Abstract
Although circadian rhythms of males and females are different in a variety of ways in many species, their mechanisms have been primarily studied in males. Furthermore, rhythms are dramatically different in diurnal and nocturnal animals but have been studied predominantly in nocturnal ones. In the present study, we examined rhythms in one element of the circadian oscillator, the PER1 protein, in a variety of cell populations in brains of diurnal female grass rats. Every 4 h five adult female grass rats kept on a 12-h light/dark (LD) cycle were perfused and their brains were processed for immunohistochemical detection of PER1. Numbers of PER1-labeled cells were rhythmic not only within the suprachiasmatic nucleus (SCN), the locus of the primary circadian clock in mammals, but also in the peri-suprachiasmatic region, the oval nucleus of the bed nucleus of the stria terminalis, the central amygdala, and the nucleus accumbens. In addition, rhythms were detected within populations of neuroendocrine cells that contain tyrosine hydroxylase. The phase of the rhythm within the SCN was advanced compared with that seen previously in male grass rats. Rhythms beyond the SCN were varied and different from those seen in most nocturnal species, suggesting that signals originating in the SCN are modified by its direct and/or indirect targets in different ways in nocturnal and diurnal species.
Keywords: extra-SCN, suprachiasmatic nucleus, tyrosine hydroxylase, diurnal rhythm, oval nucleus of the bed nucleus of the stria terminalis, central amygdala
In mammals, daily rhythms in a host of physiological and behavioral events are regulated by a biological clock located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus. These rhythms, which are entrained by the light/dark (LD) cycle to a 24 h day, are abolished by lesions of the SCN and restored by transplantation of fetal SCN back into lesioned animals (Lehman et al., 1987; Ralph et al., 1990; Klein et al., 1991). The molecular machinery of the clock itself is contained within single cells and comprises autoregulatory, transcriptional/translational feedback loops that have been elucidated in studies of mice (Reppert and Weaver, 2001; Herzog, 2007). The proteins CLOCK and BMAL1 provide positive transcriptional drives to the loop while negative feedback is mediated by the protein products of the Period (per1–2) and Cryptochrome genes (cry1–2), PER1/2 and CRY1/2, respectively. Information from this molecular clock within the SCN is transmitted to effector systems through traditional axonal outputs as well as diffusible signals (Watts et al., 1987; Silver et al., 1996).
There are now several reports of in vivo rhythms of PER1 and PER2 in brain regions outside the SCN of mammals. These regions include the oval nucleus of the bed nucleus of the stria terminalis (BNST-OV), the central amygdala (CEA) and the basolateral amygdala (BLA) (Amir et al., 2004; Lamont et al., 2005; Angeles-Castellanos et al., 2007), none of which are direct targets of SCN axonal outputs (Watts et al., 1987). But there is electrophysiological evidence that the BNST is linked to the SCN and these two brain regions exhibit synchronous rhythm in neural activity (Yamazaki et al., 1998). In the BNST-OV and the CEA of laboratory rat, the PER2 rhythms peak early in the dark phase of a 12-h LD cycle, as they do in the SCN. In the BLA and dentate gyrus (DG), the rhythms peak early in the light phase, and are thus 12 h out of phase relative to that in the SCN. Following lesions of the SCN, rhythms in the expression of clock genes in these regions damp out over a period of several days (Amir et al., 2004; Lamont et al., 2005); this is not the case for the olfactory bulbs (Abraham et al., 2005).
The vast majority of studies of clock gene expression within and beyond the SCN have been conducted with nocturnal male rodents. Although diurnal and nocturnal animals differ with respect to the temporal patterns of a host of behavioral and physiological rhythms, their basic circadian clocks appear to be quite similar (Smale et al., 2003). This is seen most directly in the phase of rhythms in expression of Per genes and their corresponding proteins within the SCN (reviewed in Smale et al., 2008). Diurnal and nocturnal species are also very similar with respect to clock outputs that have been examined within the SCN, such as mRNAs for vasopressin (VP) and prokineticin 2 (Dardente et al., 2004; Lambert et al., 2005). Differences between nocturnal and diurnal species have, however, been documented in several populations of neurons beyond the SCN, including cells that may be direct targets of SCN axonal outputs. These differences have been identified through the examination of Fos in the brains of the unstriped Nile grass rat (Arvicanthis niloticus), a diurnal rodent from East Africa. In males of this species, Fos is expressed rhythmically in the lower sub-paraventricular zone (LSPV) immediately dorsal to the SCN with a pattern quite different from that seen in nocturnal laboratory rats, and in constant darkness these rhythms persist in the former, but not the latter, species (Nunez et al., 1999; Schwartz et al., 2004). PER1 rhythms are also apparent in the LSPV of male grass rats kept in either LD (Ramanathan et al., 2006) or constant dark conditions (C. Ramanathan et al., unpublished observations). We have suggested previously that cells in this area, which project to the same regions as the SCN (Watts et al., 1987; Schwartz, 2006), may play an important role in the generation of diurnality (Nunez et al., 1999; Smale et al., 2003; Schwartz, 2006). Rhythms in a variety of other targets of the SCN, such as orexin-containing cells in the perifornical area, have also been compared in grass rats and laboratory rats (Martinez et al., 2002), as have rhythms in some populations of cells that do not receive direct input from the SCN (Novak et al., 1999, 2000). These all differ in the two species, though the nature of those differences varies from one region to another (Nunez et al., 1999; Smale et al., 2008).
Although females have also been neglected in studies of the neural substrates of circadian systems, it is clear that the fundamentals of the molecular feedback loops involved are the same in males and females (Masubuchi et al., 2000; Lincoln et al., 2002; Nakamura et al., 2005). Nevertheless, there are some sex differences in the basic properties of circadian rhythms and in how the system is influenced by internal and external variables. In some female rodents for example, there are changes in the phase and period of rhythms across the estrous cycle that are caused by changing patterns of secretion of ovarian hormones (Morin et al., 1977; Albers et al., 1981; Davis et al., 1983; Labyak and Lee, 1995). Rhythms in a variety of neuroendocrine and behavioral events are also modulated by pregnancy and lactation (Rosenwasser et al., 1987; Fewell, 1995; Zhang et al., 1995). At least some of the changes associated with reproductive state are promoted by secretion of steroids from the ovary (Morin et al., 1977; Takahashi and Menaker, 1980; Albers, 1981). The one aspect of circadian systems that has been studied more extensively in females than males involves the regulation of neuroendocrine events associated with reproduction (van der Beek, 1996; Freeman et al., 2000; de la Iglesia and Schwartz, 2006). In some species the SCN promotes the ovulatory surge in luteinizing hormone (LH), most likely through its direct projections from it to some cells containing gonadotropin releasing hormone (GnRH) and others containing estrogen receptors (de la Iglesia et al., 1995; van der Beek et al., 1997). Although there is no evidence of a molecular oscillator within GnRH cells in intact animals, rhythms in clock gene expression have been seen in GnRH cells in vitro (Gillespie et al., 2003). Prolactin also exhibits a surge around the time of ovulation that is influenced by circadian mechanisms (Freeman et al., 2000). There is some evidence that the SCN regulates this surge, and that it may do so at least partly through direct projections onto hypothalamic cells containing tyrosine hydroxylase (TH), the rate limiting enzyme in the synthesis of dopamine, an inhibitor of prolactin (Gerhold et al., 2001, 2002); rhythms in dopamine metabolism have been seen in hypothalamic areas containing these TH cells that are known to regulate prolactin secretion (Sellix and Freeman, 2003). In female rats and mice, rhythms in clock proteins have been documented in several populations of these TH cells in the periventricular nucleus and in the arcuate nucleus (Kriegsfeld et al., 2003; Sellix et al., 2006). Virtually nothing is known about rhythms of clock genes or proteins within TH cells of diurnal mammals.
The current study was designed to achieve a better understanding of the multioscillator system of female mammals, as well as to gain insight into basic mechanisms underlying diurnality. More specifically, one objective was to determine if rhythms in the SCN and peri–suprachiasmatic nucleus (pSCN) area of female grass rats are the same as those reported earlier for males (Ramanathan et al., 2006), and a second objective was to characterize extra-SCN oscillators, including those within TH-containing cells, in these diurnal females. To do this, we examined rhythms in PER1 in a variety of brain regions and cell populations of female grass rats killed at different times of day.
Experimental Procedures
Animals and housing
Adult female grass rats (Arvicanthis niloticus) (n=30) obtained from a breeding colony at MSU were singly housed and provided with access to food and water ad libitum. Animals were reproductively mature adults but there was no attempt to monitor reproductive condition as female grass rats in our colony do not show signs of estrous cycles in vaginal smears; furthermore, ovarian histology and daily tests of mating behavior reveal no evidence of a spontaneous estrous cycle (T. L. McElhinny and L. Smale, unpublished observations). Animals were maintained in a 12-h LD cycle (lights on at 06:00 h). The room was entered at irregular intervals to replenish food and water without disturbing animals. The experiment was performed in compliance with guidelines established by the Michigan State University All University Committee on Animal Use and Care, and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All efforts were made to prevent the suffering of the animals and to minimize the number of animals used in these experiments.
Specimen collection
Animals (n=5/group) were killed with an overdose of sodium pentobarbital and perfused at zeitgeber time (ZT) 2, 6, 10, 14, 18, and 22 with 0.01 M phosphate-buffered saline (PBS, pH 7.2), followed by 4% paraformaldehyde in 0.1 M phosphate buffer. Animals that were perfused during the dark phase had their heads covered with light-tight hoods to prevent acute effects of light. Brains were removed and post fixed in 4% paraformaldehyde for 4 h and then transferred into 20% sucrose in 0.1 M phosphate buffer and kept overnight. Brains were cut into three series of 30 μm coronal sections from the medial septum through the arcuate nucleus with a freezing microtome. Sections were stored in cryoprotectant solution at −20 °C until further processing.
Immunohistochemistry
The first series of sections was processed for immunohistochemical staining to detect both TH and PER1 protein. Free floating sections were rinsed in 0.01 M PBS and then incubated in 5% normal donkey serum (NDS; Jackson Laboratories, West Grove, PA, USA) in PBS with 0.3% Triton X-100 (TX) for 1 h at room temperature. After a 10 min wash, sections were incubated with a primary anti-PER1 antibody (mPER1 # 1177, made in rabbit, 1:20,000, a generous gift from Dr. David Weaver, University of Massachusetts, MA, USA), in 3% NDS and 0.3% TX-100 in PBS at 4 °C for 24 h and then with a biotinylated secondary antibody (donkey anti-rabbit; 1:200 in 3% NDS and 0.3% TX-100 in PBS) for 1 h, followed by avidin–biotin peroxidase complex (ABC; in 0.3% TX-100 with PBS) for 1 h. At this point sections were rinsed in 0.125 M acetate buffer then reacted with diaminobenzidine (DAB), nickel sulfate and 30% hydrogen peroxide. In order to label TH, the same sections were then rinsed in 0.1 M PBS followed by 0.2% phosphate-buffered saline with Triton X-100 (PBS-TX), and then incubated in 5% normal horse serum (NHS, in 0.2% PBS-TX) for 1 h after which they were placed with a primary antibody directed against TH (made in mouse; 1:20,000; Immunostar, in 3% NHS and 0.2% PBS-TX) for 24 h at 4 °C. Following a rinse in 0.2% PBS-TX they were incubated with a biotinylated horse anti-mouse secondary (in 3% NHS with 0.2% PBS-TX) for 1 h at room temperature and then in an ABC for 1 h at room temperature after which they were placed in a solution of DAB in Trizma buffer and reacting with 30% hydrogen peroxide. All sections were mounted on clean slides, dehydrated, and coverslipped.
Cell counts
We counted numbers of single labeled PER1-containing cells in the SCN and the pSCN regions. We also counted PER1-labeled cells in the shell of the nucleus accumbens (NA), the CEA, the BLA, and the BNST-OV (Fig. 1). Bilateral cell counts were done in the rostral, middle and caudal SCN. The same three sections were used to count cells in sampling boxes placed over three portions of the pSCN area as in Ramanathan et al., 2006: (1) the LSPV immediately dorsal to the SCN (215×160 μm), (2) the dorsolateral peri–suprachiasmatic nucleus area (dlpSCN) adjacent to the lateral boundary of the LSPV (160×160 μm), and (3) the ventrolateral peri–suprachiasmatic nucleus area (vlpSCN) ventral to the dlpSCN and just lateral to the SCN; the heights of the sampling boxes within the vlpSCN were 126 μm, 164 μm and 170 μm in the rostral, middle and caudal sections, respectively. PER1 was also counted bilaterally in one section through the CEA, BLA and NA, respectively, in three sections through the BNST-OV; the bilateral counts from the BNST-OV were averaged across the sections.
Fig. 1.
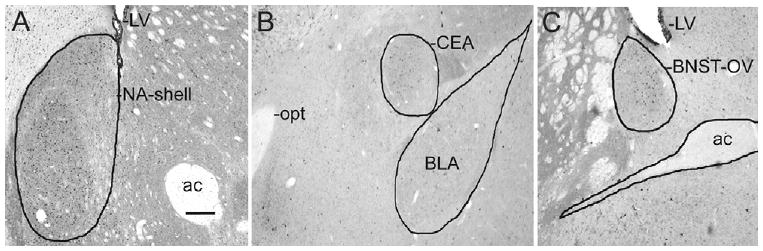
Sampling areas for the shell of the NA, the CEA and the BNST-OV. Photomicrographs showing key landmarks and the boundaries of the NA-shell (A), the CEA (B), and the BNST-OV (C). ac, Anterior commissure; opt, optic tract; LV, lateral ventricle. Scale bar=200 μm.
To evaluate PER1 in TH cells, we examined sections from the medial septum through the arcuate nucleus. We saw PER1 in four populations of TH cells (described in Mahoney et al., 2007) and in each of these we counted numbers of these cells with and without PER1. One of these was in a region dorsolateral to the rostral portion of the third ventricle; these cells represent a subset of the population referred to as periventricular hypothalamic dopaminergic neurons (PHDA), or A14, cells. The second was in the rostral arcuate nucleus; these cells are part of the A12 population and are sometimes referred to as tuberohypophysial dopaminergic neurons (THDA) cells. The third was in the dorsomedial arcuate nucleus; these cells are referred to as tuberoinfundibular dopaminergic neurons (TIDA), and are also a subset of the A12 cells. The last population of TH cells we analyzed was a magnocellular group present in the lateral hypothalamus and adjacent telencephalon (Mahoney et al., 2007). Counts were done in one section containing PHDA cells and in three sections containing each of the other populations of TH cells. All double-labeled cells were confirmed with a high power oil-immersion 100× objective.
Statistical analysis
Data were analyzed using SPSS 13 software. Numbers of single-labeled PER1+ cells in each of the three sections through the SCN and the pSCN areas were subjected to a square root transformation before analysis because of a lack of homogeneity of variance. For all figures the raw counts without transformation are presented as mean±S.E.M. For the SCN and each of the three pSCN areas, a two-way analysis of variance (ANOVA) was used to evaluate effects of 1) ZT and 2) rostro-caudal level of the section (referred to below as “level”). In the CEA, BLA, NA, and the BNST-OV the numbers of labeled-cells were analyzed with a one-way ANOVA to evaluate the effect of ZT. The percent of TH cells expressing PER1 was also analyzed using one-way ANOVAs to examine the effects of ZT; values for the A14 population were arc sine transformed prior to this analysis due to a lack of homogeneity of variance. We used the Fisher's least significant difference test for post hoc comparisons of the individual means. All values were considered significant when the P<0.05.
Results
PER1 in SCN and pSCN areas
Rhythms in PER1 were evident in the SCN as well as in all three regions around it (Fig. 2A). In the SCN there was a significant main effect of ZT on PER1 expression (F=63.04, df=5, P<0.001), but there was no effect of level (F=1.51, df=2, P=0.227) and no interaction (F=0.193, df=10, P=0.996; Fig. 2B). PER1 was lowest at ZT 2, rose progressively to a peak at ZT 14, and dropped abruptly from ZT 14 to ZT 18.
Fig. 2.
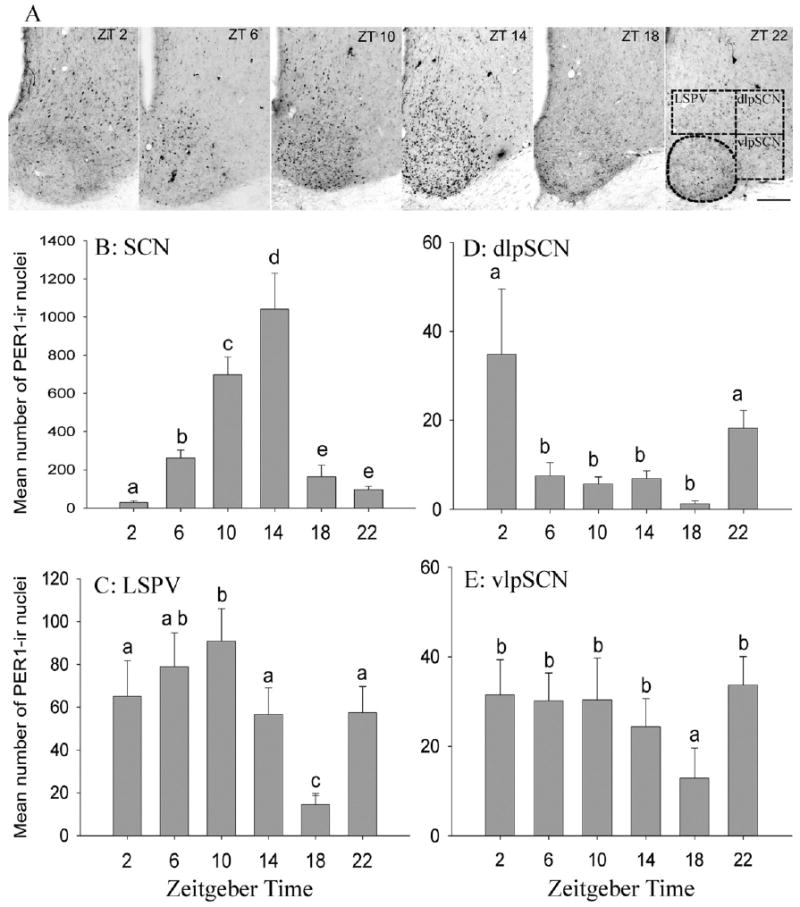
Daily rhythms of PER1 expression in the SCN and the pSCN regions. Photomicrographs of coronal sections through the SCN and the pSCN area of representative animals sampled at each of six times of day (A). Note that numbers of PER1 immunoreactive cells are elevated, and distributed across the whole SCN, at ZT 10 and ZT 14, and dorsal to it, in the LSPV, at ZT 10. Sampling boxes for the three areas of the pSCN are indicated in the photo depicting these areas at ZT 22. Scale bar=100 μm. Mean (±S.E.M.) number of PER1-containing cells in the SCN (B) and three sub-regions of the pSCN area (C–E) of female A. niloticus kept in a 12-h LD cycle and killed at 4-h intervals. Significant differences (P<0.05) are noted by different letters.
In the LSPV, there was a significant main effect of ZT on PER1 expression (F=13.55, df=5, P<0.0001) but there was no effect of level (F=1.86, df=2, P=0.17) and no interaction between ZT and level (F=0.811, df=10, P=0.62; Fig. 2C). In this region, PER1 expression was highest at ZT 10 and dropped progressively from then until ZT 18 before rising again to significantly higher levels at ZT 22, before the lights came on.
In the dlpSCN, there was a significant main effect of time on numbers of PER1-labeled cells (F=13.08, df=5, P<0.0001) as well as a significant main effect of level (F=9.627, df=2, P=0.0001) but no significant interaction (F=0.762, df=10, P=0.664; Fig. 2D). Here, PER1 expression was low from ZT 6 through ZT 18, at which point it increased sharply to significantly higher levels at ZT 22. PER1 expression remained high at ZT 2, with no significant differences seen between ZTs 22 and 2. For the effect of level, pair-wise comparisons showed that the rostral sections contained significantly more labeled cells than the other two levels (data not shown).
In the vlpSCN, there was a significant main effect of ZT (F=4.18, df=5, P<0.002) and of level (F=6.86, df=2, P<0.01) on PER1 expression, but no significant interaction (F=1.50, df=10, P=0.152; Fig. 2E). Pair-wise comparisons revealed that PER1 expression was significantly lower at ZT 18 than at all other ZTs. Pair-wise comparison across levels also revealed that PER1 expression in the rostral and middle vlpSCN was significantly higher than in the caudal level (data not shown).
PER1 in the NA, CEA, BLA and BNST-OV
In the shell of the NA there was a significant main effect of time on PER1 expression (F=4.27, df=5, P<0.01; Fig. 3A, 3B). PER1 expression was relatively high and unchanging from ZT 2 to ZT 14 and significantly lowers at ZT 18 and ZT 22 than at all other time points except ZT 10 (Fig. 3A). In the CEA there was a significant main effect of ZT on PER1 expression (F=4.66, df=5, P<0.01; Fig. 3C, 3D). Here, pair-wise comparisons revealed that numbers of PER1 labeled cells declined from ZT 2 to ZT 14 and were lowest and unchanging from ZT 14 through ZT 22. The BLA contained virtually no labeled cells (maximum=3) in animals from any time point. There was a significant main effect of ZT on PER1 expression in the BNST-OV (F=27.57, df=5, P<0.0001). In this region, numbers of labeled cells were highest at ZT 2 and decreased progressively up to ZT 10 and then remained low through ZT 22 (Fig. 3E, 3F).
Fig. 3.
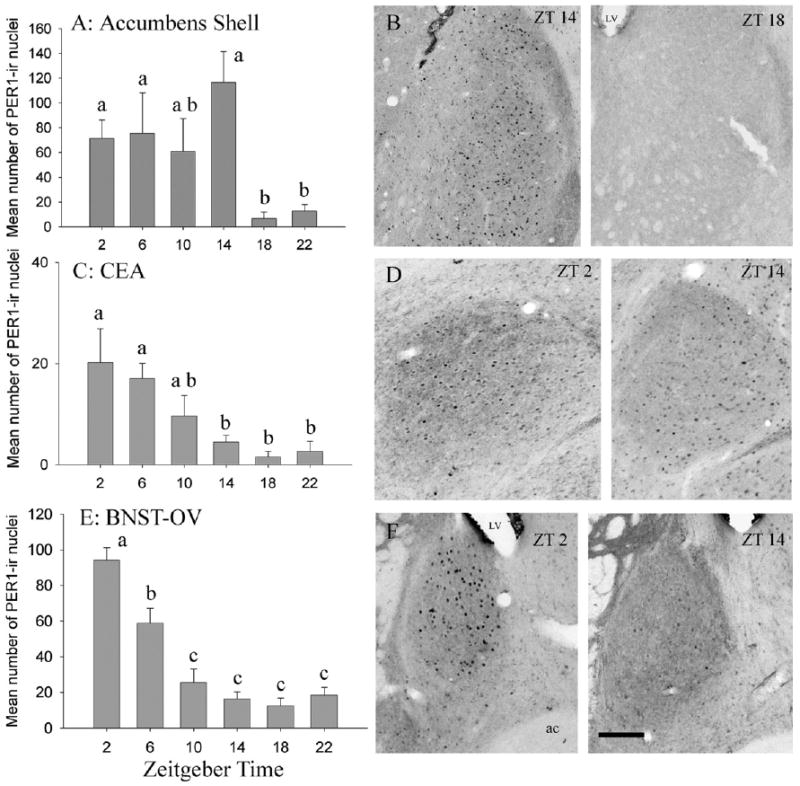
PER1 rhythm in the NA, the CEA, and the BNST-OV. Mean (±S.E.M.) numbers of cells containing PER1 and representative photomicrographs for the shell of the NA (A, B), the CEA (C, D), and the BNST-OV (E, F). LV, lateral ventricle; ac, anterior commissure. Scale bar=100 μm. Other symbols as in Fig. 2.
PER1 within TH-labeled cells
PER1 was not present in the A13 or A15 populations, but was in the four groups of TH-labeled cells analyzed here, and in all of these it varied as a function of time of day. First, in the PHDA cells there was a significant main effect of time on the percent TH-labeled cells expressing PER1 (F=3.080, df=5, P<0.05) such that double labeling was higher at ZT 22 than at all other ZTs except ZT 2 (Fig. 4A, 4B). Second, among the THDA neurons there was a significant main effect of time on the percent of TH-labeled cells expressing PER1 (F=3.92, df=5, P<0.01; Fig. 4C, 4D). Here, double labeling was low from ZT 18 to ZT 6 and then rose to intermediate levels at ZT 10 and to a peak at ZT 14. Third, among the TIDA cells there was a significant main effect of time on the percent TH-labeled cells expressing PER1 (F=21.50, df=5, P<0.0001; Fig. 4E, 4F). Pairwise comparisons revealed that double labeling was higher in these cells at ZT 10 and 14 than at all other ZTs. Finally, among the magnocellular cells of the preoptic area there was a significant main effect of time on the percent of TH-labeled cells containing PER1 (F=6.533, df=5, P<0.001; Fig. 4G, 4H) such that double labeling was significantly higher at ZT 10 and ZT 14 than at all other ZTs.
Fig. 4.
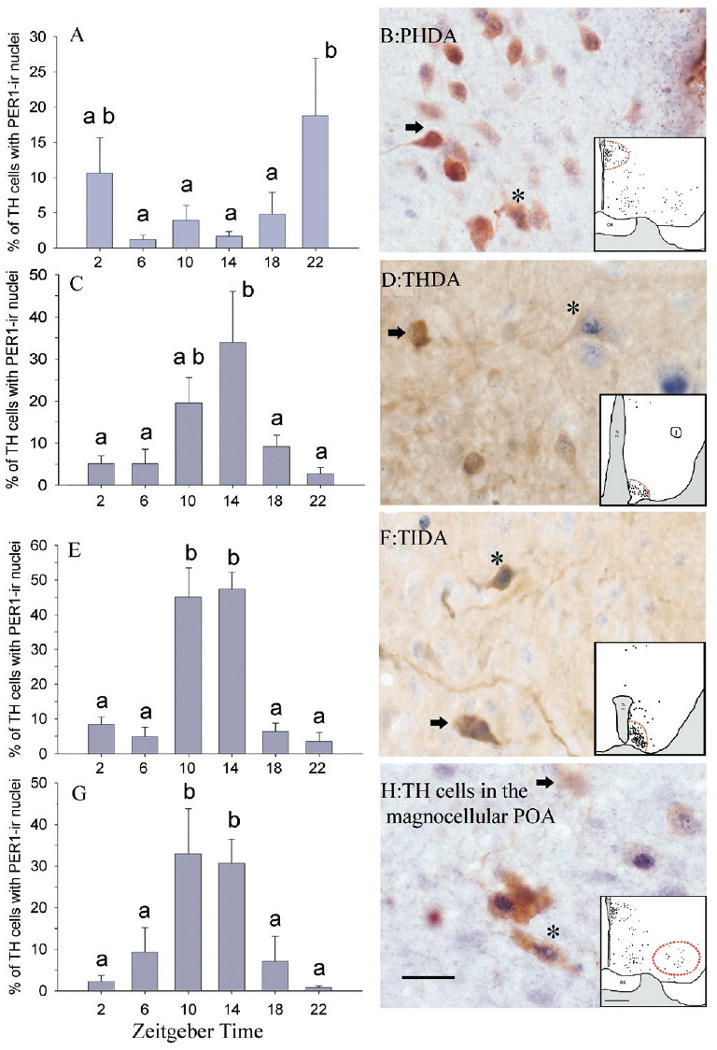
PER1 rhythm in four groups of TH-labeled cells. Mean (±S.E.M.) numbers and photomicrographs of PHDA neurons (A, B), THDA neurons (C, D), TIDA neurons (E, F) and magnocellular neurons containing TH in the preoptic area (G, H) of female grass rats. On photomicrographs, cells marked * are double labeled for TH and PER1; arrows indicate neurons expressing only TH. Dotted lines in the inserts depict areas where cells were sampled. Scale bar=20 μm for photomicrographs and 500 μm for line drawings. Other symbols as in Fig. 2.
Discussion
Diurnal rhythms in numbers of cells containing the clock protein PER1 were seen within the SCN and pSCN area, as well as in several populations of TH containing cells and a number of other regions of the forebrains of female grass rats. These rhythms were quite variable with respect to their amplitudes, waveforms and phases. For example, from ZT 18 to ZT 22 a rise in PER1 occurred in all three regions surrounding the SCN (Fig. 2) and in PHDA cells (Fig. 4), but in all other regions, including the SCN, PER1 was low and unchanging during this interval (Figs. 2–4). Across the light–dark transition from ZT 22 to ZT 2, PER1 went up in some cell populations (NA, CEA, BNST-OV; Fig. 3), did not change in others (pSCN areas, THDA and TIDA cells; Fig. 2, Fig. 4) and decreased in others (SCN and the PHDA cells; Fig. 2 and Fig. 4). Such patterns suggest that there are a variety of mechanisms involved in the coupling of extra-SCN oscillators to the LD cycle. These are likely to involve entraining influences from the SCN itself, directly or indirectly, but could theoretically include SCN-independent pathways (e.g. from the retina or the olfactory bulb).
SCN
Results from the current study add to the growing body of evidence that the patterns of coupling between rhythms in the primary molecular oscillator of the SCN and the LD cycle are very similar across species, regardless of whether they are diurnal or nocturnal (Field et al., 2000; Mrosovsky et al., 2001; Lincoln et al., 2002; Caldelas et al., 2003; Lambert et al., 2005; Novak et al., 2006). Specifically, expression of PER1 in the SCN rose progressively across the light phase to peak at the beginning of the dark phase in these female grass rats (Fig. 2B). This pattern of change in protein suggests that rhythms in mRNA for PER1 are likely to peak toward the middle of the light phase as they do in other species, both nocturnal and diurnal. These overall similarities, however, should not be interpreted to mean that more subtle differences in the molecular oscillator and its coupling to input/output systems do not exist.
Most studies of circadian rhythms in general and of clock genes and proteins in particular have been done on males, but when females have been examined some differences between the sexes and influences of ovarian hormones have been revealed (Lee and Smale, 2007). In the case of grass rats, the rhythm in PER1 of the females seen here is different from that seen previously in males (Ramanathan et al., 2006) in that it was phase advanced by approximately 4 h, the rise was more gradual and the fall in numbers of PER1-labeled cells was more precipitous. The advanced phase seen here raises the possibility that the period of the rhythm, and/or its responsiveness to light, is somewhat different in males and females. Sex differences in PER1 expression may be promoted, directly or indirectly (de la Iglesia et al., 1999), by the actions of gonadal hormones on the SCN, either during development or in adulthood. Evidence that estradiol can affect clock gene expression in the SCN of adults has recently been obtained from female laboratory rats, in which estradiol advanced the phase and increased the amplitude of Per2, but not Per1, rhythms in the SCN (Nakamura et al., 2005). However, there is no change in numbers PER2-containing cells in the SCN of laboratory rats across the estrous cycle (Perrin et al., 2006).
pSCN
Rhythms in PER1 expression were also seen in cells around the SCN of female grass rats as is the case for males of this diurnal species (Ramanathan et al., 2006). This pattern of gene expression has not been reported for nocturnal species. In the LSPV, PER1 rose toward the end of the dark period to a peak at ZT 10 and returned to trough values at ZT 18. The waveform of the rhythm was quite different from that seen in the SCN, and the amplitude was considerably lower (Fig. 2C). However, the phases of the rise and fall were only slightly shifted, both occurring 4 h earlier in the LSPV than the SCN; specifically, the rise began at ZT 18 in the LSPV and at ZT 2 in the SCN, and the fall began at ZT 10 in the LSPV and ZT 14 in the SCN. These differences (Fig. 2B, 2C) suggest that the rhythm in PER1 expression in the LSPV does not simply reflect PER1 production in displaced SCN cells. These patterns of gene expression raise the question of how the two rhythms might be coupled to each other. The LSPV rhythm could reflect an extra-SCN oscillator ultimately dependent on the SCN for maintenance of its circadian rhythmicity. However, it is also possible that the LSPV of grass rats is a component of the circadian timing system that can oscillate independently of the SCN. The pattern of the PER1 rhythm seen here in the LSPV of females was different from that seen previously in males (Ramanathan et al., 2006). Although the numbers of PER1+ cells peaked at ZT 10 in both sexes, they were relatively low at all other times of day in males. As with the SCN, the causes of the sex differences are unclear, but they are likely to be promoted by hormones, acting either early in development or in adulthood.
Rhythms of PER1 expression were also seen in the lateral portions of the pSCN area. In the most dorsal of these, the dlpSCN, PER1 expression was high at ZT 22 and ZT 2. This pattern was different from that seen previously in male grass rats, in which ZT 2 was the only time at which PER1 was above baseline levels. As with the SCN and LSPV, this pattern suggests a slight phase advance of the rhythm of females relative to that seen in males (Ramanathan et al., 2006). In the vlpSCN, lateral to the ventral part of the SCN, the effect of time on PER1 expression involved a drop in numbers of labeled cells at ZT 18 relative to all other time points. In an earlier study of male grass rats, no rhythm in PER1 expression was seen in this region (Ramanathan et al., 2006). Taken together, the present data from female grass rats reveal the existence of rhythms in PER1 expression in all three portions of the pSCN region, each with its own temporal pattern, and each of which is different from that seen in the SCN. These patterns suggest that, in this species at least, a functionally heterogeneous circadian system extends beyond the boundaries of the SCN into the surrounding regions of the pSCN area.
BNST-OV, CEA, BLA and NA
Clear rhythms of PER1 were seen here in some regions of the forebrain beyond the SCN, but not others. In the BNST-OV and the CEA, PER1 production rose sharply right after the lights came on and decreased to baseline levels between ZT 10 and 22. The initial rise could reflect a response to light, or an endogenous oscillatory process. In the BLA, by contrast, virtually no cells containing PER1 were seen at any time of day. PER1 rhythms have not been reported in these regions of females of other species. However, in male laboratory rats PER1 is not rhythmic in either the BLA or the CEA of animals provided ad libitum food (Angeles-Castellanos et al., 2007). PER2, by contrast, is rhythmic in both of these regions of both male and female laboratory rats (Amir et al., 2004; Lamont et al., 2005; Perrin et al., 2006). These patterns suggest that oscillators in the amygdala may function differently from those in the SCN with respect to relationships between PER1 and PER2. They also raise the possibility of differences between grass rats and laboratory rats, in that PER1 rhythms were seen in the grass rat CEA here, whereas no PER1 rhythms were seen in the CEA of male laboratory rats (Angeles-Castellanos et al., 2007). In the BNST-OV of female grass rats, the rhythm of PER1 expression showed a peak during the early light phase, whereas peak values occur much later (i.e. ZT 12–18) in male laboratory rats (Angeles-Castellanos et al., 2007). It is also noteworthy that rhythms in PER2 in the BNST-OV of female laboratory rats in proestrus and estrus are 180° out of phase relative to those seen here in grass rats (Perrin et al., 2006). Taken together, these patterns suggest that circadian signals setting the phase of rhythms in the BNST-OV are different or that the BNST-OV responds to such signals in opposite ways, in laboratory rats and grass rats. A fourth extra-SCN region of the forebrain in which a distinct cluster of cells containing PER1 was seen is the NA, a region heavily involved in reinforcement and addiction (Kelley et al., 2005). Here, PER1 rose when the lights came on and dropped from 2 to 6 h after the lights went out. In the laboratory rat, by contrast, PER1 rhythms dropped when the lights came on and rose when the lights went out (Angeles-Castellanos et al., 2007). Overall, the results of studies of clock protein rhythms in laboratory rats and grass rats suggest intriguing sex and species differences; there may also be changes as a function of reproductive state (Perrin et al., 2006), and further work will be needed to fully characterize these patterns and the factors modulating them. These are important questions, as the NA, BNST-OV, BLA and CEA are involved in a host of physiological and behavioral functions that fluctuate across the day (Lamont et al., 2005; Segall et al., 2006; LeDoux, 2007).
TH cells
PER1 rhythms were seen in four populations of cells containing TH. Three of these populations (TIDA, THDA and PHDA cells), serve neuroendocrine functions while the fourth, in the basal forebrain, has been less well characterized. Rhythms in the two populations of cells within the arcuate nucleus (THDA and TIDA cells) were quite similar, with peaks from ZT 10–ZT 14, and low levels at all other time points (Fig. 4). Interestingly, this pattern differed from that reported in laboratory rats (ovariectomized), but not mice (ovary intact). In laboratory rats, PER1 expression was not rhythmic in THDA cells but it did show higher levels at ZT 18 than at ZT 6 (Sellix et al., 2006). In grass rats, by contrast, levels at ZT 18 were very low, and were the same as those seen at ZT 6, and the peak occurred between these two time points (Fig. 4C). The peak of the rhythm was therefore phase advanced by approximately 4 h in grass rats relative to laboratory rats. A time of day effect on PER1 expression within TH-labeled cells in the arcuate nucleus has also been reported in transgenic Per1::GFP mice (Kriegsfeld et al., 2003). Here, although only two time points were examined, the data suggest a pattern more similar to that of grass rats than of laboratory rats, with numbers of PER1-labeled TH positive cells that were approximately fourfold higher at ZT 10 than at ZT 22. The similarities and differences between these three species therefore reveal variation in rhythms in clock gene expression in oscillators beyond the SCN, but that this variation does not seem to be related to whether an animal is diurnal or nocturnal. This is reminiscent of the picture that has emerged from studies of the SCN, where some peptides are found in some species and not others, but that variation does not distinguish diurnal from nocturnal animals (Smale et al., 2003).
These findings raise the question of what might cause the similarities and differences among species. In laboratory rats and grass rats, one pathway through which circadian information is likely to be transmitted from the SCN to the TH-labeled cells is directly via VIP-containing cells. In laboratory rats, contacts between VIP fibers and TH positive cells have been seen at the light level and synapses have been confirmed via electron microscopy (Gerhold et al., 2001), and in grass rats apparent contacts can be seen at the light level (Mahoney et al., 2007). Rhythms in VIP mRNA in the SCN are the same in female grass rats and laboratory rats (Krajnak et al., 1998; M. M. Mahoney, C. Ramanathan et al., unpublished observations). Taken together, these data suggest that the TH-labeled cells may respond differently to VIP in the two species, or that other signals converging on these cells promote differences among species.
The patterns of interspecific variation in PER1 rhythms in TH-labeled cells in the hypothalamus of mice, laboratory rats and grass rats raise the question of what their functional consequences might be. These cells are involved in the regulation of prolactin, which, among other things, exhibits a pre-ovulatory surge that is coincident with that of LH. It seems likely that prolactin also exhibits a pre-ovulatory surge in grass rats, (though it has not been possible to measure prolactin in this species). The current data raise the intriguing possibility that the coupling between PER1 rhythms and downstream intracellular processes varies across species.
TH within magnocellular neurons in the preoptic area has been seen in some species (e.g. hamsters, Vincent, 1988) but not others (e.g. rats, Mahoney et al., 2007). To our knowledge, nothing is known about the function of TH in these cells and there are no other reports suggesting that cells in this region contain elements of a molecular clock. The present data therefore raise a variety of intriguing questions about whether there is a molecular oscillator in this area in other species, and about the role that it might play in regulation of behavioral and/or physiological functions.
General conclusions
Several basic findings bearing on issues of sex and species differences in circadian mechanisms emerged from this study. First, rhythms within the SCN and pSCN area of female grass rats were very similar to those of male grass rats, but some differences were apparent. Within the SCN, PER1 expression rose and fell approximately 4 h earlier in females (current data) than in males (Ramanathan et al., 2006). Around the SCN, the waveform of the rhythm was somewhat different in the LSPV of female compared with male grass rats, and a rhythm in the ventrolateral portion of the pSCN was present in females, but not males of this diurnal species (Ramanathan et al., 2006). The functional significance of these differences is an important question for future investigation. Second, rhythms in PER1 expression are present in a variety of forebrain regions beyond the SCN in these animals, as has been seen in other species. These data also suggest that there are differences among species in the temporal patterns of PER1 expression over the course of the day. Third, rhythms in PER1 expression are present in four populations of TH positive cells in the brains of female grass rats. Some of these are phase shifted by several hours relative to those seen previously in laboratory rats (Sellix et al., 2006), but species differences in these cells may not be directly related to diurnality, as grass rats and mice appear to be more similar to each other along this dimension than are laboratory rats and mice (Kriegsfeld et al., 2003). In addition, a PER1 rhythm is present in a magnocellular population of TH-containing cells in the forebrain of female grass rats that has not been reported previously. Taken together, the data on PER1 rhythms reported here point to the fact that, although the primary circadian oscillator in the SCN is quite similar (though not identical), between the sexes and across species, the larger multioscillator system that regulates circadian rhythms can vary considerably.
With respect to the issue of diurnality, the current data support a model in which the patterns of rhythmicity in extra-SCN regions of the brain are quite different in day- and night-active animals, but these patterns of differences vary across regions. In some areas there is a complete 12 h phase inversion of rhythms in PER1 (e.g. BNST-OV), but oscillators in other cell populations are shifted by only a few hours (e.g. TIDA neurons). Thus, diurnality cannot be explained by one basic mechanism that creates a simple 12 h shift in phase of rhythms beyond the SCN. Rather, the patterns suggest that a diurnal brain emerges via different mechanisms operating at multiple points along intersecting pathways between the SCN and oscillators in other areas of the forebrain. The challenge for the future will be to determine where, and how, along these pathways the different patterns come about.
Acknowledgments
The authors wish to thank Anna Baumgras, Dr. Josh Nixon, and Dr. Gladys S. Martinez for technical assistance, and Dr. Lily Yan for her helpful comments on this manuscript and assistance with the figures. This work was supported by the National Institute of Mental Health RO1 MH53433 and National Science Foundation IBN-0130977.
Abbreviations
- ABC
avidin–biotin peroxidase complex
- ANOVA
analysis of variance
- BNST-OV
oval nucleus of bed nucleus of stria terminalis
- BLA
basolateral amygdala
- CEA
central amygdala
- DAB
diaminobenzidine
- dlpSCN
dorsolateral peri–suprachiasmatic nucleus
- GnRH
gonadotropin releasing hormone
- LD
light/dark
- LH
luteinizing hormone
- LSPV
lower sub-paraventricular zone
- NA
nucleus accumbens
- NDS
normal donkey serum
- NHS
normal horse serum
- PBS
phosphate-buffered saline
- PBS-TX
phosphate-buffered saline with Triton X-100
- PHDA
periventricular hypothalamic dopaminergic neurons
- pSCN
peri–suprachiasmatic nucleus
- SCN
suprachiasmatic nucleus
- TH
tyrosine hydroxylase
- THDA
tuberohypophysial dopaminergic neurons
- TIDA
tuberoinfundibular dopaminergic neurons
- TX
Triton X-100
- vlpSCN
ventrolateral peri–suprachiasmatic nucleus
- ZT
zeitgeber time
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- Abraham U, Prior JL, Granados-Fuentes D, Piwnica-Worms DR, Herzog ED. Independent circadian oscillations of period1 in specific brain areas in vivo and in vitro. J Neurosci. 2005;25:8620–8626. doi: 10.1523/JNEUROSCI.2225-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers HE. Gonadal hormones organize and modulate the circadian system of the rat. Am J Physiol. 1981;241:R62–R66. doi: 10.1152/ajpregu.1981.241.1.R62. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gerall AA, Axelson JF. Effect of reproductive state on circadian periodicity in the rat. Physiol Behav. 1981;26:21–25. doi: 10.1016/0031-9384(81)90073-1. [DOI] [PubMed] [Google Scholar]
- Amir S, Lamont EW, Robinson B, Stewart J. A circadian rhythm in the expression of PERIOD2 protein reveals a novel SCN-controlled oscillator in the oval nucleus of the bed nucleus of the stria terminalis. J Neurosci. 2004;24:781–790. doi: 10.1523/JNEUROSCI.4488-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Castellanos M, Mendoza J, Escobar C. Restricted feeding schedules phase shift daily rhythms of c-Fos and protein Per1 immunoreactivity in corticolimbic regions in rats. Neuroscience. 2007;144:344–355. doi: 10.1016/j.neuroscience.2006.08.064. [DOI] [PubMed] [Google Scholar]
- Caldelas I, Poirel VJ, Sicard B, Pevet P, Challet E. Circadian profile and photic regulation of clock genes in the suprachiasmatic nucleus of a diurnal mammal Arvicanthis ansorgei. Neuroscience. 2003;116:583–591. doi: 10.1016/s0306-4522(02)00654-1. [DOI] [PubMed] [Google Scholar]
- Dardente H, Menet JS, Challet E, Tournier BB, Pevet P, Masson-Pevet M. Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Brain Res Mol Brain Res. 2004;124:143–151. doi: 10.1016/j.molbrainres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Davis FC, Darrow JM, Menaker M. Sex differences in the circadian control of hamster wheel-running activity. Am J Physiol. 1983;244:R93–R105. doi: 10.1152/ajpregu.1983.244.1.R93. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Blaustein JD, Bittman EL. The suprachiasmatic area in the female hamster projects to neurons containing estrogen receptors and GnRH. Neuroreport. 1995;6:1715–1722. doi: 10.1097/00001756-199509000-00004. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Blaustein JD, Bittman EL. Oestrogen receptor-alpha-immunoreactive neurones project to the suprachiasmatic nucleus of the female Syrian hamster. J Neuroendocrinol. 1999;11:481–490. doi: 10.1046/j.1365-2826.1999.00341.x. [DOI] [PubMed] [Google Scholar]
- de la Iglesia HO, Schwartz WJ. Minireview: timely ovulation: circadian regulation of the female hypothalamo-pituitary-gonadal axis. Endocrinology. 2006;147:1148–1153. doi: 10.1210/en.2005-1311. [DOI] [PubMed] [Google Scholar]
- Fewell JE. Body temperature regulation in rats near term of pregnancy. Can J Physiol Pharmacol. 1995;73:364–368. doi: 10.1139/y95-046. [DOI] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80:1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- Gerhold LM, Horvath TL, Freeman ME. Vasoactive intestinal peptide fibers innervate neuroendocrine dopaminergic neurons. Brain Res. 2001;919:48–56. doi: 10.1016/s0006-8993(01)02993-6. [DOI] [PubMed] [Google Scholar]
- Gerhold LM, Sellix MT, Freeman ME. Antagonism of vasoactive intestinal peptide mRNA in the suprachiasmatic nucleus disrupts the rhythm of FRAs expression in neuroendocrine dopaminergic neurons. J Comp Neurol. 2002;450:135–143. doi: 10.1002/cne.10307. [DOI] [PubMed] [Google Scholar]
- Gillespie JM, Chan BP, Roy D, Cai F, Belsham DD. Expression of circadian rhythm genes in gonadotropin-releasing hormone-secreting GT1-7 neurons. Endocrinology. 2003;144:5285–5292. doi: 10.1210/en.2003-0802. [DOI] [PubMed] [Google Scholar]
- Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- Kelley AE, Baldo BA, Pratt WE. A proposed hypothalamic-thalamic-striatal axis for the integration of energy balance, arousal, and food reward. J Comp Neurol. 2005;493:72–85. doi: 10.1002/cne.20769. [DOI] [PubMed] [Google Scholar]
- Klein DC, Moore RY, Reppert SM. Suprachiasmatic nucleus: the mind's clock. New York: Oxford University Press; 1991. pp. 388–399. [Google Scholar]
- Krajnak K, Kashon ML, Rosewell KL, Wise PM. Sex differences in the daily rhythm of vasoactive intestinal polypeptide but not arginine vasopressin messenger ribonucleic acid in the suprachiasmatic nuclei. Endocrinology. 1998;139:4189–4196. doi: 10.1210/endo.139.10.6259. [DOI] [PubMed] [Google Scholar]
- Kriegsfeld LJ, Korets R, Silver R. Expression of the circadian clock gene period 1 in neuroendocrine cells: an investigation using mice with a Per1:GFP transgene. Eur J Neurosci. 2003;17:212–220. doi: 10.1046/j.1460-9568.2003.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labyak SE, Lee TM. Estrus- and steroid-induced changes in circadian rhythms in a diurnal rodent, Octodon degus. Physiol Behav. 1995;58:573–585. doi: 10.1016/0031-9384(95)00096-2. [DOI] [PubMed] [Google Scholar]
- Lambert CM, Machida KK, Smale L, Nunez AA, Weaver DR. Analysis of the prokineticin 2 system in a diurnal rodent, the unstriped Nile grass rat (Arvicanthis niloticus) J Biol Rhythms. 2005;20:206–218. doi: 10.1177/0748730405275135. [DOI] [PubMed] [Google Scholar]
- Lamont EW, Robinson B, Stewart J, Amir S. The central and basolateral nuclei of the amygdala exhibit opposite diurnal rhythms of expression of the clock protein period2. Proc Natl Acad Sci U S A. 2005;102:4180–4184. doi: 10.1073/pnas.0500901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux J. The amygdala. Curr Biol. 2007;17:R868–R874. doi: 10.1016/j.cub.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Lee T, Smale L. The neuroendocrinology of behavioral rhythms. In: Blaustein J, editor. Handbook of neurochemistry, neuroendocrinology and molecular neurobiology. 3rd. Vol. 2. New York: Plenum Press; 2007. pp. 835–868. [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626–1638. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln G, Messager S, Andersson H, Hazlerigg D. Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proc Natl Acad Sci U S A. 2002;99:13890–13895. doi: 10.1073/pnas.212517599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney MM, Ramanathan C, Smale L. Tyrosine hydroxylase positive neurons and their contacts with vasoactive intestinal polypeptide-containing fibers in the hypothalamus of the diurnal murid rodent, Arvicanthis niloticus. J Chem Neuroanat. 2007;33:131–139. doi: 10.1016/j.jchemneu.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Martinez GS, Smale L, Nunez AA. Diurnal and nocturnal rodents show rhythms in orexinergic neurons. Brain Res. 2002;955:1–7. doi: 10.1016/s0006-8993(02)03264-x. [DOI] [PubMed] [Google Scholar]
- Masubuchi S, Honma S, Abe H, Ishizaki K, Namihira M, Ikeda M, Honma K. Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. Eur J Neurosci. 2000;12:4206–4214. [PubMed] [Google Scholar]
- Morin LP, Fitzgerald KM, Zucker I. Estradiol shortens the period of hamster circadian rhythms. Science. 1977;196:305–307. doi: 10.1126/science.557840. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N, Edelstein K, Hastings MH, Maywood ES. Cycle of period gene expression in a diurnal mammal (Spermophilus tridecemlineatus): implications for nonphotic phase shifting. J Biol Rhythms. 2001;16:471–478. doi: 10.1177/074873001129002141. [DOI] [PubMed] [Google Scholar]
- Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, Shinohara K. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res. 2005;82:622–630. doi: 10.1002/jnr.20677. [DOI] [PubMed] [Google Scholar]
- Novak CM, Ehlen JC, Paul KN, Fukuhara C, Albers HE. Light and GABAA receptor activation alter period mRNA levels in the SCN of diurnal Nile grass rats. Eur J Neurosci. 2006;24:2843–2852. doi: 10.1111/j.1460-9568.2006.05166.x. [DOI] [PubMed] [Google Scholar]
- Novak CM, Smale L, Nunez AA. Fos expression in the sleep-active cell group of the ventrolateral preoptic area in the diurnal murid rodent, Arvicanthis niloticus. Brain Res. 1999;818:375–382. doi: 10.1016/s0006-8993(98)01319-5. [DOI] [PubMed] [Google Scholar]
- Novak CM, Smale L, Nunez AA. Rhythms in Fos expression in brain areas related to the sleep-wake cycle in the diurnal Arvicanthis niloticus. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1267–R1274. doi: 10.1152/ajpregu.2000.278.5.R1267. [DOI] [PubMed] [Google Scholar]
- Nunez AA, Bult A, McElhinny TL, Smale L. Daily rhythms of Fos expression in hypothalamic targets of the suprachiasmatic nucleus in diurnal and nocturnal rodents. J Biol Rhythms. 1999;14:300–306. doi: 10.1177/074873099129000713. [DOI] [PubMed] [Google Scholar]
- Perrin JS, Segall LA, Harbour VL, Woodside B, Amir S. The expression of the clock protein PER2 in the limbic forebrain is modulated by the estrous cycle. Proc Natl Acad Sci U S A. 2006;103:5591–5596. doi: 10.1073/pnas.0601310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- Ramanathan C, Nunez AA, Martinez GS, Schwartz MD, Smale L. Temporal and spatial distribution of immunoreactive PER1 and PER2 proteins in the suprachiasmatic nucleus and peri-suprachiasmatic region of the diurnal grass rat (Arvicanthis niloticus) Brain Res. 2006:1073–1074. 348–358. doi: 10.1016/j.brainres.2005.11.082. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Rosenwasser AM, Hollander SJ, Adler NT. Effects of pregnancy and parturition on free-running circadian activity rhythms in the rat. Chronobiol Int. 1987;4:183–187. doi: 10.3109/07420528709078524. [DOI] [PubMed] [Google Scholar]
- Schwartz MD. Neural substrates of diurnality in the Nile grass rat, Arvicanthis niloticus. East Lansing: Michigan State University; 2006. [Google Scholar]
- Schwartz MD, Nunez AA, Smale L. Differences in the suprachiasmatic nucleus and lower subparaventricular zone of diurnal and nocturnal rodents. Neuroscience. 2004;127:13–23. doi: 10.1016/j.neuroscience.2004.04.049. [DOI] [PubMed] [Google Scholar]
- Segall LA, Perrin JS, Walker CD, Stewart J, Amir S. Glucocorticoid rhythms control the rhythm of expression of the clock protein, period2, in oval nucleus of the bed nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neuroscience. 2006;140:753–757. doi: 10.1016/j.neuroscience.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Sellix MT, Egli M, Poletini MO, McKee DT, Bosworth MD, Fitch CA, Freeman ME. Anatomical and functional characterization of clock gene expression in neuroendocrine dopaminergic neurons. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1309–R1323. doi: 10.1152/ajpregu.00555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellix MT, Freeman ME. Circadian rhythms of neuroendocrine dopaminergic neuronal activity in ovariectomized rats. Neuroendocrinology. 2003;77:59–70. doi: 10.1159/000068334. [DOI] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Smale L, Lee T, Nunez AA. Mammalian diurnality: some facts and gaps. J Biol Rhythms. 2003;18:356–366. doi: 10.1177/0748730403256651. [DOI] [PubMed] [Google Scholar]
- Smale L, Nunez AA, Schwartz MD. Rhythms in a diurnal brain. Biol Rhythm Res. 2008;39:305–318. [Google Scholar]
- Takahashi JS, Menaker M. Interaction of estradiol and progesterone: effects on circadian locomotor rhythm of female golden hamsters. Am J Physiol. 1980;239:R497–R504. doi: 10.1152/ajpregu.1980.239.5.R497. [DOI] [PubMed] [Google Scholar]
- van der Beek EM. Circadian control of reproduction in the female rat. In: Buijs RM, Kalsbeek A, Romijn HJ, Pennartz CMA, Mirmiran M, editors. Progress in brain research. Vol. 3. Elsevier; 1996. pp. 295–320. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, Horvath TL, Wiegant VM, Van den Hurk R, Buijs RM. Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: combined tracing and light and electron microscopic immunocytochemical studies. J Comp Neurol. 1997;384:569–579. doi: 10.1002/(sici)1096-9861(19970811)384:4<569::aid-cne6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Vincent SR. Distributions of tyrosine hydroxylase-, dopamine-beta-hydroxylase-, and phenylethanolamine-N-methyltransferase-immunoreactive neurons in the brain of the hamster (Mesocricetus auratus) J Comp Neurol. 1988;268:584–599. doi: 10.1002/cne.902680408. [DOI] [PubMed] [Google Scholar]
- Watts AG, Swanson LW, Sanchez-Watts G. Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J Comp Neurol. 1987;258:204–229. doi: 10.1002/cne.902580204. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Kerbeshian MC, Hocker CG, Block GD, Menaker M. Rhythmic properties of the hamster suprachiasmatic nucleus in vivo. J Neurosci. 1998;18:10709–10723. doi: 10.1523/JNEUROSCI.18-24-10709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SQ, Kimura M, Inoue S. Sleep patterns in cyclic and pseudopregnant rats. Neurosci Lett. 1995;193:125–128. doi: 10.1016/0304-3940(95)11685-p. [DOI] [PubMed] [Google Scholar]


