Abstract
α-Synuclein is the major component of pathological inclusions characteristic of diseases like Parkinson’s disease, dementia with Lewy bodies, and multiple systems atrophy. A role for α-synuclein in neurodegenerative diseases is further supported by point mutations and duplication/triplication of the α-synuclein gene (SNCA) that are causative of these disorders. The middle hydrophobic region of the α-synuclein protein, also termed the “non-Aβ component of Alzheimer’s disease amyloid plaque (NAC)” domain, is required for α-synuclein to polymerize into amyloid filaments, which are the major components of α-synuclein pathological inclusions. In the current studies, we assessed the importance of specific stretches of hydrophobic residues in driving the intrinsic ability of α-synuclein to polymerize. Several small deletions, even one as short as 2 amino acid residues (A76 and V77), dramatically impaired the ability of α-synuclein to polymerize into mature amyloidogenic fibrils, and, instead, preferentially formed oligomers. However, this inhibition of filament assembly was clearly dependent on the spatial context, since similar and larger hydrophobic deletions in other parts of the NAC domain only reduced the rate of fibril formation, without abrogating filament assembly. Further, mutation of residue E83 to an A rescued the ability of mutant Δ76–77 α-synuclein to polymerize. These findings support the notion that while both the location and hydrophobicity of protein segments are important elements that affect the propensity to form amyloid fibrils, the intrinsic ability of a polypeptide to structural fold into amyloid is also critical.
Synucleinopathies are a group of neurodegenerative disorders sharing in common the presence of intracellular inclusions comprised predominantly of α-synuclein (α-syn) amyloidogenic fibrils (1;2). These neuronal α-syn inclusions, termed Lewy bodies and Lewy neurites, are one of the defining characteristics of the Parkinson’s disease (PD) and dementia with Lewy bodies (DLB) (1–4). Similarly, α-syn inclusions in oligodendrocytes are a hallmark of multiple system atrophy (5;6). Furthermore, the presence of brain α-syn inclusions are associated with many other neurodegenerative diseases including pure autonomic failure, neurodegeneration with brain iron accumulation type-1 (NBIA-1), Down’s syndrome, and familial and sporadic Alzheimer’s disease (1;2).
α-Syn is a highly soluble heat-stable and “natively unfolded” protein (7), predominantly highly expressed in central nervous system (CNS) neurons, where it is localized in close proximity to vesicles within presynaptic terminals (1;8;9). α-Syn is a small 140-amino acid long protein characterized by an amino-terminal region containing several imperfect KTKEGV repeats, a hydrophobic center domain and a highly negatively charged carboxy-terminus region (1) (see Fig. 1a). Although the function of α-syn is still poorly understood, several studies suggest its involvement in modulating synaptic transmission, the density of synaptic vesicles, neuronal plasticity, vesicle recycling and synaptic integrity (1;9–12).
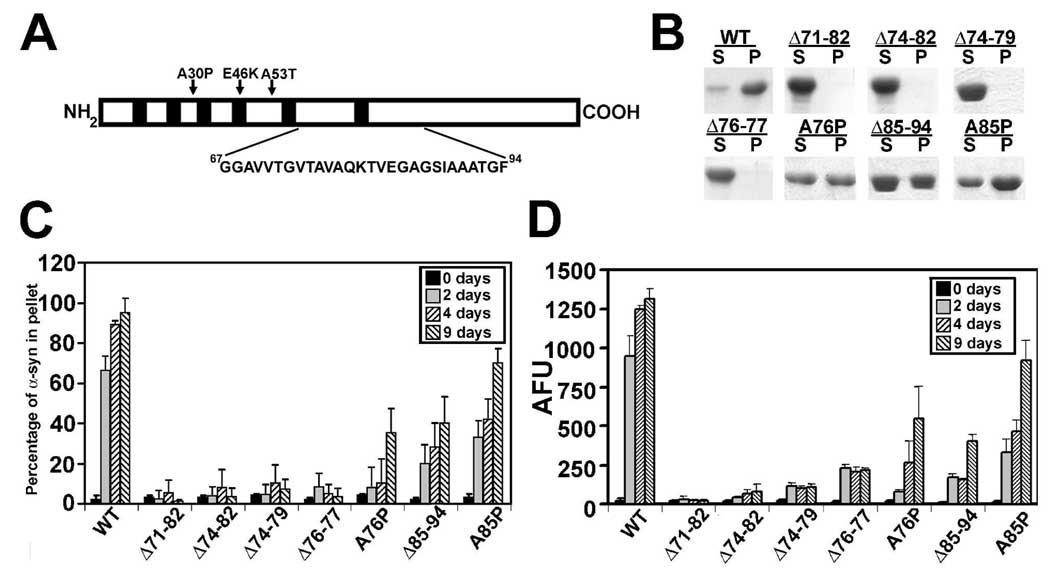
Figure 1. Studies of the effects of short deletions and P point mutations in the middle region of α-syn on polymerization.
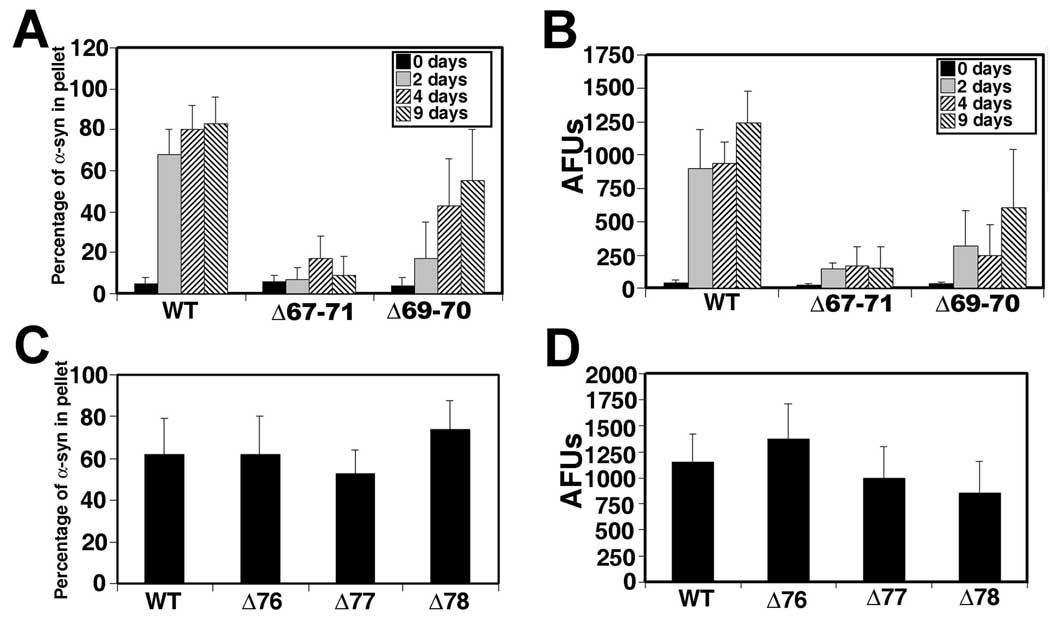
(A) Schematic of structure α-syn depicting the 6 imperfect “KTKEGV” repeats (black boxes), the three point mutations causal of disease (A30P, E46K, and A53T) and an expanded view of the amino acid sequence in the middle hydrophobic region. (B) Representative Coomassie Blue R-250 stained SDS-polyacrylamide gels showing the sedimentation state of WT α-syn or altered α-syn proteins (Δ71–82, Δ74–82, Δ74–79, Δ76–77, A76P, Δ85–94, and A85P) in supernatant (S) or pellet (P) after 9 days of assembly incubation at 5 mg/ml. (C) Quantitative analysis of the polymerization of indicated α-syn proteins assayed by sedimentation after 0, 2, 4 and 9 days of incubation under assembly conditions, as described in “Material and Methods”. (D) K114 fluorometry assessing the formation of amyloid from α-syn after incubated under assembly conditions as indicated. All proteins were incubated at 5 mg/ml. Data represent average ± SD. n = 6 for all data sets.
Disease causing alterations in the α-syn gene (SNCA) provide direct evidence for a fundamental role of α-syn in the pathogenesis of α-synucleionopathies. Genetic studies have identified three autosomal dominant missense mutations (A30P, E46K and A53T) in α-syn that are causative of PD and/or DLB (13–15). In addition, short chromosomal duplications or trisomies containing the SNCA gene, plus relatively short flanking regions on chromosome 4, were discovered in patients with PD or DLB (16;17), indicating that a 50% increase in the expression of α-syn is sufficient to cause disease.
In vitro studies have shown that recombinant soluble α-syn can readily polymerize into amyloidogenic fibrils that are structurally similar to those observed in human brains (18–21). A53T and E46K α-syn exhibit an increased rate of self-assembly and fibril formation (18;22–27), suggesting that these mutants could be pathogenic because they promote inclusion formation (28). Several studies indicate that the polymerization of α-syn progresses from unordered monomers to partially folded intermediates that assemble into oligomers/protofibrils and finally elongate into “mature” amyloid filaments (29;30). This conversion of α-syn from monomer to amyloid fibrils is associated with a dramatic conformational change from random coil to predominantly β-pleated sheet (21;30;31). Although many studies support the notion that the formation of mature fibrillar α-syn inclusions are pathological (28;32;33), some evidence also indicate that certain types of intermediate species such as protofibrils may have toxic properties (33–35).
Limited proteolysis experiments (21;36), EPR spectroscopy measurements (37;38) and hydrogen/deuterium exchange experiments (39;40) show that residues ~30–110 are buried within the core of α-syn fibrils, suggesting that these residues are important for polymerization. Consistent with these findings, deletion of amino acid residues 71–82 within the hydrophobic region abrogated the ability of human α-syn to fibrillize (21). In the current studies, we sought to further assess the importance of specific stretches of residues in the middle hydrophobic region of α-syn in driving its polymerization into fibrils.
EXPERIMENTAL PROCEDURES (MATERIALS AND METHODS)
Expression and Purification of α-Syn
The human α-syn cDNA was cloned into the Nde I and Hind III restriction sites of the bacterial expression vector pRK172. The cDNAs coding for the mutant A76P, A76G, A76V and A85P α-syn protein in the same vector were engineered by creating the corresponding nucleotide substitutions in the wild-type cDNA using complimentary sets of synthetic single stranded DNA containing the mutant sequence and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmids expressing α-syn with A76, V77, or A78 deleted were generated by using the QuikChange site-directed mutagenesis kit and oligonucleotides lacking these specific codons. 1–110 α-syn was created with the addition of a premature stop codon using QuickChange site-directed mutagenesis (41). The deletion of the nucleotide sequence coding for residues 71–82, 74–82, 74–79, 76–77, 67–71, 69–70 and 85–94 in α-syn cDNA was created using the Exsite PCR site-directed mutagenesis kit (Stratagene, La Jolla, CA) and oligonucleotides that specifically bind to the DNA sequence adjacent to the targeted deleted sequence. Deletion of nucleotide sequence coding for residues 73–83 was created by small modifications to the sequence with deletions in 71–82, using oligonucleotides which added two amino acids and removed one amino acid by QuickChange site-directed mutagenesis. The Δ76–77 α-syn construct with mutations at residues Q79A or E83A was created with oligonucleotides corresponding to the amino acid substitutions by QuickChange site-directed mutagenesis. All mutations were confirmed by DNA sequencing.
α-Syn proteins were expressed in E. coli BL21 (DE3) and purified as previously described (21;26). Briefly, bacterial pellets harvested by centrifugation were re-suspended in high-salt buffer (0.75 M NaCl, 50 mM Tris, pH 7.4, 1 mM EDTA) containing a cocktail of protease inhibitors, heated to 100°C for 10 min and centrifuged at 70,000 × g for 30 min. α-Syn proteins were purified by size-exclusion chromatography followed by ion exchange chromatography. Supernatants were dialyzed into 100 mM NaCl, 20 mM Tris, pH 7.5 and applied onto a Superdex 200 gel filtration column (GE Healthcare, Piscataway, NJ) and separated by size exclusion chromatography. The fractions were assayed for the presence of the α-syn proteins by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie Blue R-250 staining. All α-syn proteins (except for 1–110 α-syn - see below) were concentrated using Centriprep-10 units (Millipore Corp., Bedford, MA), dialyzed against 10 mM Tris, pH 7.5, applied to a Mono Q column (GE Healthcare) and eluted with a 0–0.5 M NaCl gradient. For 1–110 α-syn protein, the fractions isolated by gel filtration were dialyzed against 25 mM 2-[morpholino]ethanesulfonic acid, pH 6.25, applied to a Mono S column (GE Healthcare) and eluted with a 0–0.5 M NaCl gradient. Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce, Rockford, IL) with bovine serum albumin as a standard.
Filament Assembly and Centrifugal Sedimentation
α-Syn proteins were assembled into filaments by incubation at 37°C in 100 mM sodium acetate, pH 7.4 with continuous shaking. A fraction of each sample was set aside for K114 fluorometry and electron microscopic (EM) analysis. The remainder of each sample was centrifuged at 100,000 × g for 20 min. SDS-sample buffer (10 mM Tris, pH 6.8, 1 mM EDTA, 40 mM DTT, 1% SDS, 10% sucrose) was added to pellets and supernatants, which were heated to 100°C for 15 min. Equal volumes of α-syn proteins in the supernatants and pellets were separated by SDS-PAGE and were quantified by densitometry of Coomassie Blue R-250 stained gels.
K114 Fluorometry
α-Syn fibrils are amyloidogenic (30) and their formation can be quantified using the fluorescent amyloid binding dye K114 (42). This dye derived from the structure of Congo Red is soluble in aqueous buffers and it demonstrates a tremendous increase in fluorescence upon binding to amyloidogenic fibrils (42). This assay was conducted, as previously described (42), by incubating a fraction of each sample with the K114 (50 µM) in 100 mM glycine, pH 8.5 and measuring fluorescence (λex=380 nm, λem=550 nm, cutoff = 530 nm) with a SpectraMax Gemini fluorometer and SoftMax Pro 4.0 software.
Negative Staining EM
Assembled α-syn filaments were absorbed onto 300 mesh carbon coated copper grids, stained with 1% uranyl acetate and visualized with a JEOL 1010 transmission electron microscope (Peabody, MA). Images were captured with a Hamamatsu digital camera (Bridgewater, MA) using AMT software (Danvers, MA). For diameter determination, the width of 100 to 120 filaments was measured using Image-Pro Plus software (Media Cybernetics, Del Mar, CA).
Immuno-EM analysis
Samples were applied to 300 mesh carbon coated copper grids and following rinse with PBS, the grids were blocked with 1% BSA for 30 min. Mouse monoclonal anti-α-syn Syn214, which reacts with C-terminal residues 130–140 in α-syn (43), was diluted in PBS/1% BSA and applied to the grids for 30 min. Following extensive washes, a goat anti-mouse antibody conjugated to 6 nm colloidal gold (Electron Microscopy Sciences, Hatfield PA) diluted in PBS/1% BSA was applied to the grids for 30 min. Following washes with PBS, the samples were stained with 1% uranyl acetate.
RESULTS
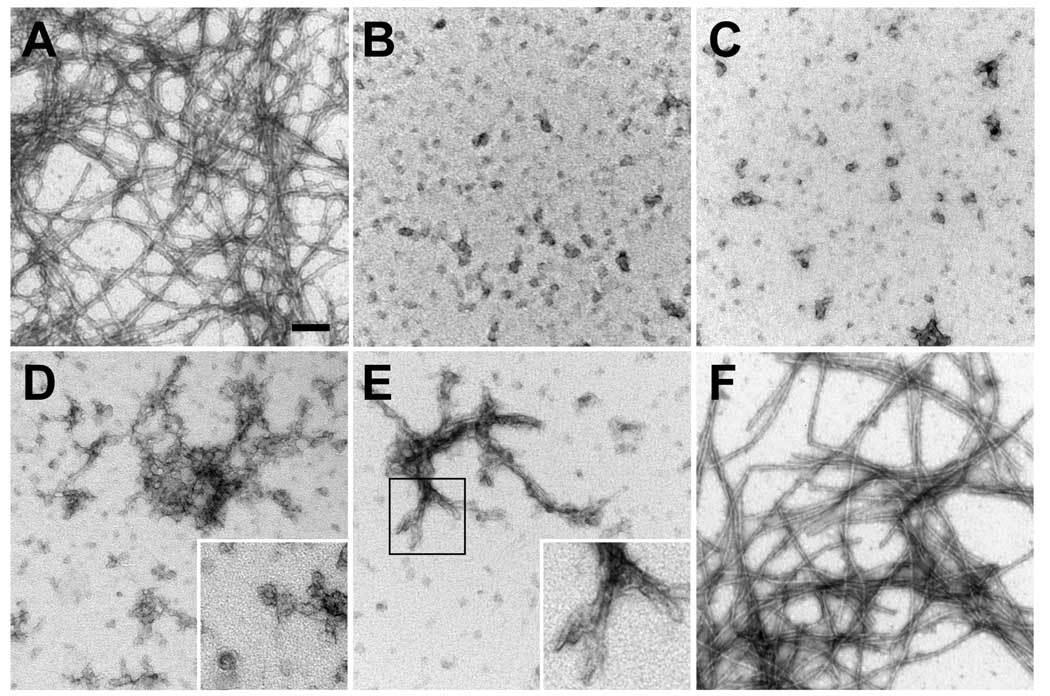
It was previously demonstrated that the deletions of residues 71–82 in α-syn impairs the ability of this protein to polymerize into amyloid fibrils (21). To better understand the molecular requirements of hydrophobic residues in fibril formation, we generated and characterized α-syn with several smaller deletions between amino acids 71–82 or with deletions in adjacent hydrophobic stretches (67GGAVV71 or 85AGSIAAATGF94). As previously reported, deletion of residues 71–82 impaired the polymerization of α-syn into amyloid fibrils as assayed by sedimentation analysis and K114 fluorometry (Fig. 1B–D). Electron microscopy (EM) analysis demonstrated that unlike wild-type (WT) α-syn that can readily form smooth negatively stained fibrils (width = 10.7 ± 2.0 nm) within 2 days, Δ71–82 α-syn predominantly formed 19.4 ± 3.2 nm wide spherical oligomers that accumulate over 9 days (Fig. 2 A, B). Smaller deletions (Δ74–82 and Δ74–79) in this region also impaired the ability of α-syn to assemble into amyloidogenic polymers as assayed by sedimentation and K114 fluorometry (Fig. 1 B–D), while EM analysis demonstrated the formation of spherical oligomers sized at 18 ± 3.2 nm and 15.6 ± 3.5 nm, respectively (data not shown; Fig. 2 C), and a paucity of fibrils. Even the deletion of only 2 amino acid residues 76AV77 (Δ76–77) dramatically impaired the polymerization of α-syn into amyloid fibrils (Fig. 1 C, D). Sedimentation analysis showed that this mutant was impaired in its ability to form large polymers, and K114 fluorometry demonstrated a small, but consistent increase in signal as early as 2 days of incubation. EM analysis revealed abundant 16.6 ± 3.4 nm spherical oligomers with Δ76–77 α-syn (Fig. 2D, E). Nevertheless, these Δ76–77 α-syn oligomers appeared to frequently coalesce (Fig. 2D), sometimes forming clusters with a fibrillar-like appearance after 9 days (Fig. 2E). However, these clusters of Δ76–77 α-syn protein never form the smooth, elongated fibrils, which are observed with WT α-syn (Fig. 2E, inset). The substitution of A76 for a P residue, an amino acid residue that strongly prevents formation of β-pleated sheet structure (44;45), reduced the propensity of α-syn to polymerize (Fig. 1 B–D). However, EM analysis showed that this mutant (A76P) could polymerize into 12.9 ± 2.4 nm wide amyloid fibrils (Fig. 2F).
Figure 2. EM analysis of the assembly of α-syn proteins.
Representative images of negative staining electron microscopy of α-syn proteins incubated at a concentration of 5 mg/ml under sedimentation conditions. The following proteins and time points were visualized: (A) WT α-syn for 2 days, (B) Δ71–82 for 9 days, (C) Δ74–79 for 9 days, (D) Δ76–77 for 4 days, (E) Δ76–77 for 9 days, (F) A76P for 9days. Bar scale = 100 nm, 50 nm for D,E-insets.
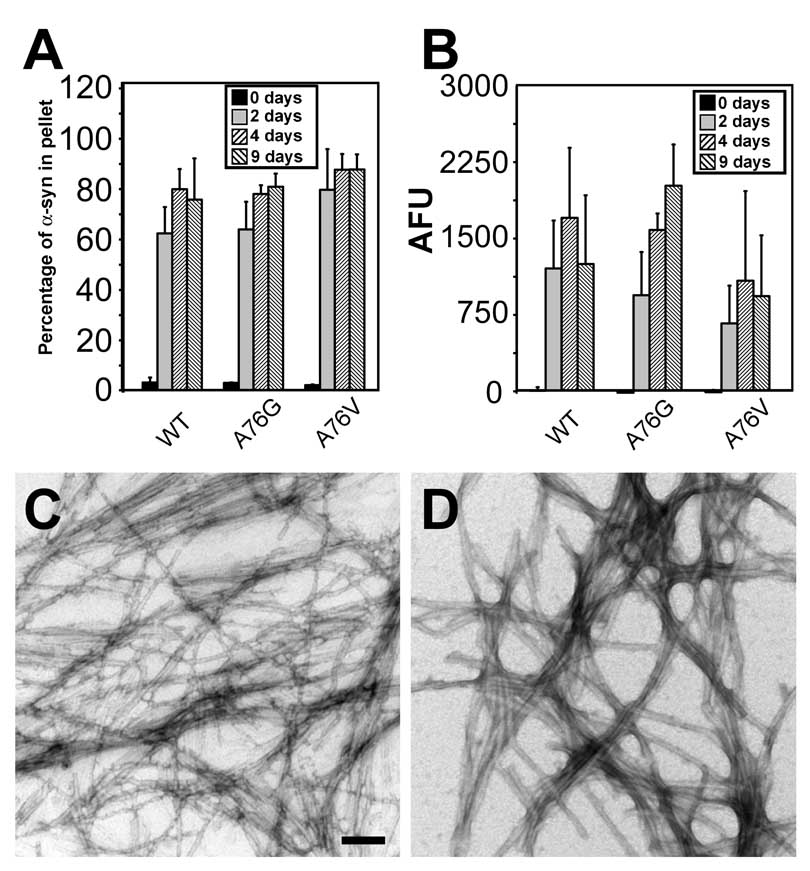
Previous studies showed that the substitution of residue A76 to charged residue (R or E) delayed fibril formation (21). To further define the preferences of this residue, A76 was mutated to a less hydrophobic, but non-polar residue (G) or more hydrophobic residue (V). Neither mutation significantly altered the rate of fibril formation (Fig. 3A,B). By EM analysis, the A76G mutation created fibrils of 12.0 ± 2.6 nm with a slightly altered morphology with fibrils that appear more directional than WT α-syn (Fig. 3C), and the A76V mutation created fibrils similar to WT α-syn (width = 10.8 ± 1.7 nm, Fig. 3D).
Figure 3. Assembly of α-syn protein with A76G or A76V mutations.
(A) Quantitative sedimentation analysis and (B) K114 fluorometry of WT α-syn verses that with the A76G or A76V mutations after 0, 2, 4, and 9 days at 5 mg/ml. Data represent average ± SD for 4 independent experiments. (C,D) Representative images of negative staining electron microscopy of α-syn with A76G (C) and A76V (D) mutations. Bar scale = 100 nm.
On the more C-terminal end of the hydrophobic domain, deletion of residues 85–94 (Δ85–94) or the substitution of P for A at position 85 (A85P) reduced the propensity of α-syn to form amyloidogenic fibrils as noted by sedimentation and K114 fluorometry (Fig. 1 B–D). Interestingly, the ultrastructure of the fibrils formed by Δ85–94 or A85P was different than typical WT α-syn fibrils (Fig. 4A,B). Instead of a smooth profile these fibrils appeared to have significant protrusions.
Figure 4. EM analysis of assembly of additional α-syn mutants.
Representative images of negative staining electron microscopy of α-syn proteins incubated at a concentration of 5 mg/ml under sedimentation conditions. The following proteins and time points were visualized: (A) Δ85–94 for 9 days, (B) A85P for 9 days, (C) Δ67–71 for 9 days, (D) Δ69–70 for 9 days, (E) Δ76 for 2 days, (F) Δ77 for 2 days. Bar scale = 100 nm.
Deletion of hydrophobic residues 67GGAVV71 (Δ67–71) also significantly reduced the ability of α-syn to form amyloidogenic fibrils (Fig. 5A,B). Albeit not frequent, clusters of Δ67–71 α-syn fibrils (width = 14.5 ± 3.0 nm) with typical negative stained smooth profiles were observed by EM at 9 days of incubation (Fig. 4C). Deletion of residues 69AV70 (Δ69–70) also reduced the rate of amyloidogenic fibril formation, but to a much lesser degree than the Δ67–71 deletion, forming fibrils of 13.2 ± 3.0 nm (Fig. 4D, 5A, B). The deletion of these 2 hydrophobic residues 69AV70 did not have the same drastic effect as deleting resides 76–77.
Figure 5. Analysis of the polymerization of α-syn deletion mutants.
(A) Quantitative sedimentation analysis and (B) K114 fluorometry of α-syn deletion mutant Δ67–71 and Δ69–70 after 0, 2, 4, and 9 days of assembly condition at 5 mg/ml. (C) Quantitative sedimentation analysis and (D) K114 fluorometry of α-syn with single amino acid deletions Δ76, Δ77 or Δ78 after 2 days of assembly condition at 5 mg/ml.
To further investigate the effect of the 76AV77 deletion mutant, we generated single amino acid deletion mutations A76 (Δ76), V77 (Δ77) and A78 (Δ78) to examine if a particular single residue was responsible for this dramatic inhibition of amyloidogenic fibrils. Δ78 α-syn was generated, since the 76AV77 deletion mutant results in a single A residue between 75 and 79. Deletion of 76AV77 is equivalent to 77VA78; therefore, the deletion of each of the three amino acids was important to investigate. However, none of these three single deletion mutations significantly affected the ability of α-syn to polymerize into amyloid fibrils (Fig. 4E, F, 5C, 5D; EM for Δ78 – data not shown).
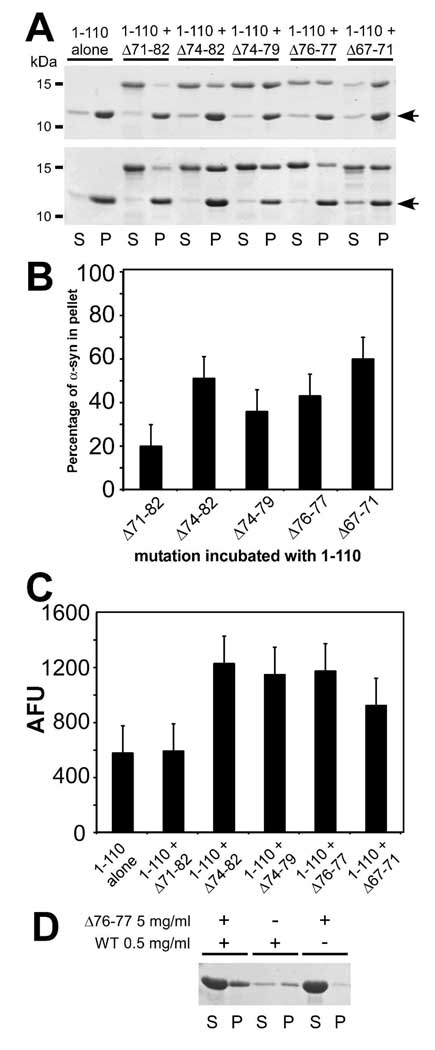
It was previously reported that WT α-syn assembly does not promote Δ71–82 α-syn fibril formation when co-incubated (21). To determine if α-syn has the ability to stimulate assembly of the other fibrillization-incompetent α-syn mutants, we co-incubated the deletion mutants with an assembly competent α-syn protein. To differentially resolve deletion mutations from fibril-competent α-syn by SDS-PAGE, we utilized C-terminally truncated α-syn 1–110, which is efficient in inducing WT α-syn amyloid formation (41). α-Syn 1–110 was co-incubated with Δ71–82, Δ74–82, Δ74–79, Δ76–77 or Δ67–71 α-syn deletion mutants. While Δ71–82 exhibited only a slight increase in sedimented protein (above that of incubation on its own), significant increases in sedimentation were observed with all of the other α-syn deletion mutants, such that between 35 and 60% of these proteins were sedimented (Fig. 6A,B). Increases in K114 fluorometry above that of 1–110 α-syn alone supported increased amyloid formation of α-syn deletion mutants Δ74–82, Δ74–79, Δ76–77, and Δ67–71 when co-incubated with 1–110 α-syn (Fig. 6C).
Figure 6. Co-assembly of 1–110 α-syn protein with deletion mutants.
(A) Representative Coomassie Blue R-250 stained SDS-polyacrylamide gels showing the sedimentation state of 1–110 α-syn with Δ71–82, Δ74–82, Δ74–79, Δ76–77 or Δ67–71 in supernatant (S) or pellet (P) after 4 days of incubation with each protein at a concentration of 2.5 mg/ml. Arrows indicate 1–110 α-syn protein. Two independent experiments are represented. (B) Quantitative sedimentation analysis of α-syn proteins with deletions in the hydrophobic region and (C) K114 fluorometry of co-assembly of deletion mutants with 1–110 α-syn at a concentration of 2.5 mg/ml after 4 days. Data represent average ± SD for 3 independent experiments. (D) Representative Coomassie Blue R-250 stained SDS-polyacrylamide gel showing sedimentation of Δ76–77 α-syn at 5 mg/ml with or without a seed of partially fibrillized WT α-syn at 0.5 mg/ml after 9 days of incubation.
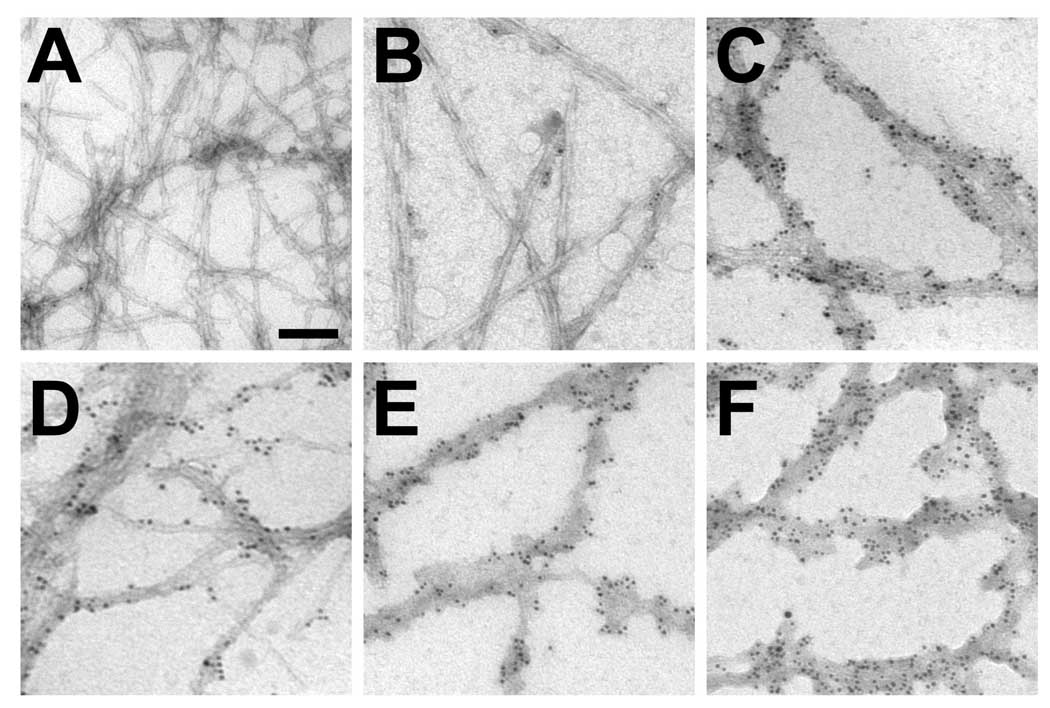
Immuno-EM analysis was performed to assess the presence and structure of the deletion mutants when incubated in the presence of 1–110 α-syn. Immuno-labeling was performed with an antibody (Syn214) specific to the extreme C-terminus of α-syn so that the labeling would reveal only proteins with deletions in the hydrophobic region, rather than 1–110 α-syn. Fibrils labeled with antibodies were decorated with 6 nm gold particles, but their morphologies were also less distinct due to the binding of antibodies to the fibrils. Samples assembled from only 1–110 α-syn revealed abundant fibrils, but no labeling with gold particles (Fig. 7A). Comparatively, 1–110 α-syn samples assembled in the presence Δ71–82 α-syn demonstrated scant labeling of fibrils (Fig. 7B), suggesting that possibly small levels of Δ71–82 protein were incorporated. Fibrils observed from samples with Δ74–82 α-syn, Δ74–79 α-syn, Δ76–77 α-syn or Δ67–71 α-syn co-incubated with 1–110 α-syn demonstrated abundant immuno-gold labeling (Fig. 7C–F), indicating that these proteins co-assembled with 1–110 α-syn.
Figure 7. Immuno-EM analysis of the ability of deletion mutant to co-assemble with 1–110 α-syn.
Immuno-EM analysis of co-assembly of α-syn with deletions in the hydrophobic region with 1–110 α-syn. Immunolabeling was performed with antibody Syn214, which reacts with the extreme C-terminus of α-syn, followed by immunogold labeling. (A) 1–110 α-syn protein assembled on its own did not show immunogold labeling, while (B) Δ71–82, (C) Δ74–81, (D) Δ74–79, (E) Δ76–77 and (F) Δ67–71 presented levels of immunogold labeling consistent with the levels of sedimentation and K114 fluorometry indicated in Fig. 6. Bar scale = 100 nm.
Δ76–77 α-syn cannot form smooth elongated amyloidogenic fibrils on its own, but can be partially incorporated into 1–110 α-syn fibrils. One possible explanation for these results is that Δ76–77 α-syn is deficient in nucleation. We therefore tested the ability of Δ76–77 α-syn to polymerize in the presence of WT α-syn seeds. WT α-syn protein was partially fibrillized at 5 mg/ml for 24 hrs, then removed and incubated at a concentration of 0.5 mg/ml with Δ76–77 α-syn at 5 mg/ml. After 9 days only 27 ± 5 % [SD] (n = 6) of a combination of Δ76–77 α-syn and the WT seeds sedimented verses 63 ± 19% [SD] of WT α-syn alone (Fig. 6D). The analysis of Δ76–77 with the WT seed was accompanied by an increase in K114 fluorometry by 484 ± 187 [SD] arbitrary fluorescent units (above that of Δ76–77 α-syn alone), verses 174 ± 49 [SD] arbitrary fluorescence units of the WT seed alone. The only modest sedimentation and incorporation of Δ76–77 α-syn, even in the presence of a seed, suggest that Δ76–77 α-syn is fundamentally impaired in the ability to polymerize into mature amyloidogenic fibrils, but similar to the data with 1–110 α-syn, it can still be partially incorporated when incubated with an assembly competent protein.
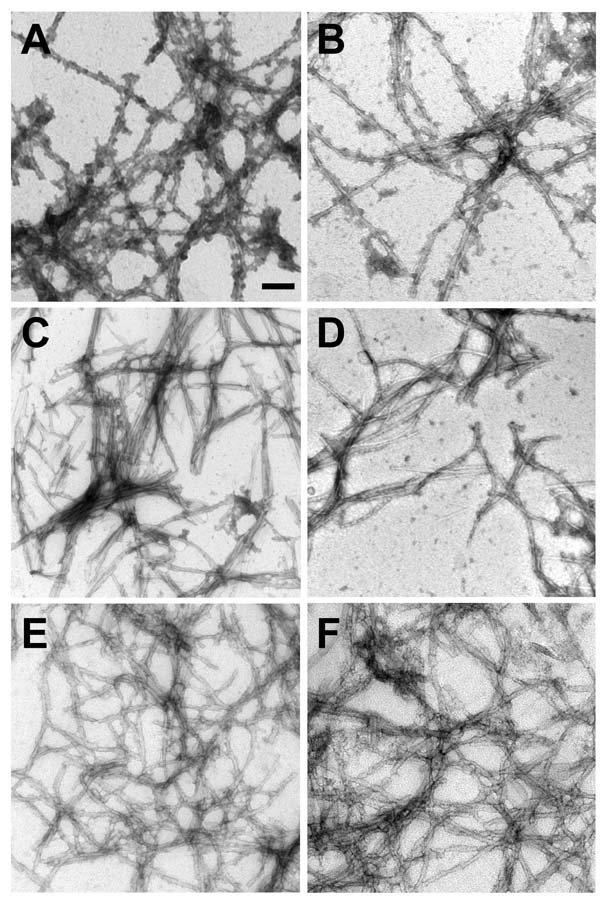
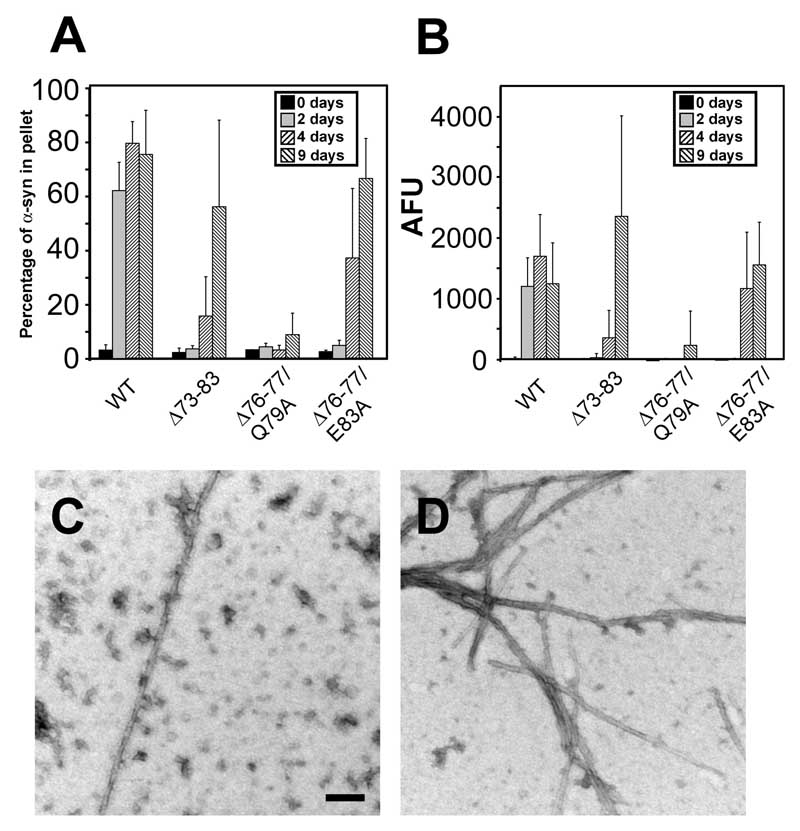
α-Syn with a deletion of amino acids 73–83 (Δ73–83) is capable of fibril formation (46), even though this alteration removes residues 76–77. We examined the ability of Δ73–83 α-syn to polymerize under our conditions and, similar to previous reports (46), found this protein competent in fibril formation, creating fibrils of 7.7 ± 1.5 nm after 4 days of incubation (Fig. 8A,B; EM – data not shown). We hypothesized that either residues Q79 (large, polar) or E83 (charged) may be preventing the polymerization of Δ76–77 α-syn, since both of these residues are absent in the Δ73–83 protein. α-Syn was created with amino acids 76–77 deleted and either Q79 or E83 mutated to an A (Δ76–77/Q79A and Δ76–77/E83A, respectively). Approximately 20% (n = 21) of Δ76–77/Q79A samples displayed increases in sedimentation and K114 fluorometry, but only after 9 days of incubation (Fig. 8A,B), and Δ76–77/E83A α-syn was capable of polymerization after 4 days of incubation (Fig. 8A,B). EM studies showed that Δ76–77/Q79A α-syn formed oligomers, similar to that observed with Δ76–77 α-syn, and in some samples incubated for 9 days, occasional smooth fibrils (width = 12.2 ± 2.9 nm) were observed (Fig. 8C). By EM, Δ76–77/E83A α-syn was observed to form abundant smooth fibrils (width = 9.3 ± 2.4 nm) by 4 days of incubation, but these fibrils had a tendency to “clump,” creating shorter fibrils than normally observed with WT α-syn (Fig. 8D). This data demonstrate that the presence of the negatively charged E83 residue contributes to the inability of Δ76–77 α-syn to polymerize into mature amyloid fibrils.
Figure 8. Study of assembly of Δ76–77 α-syn with mutation Q79A or E83A.
(A) Quantitative sedimentation analysis and (B) K114 fluorometry of WT α-syn verses α-syn with deletion of residues 73–83 (Δ73–83), or deletion of residues 76–77 with an Q79A (Δ76–77/Q79A) or E83A (Δ76–77/E83A) mutation after 0, 2, 4, and 9 days of incubation at 5 mg/ml. Data represent average ± SD for 4 independent experiments. (C,D) Representative images of negative staining electron microscopy of Δ76–77/Q79A α-syn (C) and Δ76–77/E83A α-syn (D) mutations. Bar scale = 100 nm.
DISCUSSION
The primary amino acid sequences of polypeptides that are compatible with amyloid formation can demonstrate considerable variability and flexibility, yet several basic intrinsic properties such as conformational preferences, solubility, charge state and the ability to pack into a fibrillar structure are important in influencing this process (47–50). The current study aimed to characterize the amino acid requirements for α-syn fibril formation, focusing on deletions in key hydrophobic regions and providing a more exhaustive assessment of the role that these amino acid segments have in driving α-syn amyloid formation.
Our previous work demonstrated that deletion of residues 71–82 within the hydrophobic core of α-syn abrogated the ability of this protein to form amyloid fibrils (21). To further narrow down the amino acids within this region that affect the ability of α-syn form amyloid fibrils, shorter deletion mutations (Δ74–82, Δ74–79 and Δ76–77) were generated. All these deletion mutants impaired the ability of α-syn to form typical ~ 10 nm wide unbranched amyloid fibrils as observed with WT α-syn. Instead, these mutants demonstrated a propensity to form spherical oligomers that have diameters (16–20 nm) larger than typical mature fibrils (~10 nm). Typical “on filament pathways” oligomers/protofibrils are ~ 4–5 nm in size (30;51). Therefore the oligomers generated by these deletion mutants are suggestive of “off pathway” oligomers.
Interestingly, the oligomers formed by deletions within the 71–82 region are reminiscent in size to the spherical oligomers generated when α-syn fibril formation is inhibited by treatment with dopamine, baicalein or EGCG (52–54) or to a small subpopulation of oligomers that form when α-syn is incubated under certain aqueous conditions (55;56). The inhibition of α-syn fibril formation by dopamine is due to the formation of dopamine oxidized by-products, which interact non-covalently to induce conformational changes in α-syn and form species that are unable to assemble into mature fibrils (54). It is possible that the deletion mutant proteins Δ71–82, Δ74–82, Δ74–79 and Δ76–77 α-syn form similar conformations or that they are fundamentally incompetent on their own to form the tertiary structures compatible with amyloid formation. Unlike the α-syn oligomers that are generated by dopamine treatment and that can become amyloid-compatible by denaturation/renaturation (54), mutants Δ71–82, Δ74–82, Δ74–79 and Δ76–77 α-syn intrinsically form these oligomers.
Nevertheless, co-assembly conditions with 1–110 α-syn promoted the partial co-polymerization of Δ74–82, Δ74–79 and Δ76–77 α-syn. While small amounts of fibril immunolabeling with antibody Syn214 (specific for the C-terminus of α-syn) were observed following co-incubation of 1–110 α-syn with Δ71–82 α-syn, we cannot rule out that Δ71–82 was simply non-specifically sticking to 1–110 α-syn protein. The much more robust Syn214 immunolabeling of fibrils obtained during the co-incubation of deletion mutants Δ74–82, Δ74–79, and Δ76–77 α-syn with 1–110 α-syn support that at least some of these proteins were incorporated within fibrils. The ability of these proteins to be incorporated within 1–110 α-syn fibrils is further supported by sedimentation studies and by the increase in K114 fluorescence. These data suggest that these deletion proteins are not completely incapable of amyloid formation, but they require co-assembly with a more permissive amyloidogenic protein to drive their polymerization. Additional co-assembly experiments using seeds comprised of WT α-syn demonstrate that only a small portion of Δ76–77 α-syn could be induced to fibrillize. These data indicate that the lack of amyloid formation by Δ76–77 α-syn is not predominantly due to a deficiency in seeding, and the inefficiency of filament formation in the presence of WT α-syn seeds indicated a fundamental deficiency of Δ76–77 α-syn in elongation of amyloid fibrils. Nevertheless, the small increases in K114 fluorometry of Δ76–77 α-syn after 2 days of incubation, on its own, suggest that a small portion of protein is capable of forming β-sheets, even if the protein cannot extend to form smooth amyloid fibrils.
Consistent with previous studies (21;46), we showed that while Δ71–82 α-syn was deficient in fibril formation, Δ73–83 α-syn was able to polymerize into mature amyloid, albeit at a slower rate than WT α-syn. These findings are apparently counterintuitive since both of these deletions remove a similar region and they both include residues 76 and 77, as well as other deleted segments (Δ74–79 and Δ74–82) that seem necessary for amyloid formation of α-syn. To try to explain these findings, the bulky, non-charged polar residue (Q79) and negatively charged residue (E83) in this region were mutated to an A within Δ76–77 α-syn. The Q79A amino acid substitution was able to promote the polymerization of Δ76–77 α-syn into amyloid, although inefficiently and only in a subset of samples. Conversely, the presence of the E83A mutation with Δ76–77 α-syn was able to sufficiently drive significant amyloid formation within 4 days of incubation. It was previously shown that mutating E83 to an A in WT α-syn increases the propensity of α-syn to form amyloid (26). This E residue may reduce amyloid formation due to the size of the residue and its negative charge as well as its focal inhibition of β-pleated sheets formation (26;44;45). Indeed modeling studies have suggested “a possible role of E83 as a gatekeeper residue to reduce the aggregation propensity” (57). The presence or lack of E83 likely explains some of the differences between some of the deletion proteins. The protein Δ73–83 α-syn has a significant ablation of hydrophobic residues, but this is partially compensated by the removal of E83. Furthermore, the deletion of residues 73–83 results in a new hydrophobic stretch from the adjacent upstream and downstream sequences (67GGAVVT72_84GAGSIAAATGF94). Other deletion proteins such as Δ74–82, Δ74–79 and Δ76–77 have smaller deletions of hydrophic residues, but still have the E83 residue. Our data, therefore, suggest that the presence and placement of the highly charged E83 residue plays a large role in the competency of hydrophobic deletion mutants to polymerize and can explain the differences in polymerization between the Δ71–82 and Δ73–83 deletion mutants. However, the propensity of Δ73–83 α-syn and Δ76–77/E83A α-syn to polymerize is significantly reduced when compared to WT α-syn, and Δ76–77/E83A α-syn exhibited altered morphology, as observed by EM. Therefore, the presence of the E83 residue is not the only determining factor, and conformational constraints resulting from deletion of particular hydrophobic residues also contribute to the propensity and ability of these proteins to polymerize into amyloid.
The substitution of A76 for a P residue, which is a potent β-pleated sheet inhibitor, did not result in as dramatic of an effect as the deletion of residues 76–77. The A76P mutation reduced the propensity for amyloid formation, similar to the A76E mutation (21), but it maintained the ability to form mature fibrils demonstrating a remarkable flexibility in the structure of amyloid formation at this position. These findings further suggest that deletion of residues 76–77 likely causes significant structural constraints that differ from these mutations of the A76 residue, preventing the protein from naturally folding into amyloid. Further, mutation of A76 to either G or V did not significantly alter polymerization, suggesting an overall propensity of α-syn to fibrillize despite modifications to this residue. Nevertheless, the A76G mutation exhibited slight changes to fibril morphology, appearing more directional than WT α-syn protein. While the alterations to the structure based on this mutation are currently unknown, these data suggest that a small, less hydrophobic residue may modify the packing of α-syn fibrils during amyloid formation.
Deletions of hydrophobic amino acid stretches flanking the 71–82 amino acid segment also reduce the propensity to form amyloid. Interestingly, deletion of residues 85–94 did not prevent fibril formation; Δ85–94 α-syn and the A85P mutation resulted in fibrils with abundant short protrusions rather than the typical smooth profile. This alteration in ultrastructure may be due to “stuttering” during polymerization, changes in the packing of the protein or a compensatory structure resulting from conformational constraints.
The deletion of residues 67–71 resulted in a significant reduction in amyloid formation, although filament assembly was not completely abolished. Other studies demonstrate that the deletion of residues 66–74 completely impairs amyloid formation, while deletion of residues 71–74 also significantly reduces polymerization, resulting in the formation of only short fibrils (58). The inability of Δ66–74 α-syn verses the ability of Δ67–71 α-syn (albeit significantly reduced) to polymerize may result from the conformational constraints imposed by the deletion of these particular residues. Alternatively, the deletion of the four extra hydrophobic residues in Δ66–74 α-syn may pass the threshold required to prevent fibril formation. However, the variable propensity of α-syn to form amyloid, depending on residue context and strained conditions, is further underscored by the ability of α-syn 1–74, where most of the hydrophobic middle region is removed, to still form abundant amyloid fibrils (58). In our studies, deletion of the residues 67–71 or 69–70 did not alter fibril morphology or structure as determined by EM analysis (Fig. 4C,D). Quantitative assays revealed that deletion of residues A69 and V70 resulted in abundant amyloid fibrils after 9 days of incubation (Fig. 5A,B), while deletion of the same residues in an analogous hydrophobic stretch (A76 and V77) abrogated the formation of mature amyloid. The dramatic differences in the effects of deleting these short residues highlight that it is both the nature of the residues as well as the spatial context that influence the process of amyloid formation. Single residue deletions (Δ76, Δ77 or Δ78) as well as the A76P mutation did not completely prevent amyloid formation. This suggests that even within a spatial context, inhibition of amyloid formation requires alterations to particular or minimal number of amino acids.
Our data are consistent with solid-state NMR studies and algorithms predicting β-sheet formation (59–61), which identify the major extended β-sheet region around residues 69–82. These predictions are based on an analysis of hydrophobic and charged residues in the core of the α-syn protein. While these models are informative, our data suggested considerable flexibility in the ability of α-syn to form β-amyloid.
Our studies indicate that not all hydrophobic regions of a polypeptide are equally important in determining its amyloidogenic aggregation tendency, despite the ability of very short specific amino acid stretches to facilitate amyloid fibril formation (62). Amyloid formation appears to have several influential factors, including composition of its residues and the conformational tendency to pack into amyloid tertiary and quaternary structures. However, there is also considerable flexibility in this process. While the hydrophobic core of α-syn holds the key for the ability of this protein to form amyloid, this study supports the complexity of this statement as demonstrated by the variability in amyloid formation between particular amino acid deletions. A better understanding of the basic molecular requirement and constrains associated with amyloid formation will provide important insights that may enable the generation of therapeutic agents capable of preventing the assembly of amyloid.
ACKNOWLEDGMENTS
This work was funded by grants from the National Institute on Aging (AG09215) and the National Institute of Neurological Disorders and Stroke (NS053488). E.A.W. was supported by a training grant (T32 AG00255) from the National Institute on Aging.
ABBREVIATIONS AND FOOTNOTES
- α-syn
α-synuclein
- NAC
non-Aβ component of Alzheimer’s disease amyloid plaque
- PD
Parkinson’s disease
- DLB
dementia with Lewy bodies
- NBIA-1
neurodegeneration with brain iron accumulation type-1
- CNS
central nervous system
- PAGE
polyacrylamide gel electrophoresis
- EM
electron microscopy
- WT
wild-type.
REFERENCES
- 1.Norris EH, Giasson BI, Lee VM. Alpha-synuclein: normal function and role in neurodegenerative diseases. Curr. Top. Dev. Biol. 2004;60:17–54. doi: 10.1016/S0070-2153(04)60002-0. [DOI] [PubMed] [Google Scholar]
- 2.Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 4.Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VMY, Trojanowski JQ, Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 5.Spillantini MG, Crowther RA, Jakes R, Cairns NJ, Lantos PL, Goedert M. Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson's disease and dementia with Lewy bodies. Neurosci. Lett. 1998;251:205–208. doi: 10.1016/s0304-3940(98)00504-7. [DOI] [PubMed] [Google Scholar]
- 6.Tu PH, Galvin JE, Baba M, Giasson B, Tomita T, Leight S, Nakajo S, Iwatsubo T, Trojanowski JQ, Lee VMY. Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble alpha-synuclein. Ann. Neurol. 1998;44:415–422. doi: 10.1002/ana.410440324. [DOI] [PubMed] [Google Scholar]
- 7.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 8.Iwai A, Masliah E, Yoshimoto M, Ge N, Flanagan L, de Silva HA, Kittel A, Saitoh T. The precursor protein of non-A beta component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 9.Clayton DF, George JM. Synucleins in synaptic plasticity and neurodegenerative disorders. J. Neurosci. Res. 1999;58:120–129. [PubMed] [Google Scholar]
- 10.Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, Mcllwain KL, Orrison B, Chen A, Ellis CE, Paylor R, Lu B, Nussbaum RL. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J. Neurosci. 2002;22:8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy DD, Rueter SM, Trojanowski JQ, Lee VMY. Synucleins are developmentally expressed, and alpha-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J. Neurosci. 2000;20:3214–3220. doi: 10.1523/JNEUROSCI.20-09-03214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra S, Gallardo G, Fernández-Chacón R, Schlüter OM, Südhof TC. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123:359–361. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 14.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat. Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 15.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez TE, del Ser T, Munoz DG, de Yebenes JG. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 16.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 17.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 18.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat. Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 19.Giasson BI, Uryu K, Trojanowski JQ, Lee VMY. Mutant and wild type human alpha-synucleins assemble into elongated filaments with distinct morphologies in vitro. J. Biol. Chem. 1999;274:7619–7622. doi: 10.1074/jbc.274.12.7619. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto M, Hsu LJ, Sisk A, Xia Y, Takeda A, Sundsmo M, Masliah E. Human recombinant NACP/alpha-synuclein is aggregated and fibrillated in vitro: relevance for Lewy body disease. Brain Res. 1998;799:301–306. doi: 10.1016/s0006-8993(98)00514-9. [DOI] [PubMed] [Google Scholar]
- 21.Giasson BI, Murray IV, Trojanowski JQ, Lee VMY. A hydrophobic stretch of 12 amino acid residues in the middle of alpha- synuclein is essential for filament assembly. J. Biol. Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Uversky VN, Fink AL. Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- 23.Narhi L, Wood SJ, Steavenson S, Jiang Y, Wu GM, Anafi D, Kaufman SA, Martin F, Sitney K, Denis P, Louis JC, Wypych J, Biere AL, Citron M. Both familial Parkinson's disease mutations accelerate alpha-synuclein aggregation. J. Biol. Chem. 1999;274:9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 24.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U. S. A. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway KA, Lee SJ, Rochet JC, Ding TT, Harper JD, Williamson RE, Lansbury PT. Accelerated oligomerization by Parkinson's disease linked alpha-synuclein mutants. Ann. N. Y. Acad. Sci. 2000;920:42–45. doi: 10.1111/j.1749-6632.2000.tb06903.x. [DOI] [PubMed] [Google Scholar]
- 26.Greenbaum EA, Graves CL, Mishizen-Eberz AJ, Lupoli MA, Lynch DR, Englander SW, Axelsen PH, Giasson BI. The E46K mutation in alpha -synuclein increases amyloid fibril formation. J. Biol. Chem. 2005;280:7800–7807. doi: 10.1074/jbc.M411638200. [DOI] [PubMed] [Google Scholar]
- 27.Choi W, Zibaee S, Jakes R, Serpell LC, Davletov B, Crowther RA, Goedert M. Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS Lett. 2004;576:363–368. doi: 10.1016/j.febslet.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Giasson BI, Lee VMY. Are ubiquitination pathways central to Parkinson's disease? Cell. 2003;114:1–8. doi: 10.1016/s0092-8674(03)00509-9. [DOI] [PubMed] [Google Scholar]
- 29.Uversky VN, Li J, Fink AL. Evidence for a Partially-Folded Intermediate in α-Synuclein Fibril Formation. J. Biol. Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 30.Conway KA, Harper JD, Lansbury PT. Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 31.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid- like cross-beta conformation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazzulli JR, Mishizen AJ, Giasson BI, Lynch DR, Thomas SA, Nakashima A, Nagatsu T, Ota A, Ischiropoulos H. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J. Neurosci. 2006;26:10068–10078. doi: 10.1523/JNEUROSCI.0896-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waxman EA, Giasson BI. Molecular mechanisms of alpha-synuclein neurodegeneration. Biochim. Biophys. Acta. 2008;1792:616–624. doi: 10.1016/j.bbadis.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volles MJ, Lee S-J, Rochet J-C, Shtilerman MD, Ding TT, Kessler JC, Lansbury PT. Vesicle permeabilization by protofibrillar α-synuclein: implications for the pathogenesis and treatment of Parkinson's disease. Biochemistry. 2001;40:7812–7819. doi: 10.1021/bi0102398. [DOI] [PubMed] [Google Scholar]
- 35.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 36.Miake H, Mizusawa H, Iwatsubo T, Hasegawa M. Biochemical characterization of the core structure of alpha-synuclein filaments. J. Biol. Chem. 2002;277:19213–19219. doi: 10.1074/jbc.M110551200. [DOI] [PubMed] [Google Scholar]
- 37.Der-Sarkissian A, Jai CC, Chen J, Langen R. Structral organization of alpha-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 39.Del MC, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Structure and properties of alpha-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilar M, Chou HT, Luhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. The fold of alpha-synuclein fibrils. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, Trojanowski JQ, Lee VM. Role of alpha-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 2003;42:8530–8540. doi: 10.1021/bi027363r. [DOI] [PubMed] [Google Scholar]
- 42.Crystal AS, Giasson BI, Crowe A, Kung MP, Zhuang ZP, Trojanowski JQ, Lee VM. A comparison of amyloid fibrillogenesis using the novel fluorescent compound K114. J. Neurochem. 2003;86:1359–1368. doi: 10.1046/j.1471-4159.2003.01949.x. [DOI] [PubMed] [Google Scholar]
- 43.Giasson BI, Jakes R, Goedert M, Duda JE, Leight S, Trojanowski JQ, Lee VMY. A panel of epitope-specific antibodies detects protein domains distributed throughout human alpha-synuclein in Lewy bodies of Parkinson's disease. J. Neurosci. Res. 2000;59:528–533. doi: 10.1002/(SICI)1097-4547(20000215)59:4<528::AID-JNR8>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 44.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 45.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 46.Zibaee S, Jakes R, Fraser G, Serpell LC, Crowther RA, Goedert M. Sequence Determinants for Amyloid Fibrillogenesis of Human alpha-Synuclein. J. Mol. Biol. 2007;374:454–464. doi: 10.1016/j.jmb.2007.09.039. [DOI] [PubMed] [Google Scholar]
- 47.Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of "aggregation-prone" and "aggregation-susceptible" regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 49.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Bemporad F, Calloni G, Campioni S, Plakoutsi G, Taddei N, Chiti F. Sequence and structural determinants of amyloid fibril formation. Acc. Chem. Res. 2006;39:620–627. doi: 10.1021/ar050067x. [DOI] [PubMed] [Google Scholar]
- 51.Apetri MM, Maiti NC, Zagorski MG, Carey PR, Anderson VE. Secondary structure of alpha-synuclein oligomers: characterization by raman and atomic force microscopy. J. Mol. Biol. 2006;355:63–71. doi: 10.1016/j.jmb.2005.10.071. [DOI] [PubMed] [Google Scholar]
- 52.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 53.Zhu M, Rajamani S, Kaylor J, Han S, Zhou F, Fink AL. The flavonoid baicalein inhibits fibrillation of alpha -synuclein and disaggregates existing fibrils. J. Biol. Chem. 2004;279:26846–26857. doi: 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- 54.Norris EH, Giasson BI, Hodara R, Xu S, Trojanowski JQ, Ischiropoulos H, Lee VM. Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J. Biol. Chem. 2005;280:21212–21219. doi: 10.1074/jbc.M412621200. [DOI] [PubMed] [Google Scholar]
- 55.Lashuel HA, Petre BM, Wall J, Simon M, Nowak RJ, Walz T, Lansbury PT., Jr Alpha-synuclein, especially the Parkinson's disease-associated mutants, forms pore-like annular and tubular protofibrils. J. Mol. Biol. 2002;322:1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- 56.Fredenburg RA, Rospigliosi C, Meray RK, Kessler JC, Lashuel HA, Eliezer D, Lansbury PT., Jr The impact of the E46K mutation on the properties of alpha-synuclein in its monomeric and oligomeric states. Biochemistry. 2007;46:7107–7118. doi: 10.1021/bi7000246. [DOI] [PubMed] [Google Scholar]
- 57.Rivers RC, Kumita JR, Tartaglia GG, Dedmon MM, Pawar A, Vendruscolo M, Dobson CM, Christodoulou J. Molecular determinants of the aggregation behavior of alpha- and beta-synuclein. Protein Sci. 2008;17:887–898. doi: 10.1110/ps.073181508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du HN, Tang L, Luo XY, Li HT, Hu J, Zhou JW, Hu HY. A peptide motif consisting of glycine, alanine, and valine is required for the fibrillization and cytotoxicity of human alpha-synuclein. Biochemistry. 2003;42:8870–8878. doi: 10.1021/bi034028+. [DOI] [PubMed] [Google Scholar]
- 59.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci U. S. A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heise H, Celej MS, Becker S, Riedel D, Pelah A, Kumar A, Jovin TM, Baldus M. Solid-state NMR reveals structural differences between fibrils of wild-type and disease-related A53T mutant alpha-synuclein. J. Mol. Biol. 2008;380:444–450. doi: 10.1016/j.jmb.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 61.Zibaee S, Makin OS, Goedert M, Serpell LC. A simple algorithm locates beta-strands in the amyloid fibril core of alpha-synuclein, Abeta, and tau using the amino acid sequence alone. Protein Sci. 2007;16:906–918. doi: 10.1110/ps.062624507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ventura S, Zurdo J, Narayanan S, Parreno M, Mangues R, Reif B, Chiti F, Giannoni E, Dobson CM, Aviles FX, Serrano L. Short amino acid stretches can mediate amyloid formation in globular proteins: the Src homology 3 (SH3) case. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]