Ortuno D, Vaidya A, Jessell TM, Pfaff SL, O’Leary DD, Lemke G. Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell. 2000;102:77–88. doi: 10.1016/s0092-8674(00)00012-x. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–44. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–95. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Du G, Fonseca M, Gillespie LA, Turk WJ, Rubenstein JL, Eisenstat DD. Dlx1 and Dlx2 function is necessary for terminal differentiation and survival of late-born retinal ganglion cells in the developing mouse retina. Development. 2005;132:311–22. doi: 10.1242/dev.01560. [DOI] [PubMed] [Google Scholar]
- Demyanenko GP, Maness PF. The L1 cell adhesion molecule is essential for topographic mapping of retinal axons. J Neurosci. 2003;23:530–8. doi: 10.1523/JNEUROSCI.23-02-00530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–19. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007a;503:182–97. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007b;27:12707–20. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O’Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–6. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O’Connell SM, Krones AI, Kirby MA, Rapaport DH, Bermingham JR, et al. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28:779–92. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Farah MH, Easter SS., Jr Cell birth and death in the mouse retinal ganglion cell layer. J Comp Neurol. 2005;489:120–34. doi: 10.1002/cne.20615. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie X, Joshi PS, Yang Z, Shibasaki K, Chow RL, Gan L. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133:4815–25. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–2. [PubMed] [Google Scholar]
- Galli-Resta L, Resta G, Tan SS, Reese BE. Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J Neurosci. 1997;17:7831–8. doi: 10.1523/JNEUROSCI.17-20-07831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–80. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–5. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CA, Rhee JM, Gleiberman A, Turner EE. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol. 1997;17:2391–400. doi: 10.1128/mcb.17.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–9. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, Xiang M. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128:2421–32. doi: 10.1242/dev.128.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–42. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Jensen AM, Wallace VA. Expression of Sonic hedgehog and its putative role as a precursor cell mitogen in the developing mouse retina. Development. 1997;124:363–71. doi: 10.1242/dev.124.2.363. [DOI] [PubMed] [Google Scholar]
- Jin Z, Zhang J, Klar A, Chedotal A, Rao Y, Cepko CL, Bao ZZ. Irx4-mediated regulation of Slit1 expression contributes to the definition of early axonal paths inside the retina. Development. 2003;130:1037–48. doi: 10.1242/dev.00326. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem. 1998;273:3152–7. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–96. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–73. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Johnson WA, Luo L, Jefferis GS. From lineage to wiring specificity. POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;112:157–67. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–91. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–45. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. Embo J. 1995;14:3937–45. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron. 2004;43:475–85. doi: 10.1016/j.neuron.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lin L, Bu L, Cai CL, Zhang X, Evans S. Isl1 is upstream of sonic hedgehog in a pathway required for cardiac morphogenesis. Dev Biol. 2006;295:756–63. doi: 10.1016/j.ydbio.2006.03.053. [DOI] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci U S A. 2001;98:1649–54. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Mason CA. The first retinal axon growth in the mouse optic chiasm: axon patterning and the cellular environment. J Neurosci. 1995;15:6389–402. doi: 10.1523/JNEUROSCI.15-10-06389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Beremand PD, Zhao S, Pershad R, Sun H, Scarpa A, Liang S, Thomas TL, Klein WH. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004;131:1197–210. doi: 10.1242/dev.01010. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005a;280:467–81. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, Klein WH. Ganglion cells are required for normal progenitor- cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Curr Biol. 2005b;15:525–30. doi: 10.1016/j.cub.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Lewis SE, Dahl D, Marotta CA, Drager UC. Early posttranslational modifications of the three neurofilament subunits in mouse retinal ganglion cells: neuronal sites and time course in relation to subunit polymerization and axonal transport. Brain Res Mol Brain Res. 1989;5:93–108. doi: 10.1016/0169-328x(89)90001-6. [DOI] [PubMed] [Google Scholar]
- Pak W, Hindges R, Lim YS, Pfaff SL, O’Leary DD. Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell. 2004;119:567–78. doi: 10.1016/j.cell.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Pan L, Yang Z, Feng L, Gan L. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132:703–12. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Perry VH, Henderson Z, Linden R. Postnatal changes in retinal ganglion cell and optic axon populations in the pigmented rat. J Comp Neurol. 1983;219:356–68. doi: 10.1002/cne.902190309. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–20. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O’Leary DD, Goulding M, Turner EE. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci. 2005;25:11595–604. doi: 10.1523/JNEUROSCI.2837-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–21. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–36. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–9. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–28. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–81. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Sohal GS, Bockman DE, Ali MM, Tsai NT. DiI labeling and homeobox gene islet-1 expression reveal the contribution of ventral neural tube cells to the formation of the avian trigeminal ganglion. Int J Dev Neurosci. 1996;14:419–27. [PubMed] [Google Scholar]
- Suh LH, Oster SF, Soehrman SS, Grenningloh G, Sretavan DW. L1/Laminin modulation of growth cone response to EphB triggers growth pauses and regulates the microtubule destabilizing protein SCG10. J Neurosci. 2004;24:1976–86. doi: 10.1523/JNEUROSCI.1670-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Trieu M, Ma A, Eng SR, Fedtsova N, Turner EE. Direct autoregulation and gene dosage compensation by POU-domain transcription factor Brn3a. Development. 2003;130:111–21. doi: 10.1242/dev.00194. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–70. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–9. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Mu X, Bowers WJ, Kim DS, Plas DJ, Crair MC, Federoff HJ, Gan L, Klein WH. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002a;129:467–77. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–13. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Wang YP, Dakubo G, Howley P, Campsall KD, Mazarolle CJ, Shiga SA, Lewis PM, McMahon AP, Wallace VA. Development of normal retinal organization depends on Sonic hedgehog signaling from ganglion cells. Nat Neurosci. 2002b;5:831–2. doi: 10.1038/nn911. [DOI] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–69. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–85. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Finney M, Ruvkun G, Chalfie M. Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. Embo J. 1992;11:4969–79. doi: 10.1002/j.1460-2075.1992.tb05604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Tu Y, Chalfie M. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science. 1993;261:1324–8. doi: 10.1126/science.8103239. [DOI] [PubMed] [Google Scholar]
- Yan RT, Ma W, Liang L, Wang SZ. bHLH genes and retinal cell fate specification. Mol Neurobiol. 2005;32:157–71. doi: 10.1385/MN:32:2:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–54. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Zhang F, Lu C, Severin C, Sretavan DW. GAP-43 mediates retinal axon interaction with lateral diencephalon cells during optic tract formation. Development. 2000;127:969–80. doi: 10.1242/dev.127.5.969. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Yang XJ. Regulation of retinal ganglion cell production by Sonic hedgehog. Development. 2001;128:943–57. doi: 10.1242/dev.128.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.