Summary
Among the four DNA bases, guanine is particularly vulnerable to oxidative damage and the most common oxidative product, 7,8 dihydro-8-oxoguanine (8-oxoG), is the most prevalent lesion observed in DNA molecules. Fortunately, 8-oxoG is recognized and excised by the 8-oxoguanine DNA glycosylase (Ogg) of the base excision repair pathway. Ogg enzymes are divided into three separate families, namely Ogg1, Ogg2, and AGOG. To date, structures of members of both Ogg1 and AGOG families are known but no structural information is available for members of Ogg2. Here we describe the first crystal structures of two archaeal Ogg2: Methanocaldococcus janischii Ogg (MjaOgg) and Sulfolobus solfataricus Ogg (SsoOgg). A structural comparison with OGG1 and AGOG suggested that the C-terminal lysine of Ogg2 may play a key role in discriminating between guanine and 8-oxoG. This prediction was substantiated by measuring the glycosylase/lyase activity of a C-terminal deletion mutant of MjaOgg.
Introduction
DNA molecules are subjected to a wide variety of damage caused by oxidizing agents from an organism’s own metabolism, from exogenous sources, and by ionizing radiation. Among the four bases, guanine is particularly prone to oxidation and its oxidative product, 7,8-dihydro-8-oxoguanine (8-oxoG), is the most frequent lesion observed in DNA (Grollman and Moriya, 1993). 8-oxoG can form a normal Watson-Crick base pair with cytosine (8-oxoG:C); however it has also been shown to form a stable Hoogsteen pair with adenine (8-oxoG:A) (Kuchino, et al., 1987, Shibutani, et al., 1991), which can lead to a G:C→T:A transversion after replication (Kuchino, et al., 1987, Wood, et al., 1990). 8-oxoG is repaired by the base excision repair (BER) pathway, a process initiated by either of two DNA glycosylases, formamidopyrimidine-DNA glycosylase (Fpg) or 8-oxoguanine DNA glycosylase (Ogg) (Barnes and Lindahl, 2004, David, et al., 2007). Fpg is found mostly in bacteria whereas Ogg is the primary DNA glycosylase to excise 8-oxoG in eukaryotes and archaea. Both Fpg and Ogg are bifunctional glycosylases: These enzymes catalyze the excision of the oxidized base by cleaving the N-glycosylic bond between the base and the deoxyribose moiety (glycosylase activity) and subsequently cleave the DNA backbone (lyase activity).
In recent years, tremendous progress has been accomplished on the front of genome sequencing and a multitude of complete genomes from a wide variety of organisms are now available. Among these, the first archaeal genome to be sequenced in its entirety was that of Methanocaldococcus janischii. Inspection of its genome revealed an open reading frame containing the helix-hairpin-helix (HhH) motif typical of DNA glycosylases of the HhH-GPD superfamily (Bult, et al., 1996, Gogos and Clarke, 1999). Characterization of the gene product revealed this protein to be an 8-oxoguanine DNA glycosylase (Gogos and Clarke, 1999). Similar Oggs were subsequently identified in archaea such as Sulfolobus solfataricus (Brock, et al., 1972, She, et al., 2001) and Archeoglobus fulgidus (Chung, et al., 2001) and eubacteria such as Thermotoga maritima (Im, et al., 2005, Nelson, et al., 1999).
The 8-oxoguanine DNA glycosylases belong to three distinct families. The Ogg1 family encompasses the largest number of members including human OGG1 (hOGG1) (Aburatani, et al., 1997, Arai, et al., 1997, Bjørås, et al., 1997, Nagashima, et al., 1997, Radicella, et al., 1997, Roldan-Arjona, et al., 1997, Rosenquist, et al., 1997) and bacterial Oggs such as Clostridium acetobutylicum Ogg (CacOgg) (Robey-Bond, et al., 2008). The Ogg2 family comprises archaeal species such as M. jannaschii Ogg (MjaOgg) (Gogos and Clarke, 1999), S. solfataricus Ogg (SsoOgg), A. fulgidus Ogg (AfuOgg) (Chung, et al., 2001) and eubacteria, e.g., T. maritima Ogg (Tm1821) (Im, et al., 2005). Ogg2 enzymes lack the A domain of Ogg1 family members, share a very low sequence identity with hOGG1 (13–19%) and display a reduced specificity for the base opposite the lesion. AGOG (Archaeal GO Glycosylase) (Gondichatnahalli, et al., 2004, Sartori, et al., 2004) constitute the third family and the crystal structure of Pyrobaculum aerophilum AGOG (Pa-AGOG) was published recently (Lingaraju, et al., 2005). It is noteworthy that crystal structures have been reported for both Ogg1 and AGOG family members whereas, to our knowledge, no structure of Ogg2 is currently available.
Previous structural and functional studies on hOGG1 (Bjørås, et al., 2002, Bruner, et al., 2000) pointed out that the conserved glycine at position 42 on the αA-βB loop in the A domain of OGG1 is essential for the discrimination between guanine and 8-oxoguanine. Since Ogg2 enzymes are devoid of this domain, it was unclear until now how these enzymes discriminate between guanine and 8-oxoG. We report here the first crystal structures of two members of the Ogg2 family, MjaOgg and SsoOgg. Superposition of these structures with hOGG1 identified the well conserved C-terminal lysine of Ogg2 as a major molecular determinant for the distinction between guanine and 8-oxoG. Our prediction was confirmed by generating a C-terminal deletion mutant of Ogg2, which unambiguously showed that the C-terminal lysine of Ogg2 is essential for the recognition of the oxidized guanine.
Results and discussion
Crystallization and structure determination of MjaOgg and SsoOggK128Q in their apo-form and overall description of the structures
Crystals of apo-MjaOgg were obtained by the vapor diffusion method. Crystals grew rapidly at 12°C (See experimental procedures for crystallization conditions). Since MjaOgg contains 2 methionines for a total of 207 residues, we crystallized a selenomethionyl derivative to obtain multiwavelength anomalous diffraction (MAD) phases. A single crystal was used to collect data at three different wavelengths at the APS synchrotron (See Table 1 for diffraction statistics). The resulting model was refined at 2.0Å to an Rfree of 0.251 and Rcryst of 0.202 with good stereochemistry. The final model comprises the totality of MjaOgg residues (1–207). Furthermore, seven additional residues were observed at the N-terminal end (residues −6 to 0), extending helix αA (see Figure 1A). These residues originated from the cloning vector and include a penta-histidine tag. The catalytically inactive variant (K128Q) of SsoOgg was crystallized with a DNA duplex containing the 8-oxoG lesion. A complete X-ray diffraction data set was collected at the APS synchrotron (See Table 1 for diffraction statistics). After molecular replacement using the coordinates of MjaOgg (with which it shares 39.1% sequence identity), we were unable to locate any electronic density corresponding to the DNA molecule. The SsoOgg structure was therefore refined as an apo-form. A closer look at the crystal packing revealed that helix αM of one of the symmetry related molecules would hinder the positioning of a DNA duplex in the active site and confirmed that our model is indeed unliganded. The final model (Figure 1B) comprises every residue of SsoOgg from 1 to 207.
Table 1.
Summary of data collection and refinement statistics.
| MjaOgg Se-Peak | Se-Inflection | Se-Remote | SsoOgg | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.9794 | 0.9796 | 0.9717 | 1.0332 |
| Resolution (Å) | 20-2.0 (2.1-2.0) | 20-2.0 (2.1-2.0) | 20-2.0 (2.1-2.0) | 19.7-1.9 (2.0-1.9) |
| Space group | C2 | C2 | C2 | P6 |
| Unit-cell parameters | ||||
| a,b,c (Å) | 89.32 37.78 74.80 | 89.32 37.78 74.80 | 89.32 37.78 74.80 | 110.96, 110.96, 34.41 |
| α, β, γ (°) | 90 116.063 90 | 90 116.063 90 | 90 116.063 90 | 90, 90, 120 |
| Total reflection | 54731 (7074) | 55051 (6937) | 54686 (6886) | 206107 (26109) |
| Unique reflection | 28167 (3647) | 28272 (3570) | 28081 (3519) | 35849 (4583) |
| Completeness (%) | 94.7 (90.2) | 94.8 (88.3) | 93.9 (86.5) | 95.9 (85.8) |
| I/σ(I) | 10.7 (6.0) | 10.7 (5.2) | 10.1 (4.1) | 17.6 (2.7) |
| Rmerge (%)b | 11.8 (37.4) | 12.5 (43.0) | 13.7 (51.1) | 8.5 (53.6) |
| Redundancy | 1.9 (1.9) | 1.9 (1.9) | 1.9 (2.0) | 7.9 (7.8) |
| Selenium sites | 2 | 2 | 2 | |
| Rcullis c | 0.85 | 0.91 | 0.94 | |
| Phasing power | 0.8 | 0.54 | 0.46 | |
| Overall mean FOMd | 0.18/0.63 | |||
|
| ||||
| Refinement | ||||
| Rcryst (%)e | 20.2 | 20.7 | ||
| Rfree (%)e | 25.1 | 24.8 | ||
| Rmsd from ideal bond length(Å)/angles (°) | 0.006/1.1 | 0.005/1.1 | ||
| Non-hydrogen atoms | ||||
| All atoms | 2008 | 1917 | ||
| Protein | 1855 | 1724 | ||
| Water | 153 | 114 | ||
| Heterogeneous atoms | 79 | |||
| Average B factors (Å2) | 24.7 | 33.3 | ||
|
| ||||
| Ramachandran plot (%) | ||||
| Most favored regions | 94.9 | 92.3 | ||
| Allowed regions | 4.6 | 7.7 | ||
| Disallowed regions | 0.5 | 0 | ||
High resolution shell is shown in parentheses.
Rmerge=P|I_<I>|/PI, where <I> is the average intensity from multiple observations of symmetry-related reflections.
RCullis=[<(LOC)2>]1/2 (<|_F|2>)1/2, where LOC is the lack-of-closure error.
Before and after density modification.
Rwork and Rfree=Ph|||Fo|_|Fc||/Ph|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree was calculated with 5% of the reflections not used in refinement
Figure 1. Overall fold of A) MjaOgg and B) SsoOgg.
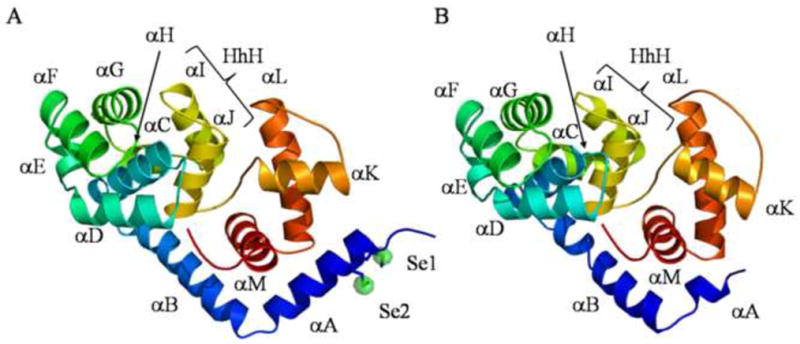
Ribbon diagrams of A) MjaOgg and B) SsoOgg. The helix-hairpin-helix (HhH) motif is composed of helices I and J in both proteins. Proteins are colored according to the amino acid sequence going from cold blue to warm red from N- to C-terminal. In panel A, an anomalous difference Fourier map (green) contoured at 6σ pinpoints the two selenium atoms (Se1 and Se2) in MjaOgg. All figures were prepared using PYMOL (DeLano, 2008).
MjaOgg and SsoOgg are members of the Ogg2 family and share a common two-domain architecture. These two domains are separated by the central HhH motif considered the fingerprint of DNA repair glycosylases of this superfamily (Denver, et al., 2003). The N-terminal domain encompasses helices αB to αJ arranged in an orthogonal bundle and contains the HhH motif whereas the C-terminal domain comprises helices αA and αK to αM. The catalytic lysine (MjaOggLys129 and SsoOggLys128) and the other important strictly conserved catalytic aspartate residue (MjaOggAsp147 and SsoOggAsp146) belong to helix αJ and the αJ-αK loop, respectively. The overall RMSD for the 207 Cα of MjaOgg and SsoOgg is 1.52Å. Most of the helices superimpose very well in both structures. One difference we observed is that helix αK and the loop between αJ and αK adopt a different orientation in each structure. This region appears to be somewhat mobile in hOGG1 where it undergoes some reorganization upon binding 8-oxoG (Bjørås, et al., 2002, Bruner, et al., 2000).
Structural comparison of Ogg2 enzymes with Ogg1 and AGOG
The most notable difference between Ogg2 and hOGG1 is the lack of the A domain of hOGG1 in the Ogg2 enzymes. As shown in Figure 2a, the anti-parallel twisted β-sheet forming the A domain of hOGG1 is totally absent in MjaOgg and SsoOgg. Apparently, the A domain of hOGG1 is replaced by helix αB in MjaOgg and SsoOgg. It is noteworthy that AGOG enzymes are also devoid of the A domain of Ogg1 (see Figure 2b) (Lingaraju, et al., 2005). Since Ogg2 and AGOG enzymes can function without the A domain what, then, is its role in Ogg1? Previous studies revealed that hOGG1 associates with the nuclear matrix and chromatin in vivo, depending on its phosphorylation state (Dantzer, et al., 2002). Importantly phosphorylation does not affect the glycosylase activity. One of the two possible serine protein kinase C phosphorylation sites is located in the A domain (Ser44PheArg), suggesting that it might play a part in the localization of hOGG1. Alternatively the function of the A domain might be to interact with a specific protein binding partner. The Human Protein Reference Database lists several protein interactors for hOGG1, in addition to protein kinase C, such as XRCC1, Chromogranin B, and small nuclear ribonucleoprotein polypeptide F (Mishra, et al., 2006). AlkA, a DNA glycosylase of the same family, harbors a similar domain with a topology reminiscent of TATA-binding protein (Hollis, et al., 2000, Labahn, et al., 1996), which might be indicative that AlkA may partake in transcription regulation but has yet to be demonstrated
Figure 2. Superposition of MjaOgg on A) hOGG1 and B) Pa-AGOG.
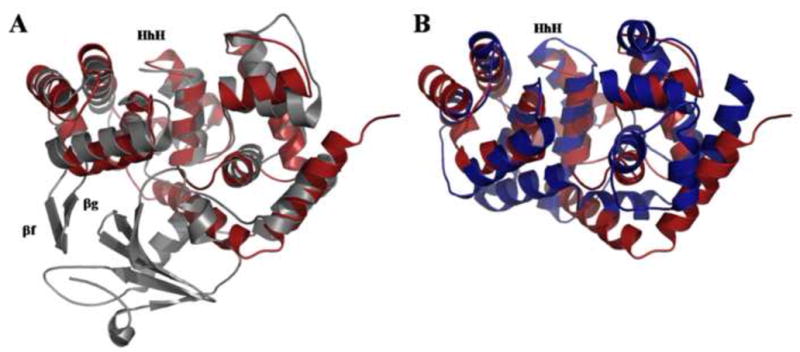
A) Structural superposition of unliganded hOGG1 (PDB ID code: 1KO9) (Ogg1; gray) on MjaOgg (Ogg2; red) illustrating the similar architecture of domains B and C of hOGG1 to the corresponding domains (N- and C-terminal) in Ogg2 and the absence of the hOGG1 A domain in Ogg2. B) Structural superposition of Pa-AGOG (PDB ID code: 1XQO) (AGOG; blue) onto MjaOgg (Ogg2; red) showing a similar fold for the two enzymes.
Despite the absence of an A domain, the two protein families (Ogg1 and Ogg2) share some similarity. For example, the N-terminal domain of MjaOgg superimposes very well on domain B of hOGG1 (Figure 2a). MjaOgg (and SsoOgg), however, lack the two extra β-strands (βF andβG) observed in hOGG1. These two β-strands in hOGG1 (Bjørås, et al., 2002) have no known biological function and do not interact with either the damaged base or the DNA backbone (Bruner, et al., 2000). Moreover, the N-terminal domain of Ogg2 overlays very well onto the N-terminal domain of Pa-AGOG (Figure 2b), where most of the secondary structure elements are conserved.
The central element of the Ogg enzymes is the helix-hairpin-helix motif, a ubiquitous DNA binding motif (Doherty, et al., 1996), which is considered to be the fingerprint of DNA glycosylases of the HhH-GPD superfamily (Denver, et al., 2003, Sartori, et al., 2004). It is therefore not surprising that the HhH motifs of hOGG1 (Ogg1) and MjaOgg/SsoOgg (Ogg2) superimpose so well (Figure 3). The second helix of the HhH motif harbors the conserved catalytic Lys while the strictly conserved catalytic Asp is located on a long loop on the C-terminal end of that same helix. Both helices and the hairpin loop of the HhH motif are very similar in size and in position in Ogg1 and Ogg2. In contrast the HhH observed in AGOG is markedly different (See Figure 3): The two α-helices, especially the C-terminal one, are longer than those of Ogg1 and Ogg2. The most striking difference between Ogg1-2 and AGOG, however, is the position of the hairpin loop. The hairpin structure is facing the orthogonal helix bundle domain in Ogg1-2, whereas the hairpin loop is facing the C-terminal domain in AGOG. This particular HhH motif in AGOG is associated with a different binding mode of 8-oxoG and a locally modified DNA contact compared to hOGG1 (Lingaraju, et al., 2005).
Figure 3. Sequence alignment and superposition of HhH motif of various OGG.
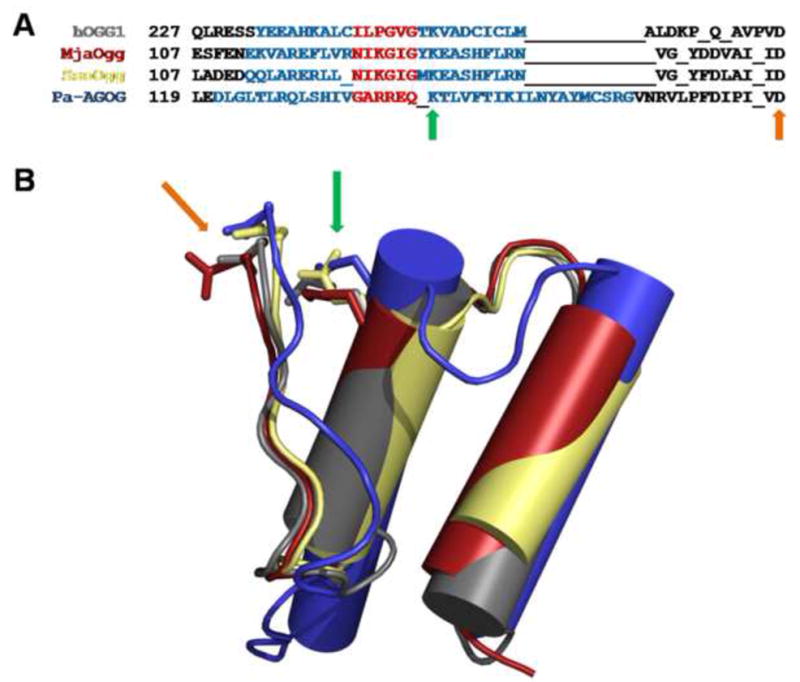
A) Structure based sequence alignment of hOGG1, MjaOgg, SsoOgg, and Pa-AGOG, where residues of the two helices and the hairpin of the HhH motif are highlighted in blue and red, respectively. B) Superposition of the HhH motif of hOGG1 (gray) (PDB ID code: 1KO9 (Bjørås, et al., 2002) ), MjaOgg (red), SsoOgg (pale yellow) and Pa-AGOG (blue) (PDB ID code: 1XQO (Lingaraju, et al., 2005)), illustrating the different orientation of HhH of Pa-AGOG compared to the other Ogg1-2 enzymes. The green arrows indicate the position of the conserved catalytic lysine whereas the orange arrows point to the conserved catalytic aspartate.
Finally, the C-terminal domain of Ogg2 (MjaOgg and SsoOgg) and the C domain of hOGG1 appear to be topologically conserved whereas AGOG harbors two additional helices (α11 and α14) (Figure 2a and b). Previous studies on hOGG1 (Bjørås, et al., 2002) demonstrated a structural reorganization upon binding of 8-oxoG of three of the α-helices (αD, αM and αO) of the C domain. In addition several residues of the C-terminal α-helix of hOGG1 are implicated in the substrate binding. The most pronounced movement is performed by Phe319, which goes from a distal position in the unliganded hOGG1 (PDB ID code: 1KO9) to a proximal position in the hOGG1/8-oxoG complex (PDB ID code: 1EBM), to stack against the six-membered ring of 8-oxoG (see Figure 4) (Bjørås, et al., 2002, Bruner, et al., 2000). This OGG1 conserved Phe appears to be replaced by a conserved Trp in Ogg2 (Trp198 in the MjaOgg and SsoOgg sequences), a residue which can also participate in a stacking interaction with 8-oxoG (Figure 4).
Figure 4. Close-up view of the C-terminal loop of Ogg2 and the αA-βB loop of hOGG1.
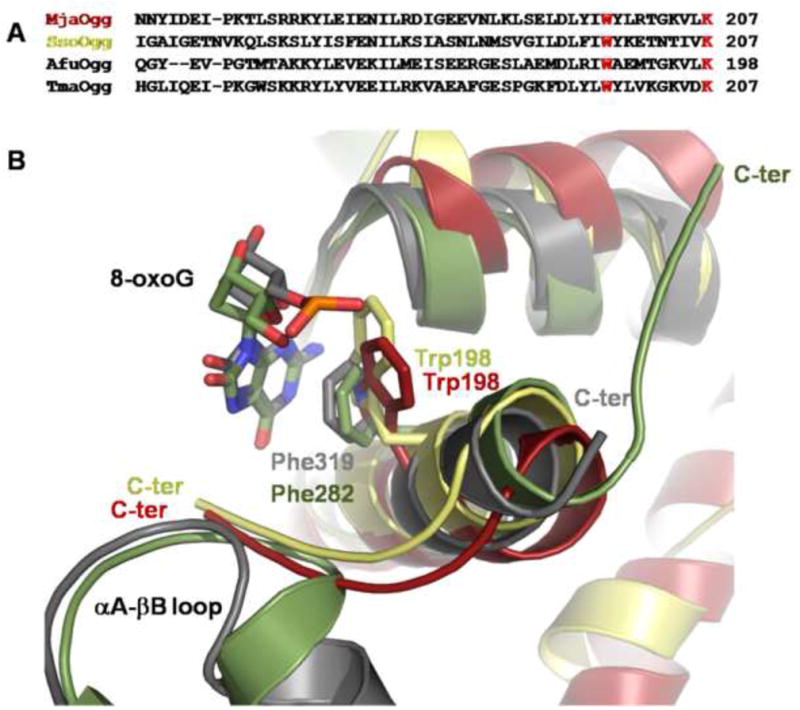
A) Sequence alignment of the C-terminal region of MjaOgg, SsoOgg, AfuOgg (Archaeoglobus fulgidus) (Chung, et al., 2001) and TmaOgg (Thermotoga maritima) (Im, et al., 2005), showing the conserved “stacking” tryptophan and the C-terminal lysine of Ogg2 (in red) B) Ribbon diagram showing a superposition of the C-terminal loop of MjaOgg (red), SsoOgg (pale yellow) and the αA-βB loop of hOGG1 (gray) (PDB ID code: 1EBM (Bruner, et al., 2000)). The superposition indicates that Trp198 of Ogg2 may be involved in a similar stacking interaction with 8-oxoG as Phe319 in hOGG1. Some structural elements were omitted for clarity.
A notable difference resides in the C-terminal loop of Ogg2. As seen in Figure 4b, the position of the C-terminal loop adopts a very different conformation in MjaOgg/SsoOgg and hOGG1. The C-terminal loop of Ogg2 makes a sharp turn at the end of the α-helix and points towards the active site, whereas the corresponding loop in hOGG1 is pointing towards the surface of the protein, some 180° away. The presence of the αA-βB loop from the A domain of hOGG1 undoubtedly contributes to force the C-terminal loop to adopt a conformation which differs from that observed in Ogg2. It is noteworthy that the αA-βB loop in hOGG1 contains the glycine residue involved in the discrimination between guanine and 8-oxoG (Bruner, et al., 2000). The fact that the C-terminal loop of Ogg2 overlaps with the αA-βB loop of hOGG1 urged us to investigate its role further (See below).
Opposite base specificity
Previous work showed that MjaOgg displays little preference for the base opposite the lesion in contrast to hOGG1, which removes 8-oxoG opposite C but not paired to G or A (Gogos and Clarke, 1999, Bruner, et al., 2000). In the structure of hOGG1 in complex with DNA containing 8-oxoG:C (PDB ID code: 1EBM) residues Asn149, Arg154, and Arg204 interact with the estranged cytosine (Bruner, et al., 2000). The H-bond network is composed of hydrogen donors and acceptors that select for cytosine. The structures of MjaOgg and SsoOgg allow us to speculate about the putative interactions these enzymes make with the base opposite the lesion. A structure based sequence alignment reveals that two of the three interacting residues of hOGG1 are conserved in Ogg2: hOGG1 Asn149 and Arg204 appear to be structurally equivalent to MjaOgg Asn49 and Arg89 (see Figure 5) while no structural equivalent is observed for hOGG1 Arg154. The role of Arg154 in the specificity for the estranged base was studied in hOGG1 and a bacterial Ogg, Clostridium acetobutylicum Ogg (CacOGG). The hOGG1 Arg154His mutation has been identified in some human cancers. While the glycosylase activity of the Arg154His variant is similar to the wild type enzyme its specificity for C is relaxed (Bruner, et al., 2000). CacOgg, which displays little preference for the base opposite the damage (Robey-Bond, et al., 2008), differs in sequence from hOGG1 at two sequence positions, Met132 and Phe179, which correspond to amino acids Arg154 and Tyr203 in the human enzyme. When the CacOgg residues were mutated into their human counterparts the resulting variants showed a substantial increase in specificity for 8-oxoG·C over 8-oxoG·A (Robey-Bond, et al., 2008). In the crystal structure of hOGG1 Tyr203 interacts with Asn149, which in turn participates in a H-bond with the N4 amine of C. The role of Tyr203 in determining opposite base specificity is indirect and may be to stabilize Asn149. In CacOgg the tyrosine is replaced by a Phe (Phe179); the H-bond with the asparagine residue is lost and Asn127 (which corresponds to hOGG1 Asn149) does not appear to interact with C (Faucher, et al., 2009). In the Ogg2 enzymes the residue corresponding to hOGG1 Ty203 is also a phenylalanine (Phe85 in MjaOgg and Phe81 in SsoOgg). These structural and biochemical data taken together strongly suggest a crucial role for Arg154 in determining the specificity for the estranged base, with an additional indirect contribution from Tyr203. The absence of these residues in CacOgg and Ogg2 provides an explanation for their lack of opposite base specificity.
Figure 5. Putative interactions of MjaOgg with the cytosine opposite the lesion.
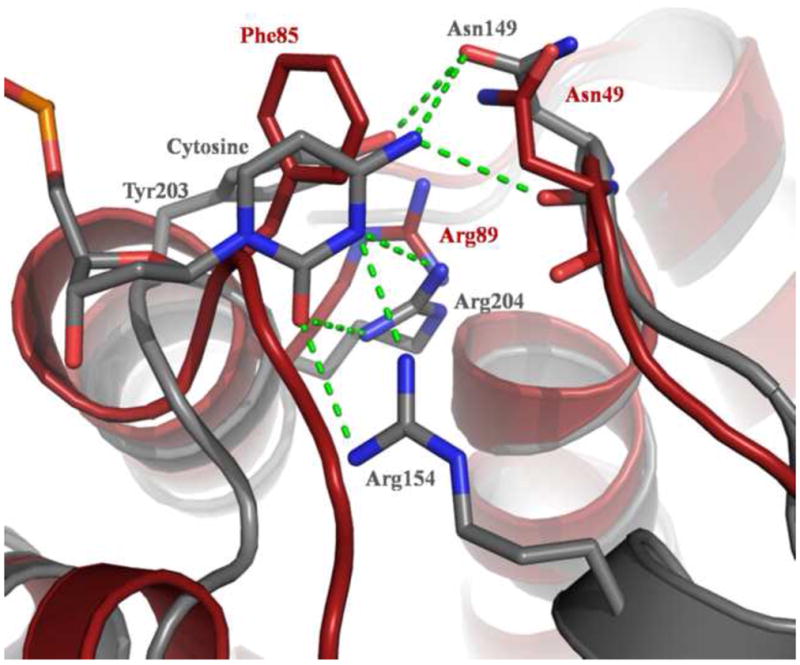
The figure shows the interactions made by hOGG1 (PDB ID code 1EBM) with the estranged C (shown in gray) and putative interacting residues in MjaOgg (red). While hOGG1 Asn149 and Arg204 appear to have structural equivalents in MjaOgg (Asn49 and Arg89) Arg154 does not.
Structural basis for the discrimination between guanine and 8-oxoguanine by Ogg2
As with other DNA glycosylases, Ogg2 enzymes are presumed to bind their substrate in an extrahelical conformation (Bruner, et al., 2000, Fromme and Verdine, 2003, Fromme, et al., 2004, Labahn, et al., 1996). The damaged base is rotated out of the DNA helix and bound in a cavity lined with specific binding residues (Huffman, et al., 2005) in a deep groove located at the junction of the two domains and the HhH motif. Although the presence of a keto moiety on the C8 atom of 8-oxoG might appear to be the major determinant to discriminate between guanine and 8-oxoG, earlier structural studies (Bjørås, et al., 2002, Bruner, et al., 2000) revealed that the C8 carbonyl group is completely devoid of any direct hydrophilic interactions with hOGG1. In contrast, in Pa-AGOG a H-bond was observed between the 8-oxygen atom of 8-oxoG and the nitrogen of the indole group of Trp69 (Lingaraju, et al., 2005).
One consequence of the oxidation of the C8 atom of guanine is the electron delocalization of the double bond between the N7 and C8 atoms of the 8-oxygen and the protonation of N7. The N7 proton allows 8-oxoG to establish a crucial hydrophilic contact with the main chain carbonyl of the conserved glycine of the αA-βB loop (Gly42 in hOGG1) (Gray dashed line on Figure 6), which cannot occur with G. Interestingly, this interaction was observed even if the glycine residue is mutated to an alanine. The protein fold seems to adapt to the structural stress induced by the introduction of a methyl group (Radom, et al., 2007). Previous work demonstrated that the hydrophilic bond between the conserved Gly and the 8-oxoG molecule was not the only factor hOGG1 uses to distinguish G and 8-oxoG. Banarjee et al. showed that in the active site of hOGG1 a dipole formed by Lys249-NH3+ and Cys253-S− complements perfectly the dipole observed on the 8-oxoG molecule while creating a repulsive force on G (Banerjee, et al., 2005). This pair of residues is conserved in CacOgg and yeast OGG1, but the cysteine is replaced by a histidine in MjaOgg/SsoOgg, suggesting that such dipole effect is less critical in Ogg2 enzymes. In addition, because Ogg2 glycosylases lack the A domain of hOGG1 and therefore the αA-βB loop and the glycine responsible for the G/8-oxoG discrimination, the mechanism for distinction of G/8-oxoG must reside elsewhere. As seen in Figure 4, the C-terminal loop of MjaOgg/SsoOgg overlaps with the αA-βB loop precisely at the position of the glycine responsible for the recognition of 8-oxoG in OGG1. A close-up view of this region (Figure 6) shows that the C-terminal lysine of MjaOgg/SsoOgg superimposes very well onto hOGG1 Gly42. In this position, the carboxyl group of the C-terminal Lys207 in Ogg2 can easily form an H-bond with the N7 hydrogen of 8-oxoG (green dashed line on Figure 6). The side chain of the C-terminal Lys207 is stabilized by a hydrophilic network involving several conserved residues. The amino group of MjaOgg-Lys207 interacts with the side chains of Glu40 and Gln59 while SsoOgg-Lys207-NH3+ interact with Glu21 and Glu36. Among the five residues interacting via main chain or side chain atoms with the C-terminal lysine, only MjaOgg-Gln59 appears to be not strictly conserved in Ogg2. Taken together these observations provide a strong indication that the C-terminal lysine of MjaOgg/SsoOgg (and other Ogg2 enzymes) may be a critical molecular determinant for the discrimination between G and 8-oxoG.
Figure 6. Interactions involving the C-terminal lysine of Ogg2.
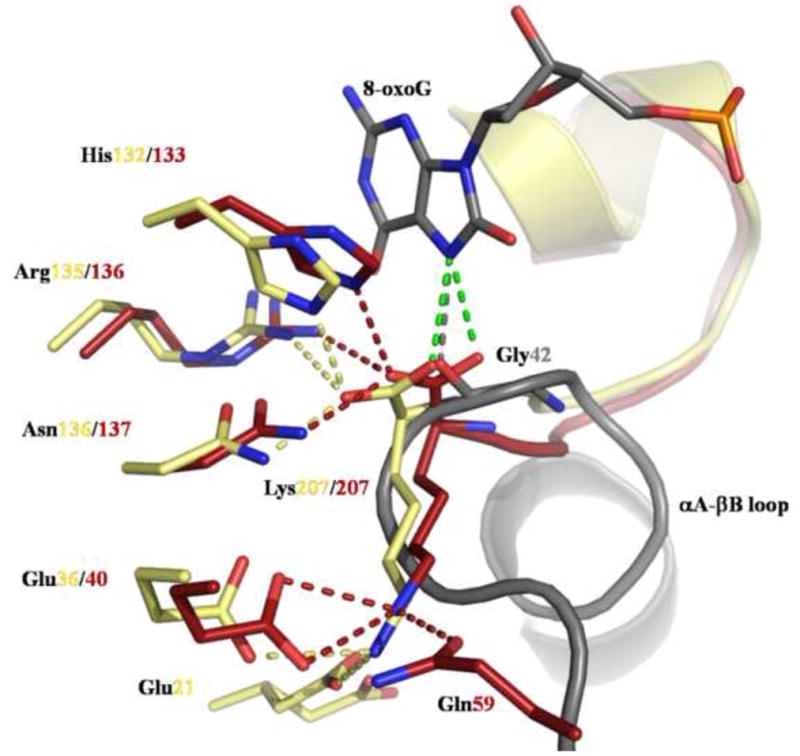
The figure shows the hydrophilic interactions made by the conserved glycine in hOGG1 with 8-oxoG (gray) (PDB ID code: 1EBM (Bruner, et al., 2000)) and by the C-terminal lysine of MjaOgg (red) and SsoOgg (pale yellow) with several protein residues. The putative interaction of the carboxyl group of Lys207 of MjaOgg/SsoOgg with 8-oxoG is shown as a green dashed line. Glu21 adopts an alternate conformation in SsoOgg. Some structural elements were omitted for clarity.
Role of the Ogg2 C-terminal lysine in the 8-oxoguanine glycosylase activity
Previous attempts have been made to break the interaction between the main chain carbonyl of glycine (Gly42 in hOGG1) and 8-oxoG-N7-H (Radom, et al., 2007) by mutating the glycine to an alanine. Despite the introduction of a methyl group, the main chain carbonyl of Ala was still able to interact with 8-oxoG. Apparently, it is impossible to break this interaction without deleting the entire αA-βB loop, which would have drastic consequences on protein integrity. Ogg2 enzymes, on the other hand, provide us with a unique opportunity to investigate the role of the main chain carbonyl/8-oxoG-N7-H interaction, because the residue making this interaction is located at the extreme C-terminus of the enzyme allowing us to construct a deletion mutant without dire consequences for the protein fold.
In order to investigate the role of the C-terminal lysine of Ogg2, we created a truncated form of MjaOgg by deleting the last three amino acid residues. The resulting MjaOggΔ3 variant was assayed for glycosylase activity using a 35mer double stranded DNA containing the 8-oxoG lesion opposite each of the four bases. Unlike hOGG1, which displays a strong preference for C as the opposite base, MjaOgg is known to cleave 8-oxoG across all four bases (Gogos and Clarke, 1999). As seen in Figure 7A, only the wild-type MjaOgg is able to excise 8-oxoG opposite each of the four bases while MjaOggΔ3 is unable to excise the damaged base. To make sure that the deletion of the three C-terminal residues did not significantly alter the proper folding of the enzyme a lyase activity assay was performed on both wild-type and truncated MjaOgg using a DNA oligomer containing an abasic site. Panel B on Figure 7 shows that both wild-type MjaOgg and MjaOggΔ3 are able to process the second step of the glycosylase/lyase reaction. MjaOggΔ3 displays a slightly less robust lyase activity compared to wild-type MjaOgg, which could be explained by a possible destabilization of the DNA backbone due to the lack of some DNA interactions. The activity assay confirms our hypothesis and underscores the paramount importance of the C-terminal lysine of Ogg2 in the discrimination between G and 8-oxoG.
Figure 7. Glycosylase/lyase activity assay of MjaOgg and MjaOggΔ3.
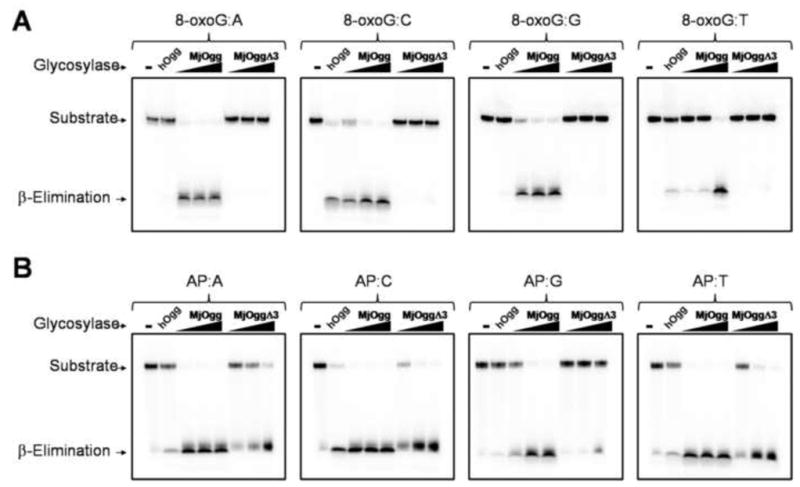
The figure shows a comparison of A) the glycosylase activity of MjaOgg and MjaOggΔ3 and B) the lyase activity for the same enzymes. The glycosylase assay was performed on an oligonucleotide containing 8-oxoG opposite each of the four bases whereas the lyase activity assay was performed with an oligonucleotide containing an abasic site opposite each of the four bases. The 32P-labeled 8-oxoG and AP substrates (10 nM) were incubated with 20 nM hOGG1 for positive control or increasing concentrations (20 nM, 50 nM, and 100 nM) of MjaOgg and MjaOggΔ3 in their respective reaction buffers.
Concluding remarks
Because hOGG1 and Fpg can recognize methyl-formamidopyrimidine (Me-Fapy) as well as 8-oxoG and Me-Fapy lacks the N7-hydrogen the role of the protonated N7 in the recognition of 8-oxoG has been subject of debate (David, et al., 2007, Hamm, et al., 2007). A recent study which focused on guanine and 8-oxoguanine analogues that varied at the N7 and/or C8 positions suggested that discrimination between the two bases stems from the absence of a fully sp2-hydridized N7 rather than the presence of a N7-hydrogen or a C8-oxygen in 8-oxoG (David, et al., 2007, Hamm, et al., 2007). Here we present the first crystal structures of two archaeal Ogg2 enzymes and demonstrate the importance of the C-terminal lysine in the discrimination between guanine and its oxidized product. A structural analysis of a complex of Ogg2 with 8-oxoG-containing DNA will be required to decipher the detailed interactions between the enzyme and its substrate.
Experimental procedures
Recombinant SeMet-MjaOgg expression and purification
The open reading frame of MjaOgg was cloned using Nde1-Xho1 restriction sites of a pET-22b vector (Novagen). A penta-histidine-tag sequence was inserted before the first ATG codon of MjaOgg while the stop codon of MjaOgg was conserved. Recombinant SeMet-MjaOgg was engineered by inhibiting the methionine biosynthesis pathway (Doublié, 2007). Briefly, SeMet-MjaOgg was expressed in M9 minimal medium supplemented with select amino acids and selenomethionine (60 mg/L) 15 minutes prior to induction with IPTG. After 4 hours at 37°C, the bacterial pellet was sonicated in lysis buffer (50 mM Na-Phosphate pH 8.0, 100 mM NaCl, 10 mM imidazole and 5 mM β-mercaptoethanol). The centrifuged lysate was loaded on a Ni-NTA column (GE Healthcare) and eluted using lysis buffer supplemented with 500 mM imidazole. Pooled fractions of MjaOgg were dialyzed in 20 mM Hepes pH 7.6, 150 mM NaCl, 10% (v/v) glycerol and 5 mM β-mercaptoethanol then loaded on an SP column (GE Healthcare) and eluted with a salt gradient. The purified protein was concentrated to 0.8 mg/ml prior to crystallization.
Recombinant SsoOggK128Q expression and purification
SsoOgg was cloned in a manner identical to that used for MjaOgg and the K128Q glycosylase deficient mutant was generated by site directed mutagenesis. After overnight expression at 16°C with 1mM IPTG, the bacterial pellet was sonicated in lysis buffer (50 mM Na-Phosphate pH 8.0, 100 mM NaCl, 10 mM imidazole and 5 mM β-mercaptoethanol). The centrifuged lysate was loaded on a Ni-NTA column (GE Healthcare) and eluted using an imidazole gradient. Pooled fractions of SsoOgg were dialyzed in 20 mM HEPES pH 7.6, 150 mM NaCl, 10% (v/v) glycerol and 5 mM β-mercaptoethanol then loaded on an SP column (GE Healthcare) and eluted with a salt gradient. The purified protein was concentrated to 5 mg/ml prior to crystallization.
Crystallization, X-ray analysis, and structure determination of the recombinant SeMet-MjaOgg
Crystals of SeMet-MjaOgg were obtained by hanging-drop vapor diffusion at 12 °C in 2 L drops of a 1:1 ratio of purified protein and well solution (30% (w/v) PEG-4000, 0.1 M Na-Citrate pH 5.9 and 1% β-mercaptoethanol). Crystals grew to dimensions suitable (80 × 80 × 80 μm3) for X-ray diffraction experiments. Crystals were flash cooled directly from the crystallization drop into a stream of nitrogen kept at 100K. Since molecular replacement attempts failed to produce a clear solution, we collected a MAD data set at three wavelengths corresponding to the peak, inflection, and high energy remote of the K edge of selenium (See Table 1 for data collection statistics) at beamline 23-ID B at the Argonne Advanced Photon Source. The diffraction images were integrated using XDS (Kabsch, 1993) and merged/scaled with CCP4. (Bailey, 1994) The program SOLVE (Terwilliger and Berendzen, 1999) identified the two selenium sites. AutoSHARP (Vonrhein, et al., 2007) was then used for refinement of the selenium parameters. The phasing information was then used in ARP/wARP (Perrakis, et al., 1999) for density modification and iterative model building. A MAD map after density modification and a simulated annealing omit map are shown in Supplementary Figure S1. The refinement procedure was performed with CNS (Brunger, et al., 1998). The initial model issued from ARP/wARP was submitted to a cycle of simulated annealing at 3000 K followed by energy minimization and B-factor refinement cycle. Afterwards, the model was refined by simple energy minimization followed by isotropic B-factors refinement (restrained and individual) and corrected by manual rebuilding using O (Jones, et al., 1991). Missing parts of the model, glycerol, and water molecules were progressively added during the refinement procedure. Finally, the quality of the model was verified with PROCHECK (Laskowski, et al., 1993). There is only one residue (Lys165) in the disallowed region of the Ramachandran plot probably caused by symmetry interactions. Residues Lys100, Asn187, and Ile197 appear to adopt alternate conformations.
Crystallization, X-ray analysis, and structure determination of the recombinant SsoOggK128Q
Crystals of SsoOgg were obtained by hanging-drop vapor diffusion at 12 °C. DNA oligonucleotides (16-mer) were ordered from Midland Certified Reagent Co. (Midland, TX) and purified on a Mono Q column (GE Healthcare). The sequences were as follows: 5′-AGC-GTC-CAX-GTC-TAC-C-3′ and 5′-GGT-AGA-CCT-GGA-CGC-T-3′ where X is 8-oxoG. Protein and duplex DNA were mixed in a 1:1.2 molar ratio. Crystals grew in 2 μL drops containing a 1:1 ratio of protein/DNA mix and well solution (2 M (NH4)2SO4, 50 mM NaCacodylate pH 6.5 and 10 mM MgSO4). This procedure yielded crystals with dimensions suitable (150 × 30 × 30 μm3) for X-ray diffraction experiments. A complete data set was collected at 100 K and a wavelength of 1.0332 Å at beamline 23-ID B at the Argonne Advanced Photon Source on a single four-year old crystal. The supersaturation in the protein drop was such that there was no need to add a cryoprotectant and the crystal was flash cooled directly from the drop. Diffraction data were indexed using XDS (Kabsch, 1993) and scaled with XSCALE. The structure of SsoOggK128Q was solved by molecular replacement with MOLREP from the CCP4 suite (Bailey, 1994) using the MjaOgg coordinates as a model. Refinement procedure was performed as stated above for MjaOgg. A simulated annealing omit map is shown in Supplementary Figure S2. The quality of the model was verified with PROCHECK. (Laskowski, et al., 1993). There are no residues in the disallowed region of the Ramachandran plot. Residues Glu21 and Leu42 seem to adopt alternate conformations.
MjaOgg C-terminal deletion mutant
The MjaOgg deletion mutant was generated by PCR cloning using the following primers: 5′-GGG-CAT-ATG-GGC-AAC-CAT-CAT-CAT and 5′-GGG-CTC-GAG-TTA-TTT-GCC-GGT-ACG-TAA-T and the pET22b-MjaOgg plasmid. The PCR products and an empty pET22b vector were digested with Xho1-Nde1 and purified on an agarose gel. Vector and insert were ligated using a Rapid DNA ligation Kit (Roche). A positive clone was sequenced in its entirety to confirm the deletion.
Glycosylase/lyase activity assays
The DNA substrates used for glycosylase and lyase assays were purchased from Midland Certified Reagent Co. (Midland, TX). The DNA sequences were 5′-TGT-CAA-TAG-CAA-GXG-GAG-AAG-TCA-ATC-GTG-AGT-CT-3′ for the damage-containing strand, where X was either 8-oxoG or uracil, and 5′-AGA-CTC-ACG-ATT-GAC-TTC-TCC-YCT-TGC-TAT-TGA-CA-3′ for the complementary strand, where Y was either one of the four bases. 1 pmole of the damage-containing oligonucleotide was phosphorylated with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) using [γ-32P] dATP. The labeled DNA strand was annealed with the complementary oligonucleotide in buffer containing 10 mM Tris-HCl pH 8.0 and 50 mM NaCl. Abasic sites were generated by treating the 32P-labeled double-stranded oligonucleotide containing uracil with 2 U of E. coli uracil DNA glycosylase (New England Biolabs, Ipswich, MA) for 30 min at 37°C, directly in the annealing buffer.
The assays were performed in 10 μl reaction by incubating 10 nM substrates containing 8-oxoG or an AP site with either 20 nM of the control enzyme hOGG1 or increasing concentrations (20, 50, and 100 nM) of MjaOgg or the deletion mutant, MjaOggΔ3. Reactions involving hOGG1 were incubated for 30 min at 37°C in 50 mM Tris-HCl pH 7.4, 50 mM NaCl, 2 mM EDTA, 0.1 mg/ml BSA, and 5% (v/v) glycerol while MjaOgg and MjaOggΔ3 reactions were conducted as described previously (Gogos and Clarke, 1999) for 30 min at 50°C in 20 mM Tris-HCl pH 8.5, 80 mM NaCl, 1 mM DTT, 1 mM EDTA, and 0.1 mg/ml BSA. Reactions were stopped by adding 10 μl of gel-loading dye (99% formamide, 5 mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue) and heated at 95°C for 3 min before loading on a 12% denaturing polyacrylamide gel.
Supplementary Material
Acknowledgments
We thank Wendy Cooper, Alicia S. Holmes, and April Averill for help with protein purification and Dr. Pierre Aller and Karl Zahn for collecting the MAD data set at the APS synchrotron. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. This research was supported by National Institutes of Health grants R01CA33657 and P01CA098993 awarded by the National Cancer Institute.
Abbreviations
- 8-oxoG
7,8-dihydro-8-oxoguanine
- AGOG
archaeal GO glycosylase
- AP site
apurinic/apyrimidinic site
- BER
base excision repair
- Fpg
formamidopyrimidine-DNA glycosylase
- hOGG1
human OGG1
- Me-Fapy
methyl-formamidopyrimidine
- HhH
helix-hairpin-helix
- Ogg
8-oxoguanine DNA glycosylase
- PKC
protein kinase C
- RMSD
root mean square deviation
- ROS
reactive oxygen species
Footnotes
Supplemental Data include two figures and can be found at http://www.structure.org
Accession numbers
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank and are available under the following accession codes: 3FHF for MjaOgg and 3FHG for SsoOggK128Q
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Bjørås M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. Embo J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørås M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal “flipping” underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J Mol Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutat Res. 2001;486:99–111. doi: 10.1016/s0921-8777(01)00081-7. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Luna L, Bjørås M, Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos; CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. In: Doublié S, editor. Macromolecular Crystallography Protocols. Vol. 363. Totowa, NJ: Humana Press; 2007. pp. 91–108. [DOI] [PubMed] [Google Scholar]
- Faucher F, Duclos S, Bandaru V, Wallace SS, Doublié S. Structural characterization of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase in its apo-form and in complex with 8-oxodeoxyguanosine. Journal of Molecular Biology. 2009 doi: 10.1016/j.jmb.2009.01.067. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. Embo J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J Biol Chem. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- Gondichatnahalli L, Sartori A, Hunziker P, Kostrewa D, Prota A, Jiricny J, Winkler F. Pa-AGOG, a novel 8-oxoguanine DNA glycosylase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Faseb Journal. 2004;18:C177–C177. [Google Scholar]
- Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- Hamm ML, Gill TJ, Nicolson SC, Summers MR. Substrate specificity of Fpg (MutM) and hOGG1, two repair glycosylases. J Am Chem Soc. 2007;129:7724–7725. doi: 10.1021/ja0716453. [DOI] [PubMed] [Google Scholar]
- Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Im EK, Hong CH, Back JH, Han YS, Chung JH. Functional identification of an 8-oxoguanine specific endonuclease from Thermotoga maritima. J Biochem Mol Biol. 2005;38:676–682. doi: 10.5483/bmbrep.2005.38.6.676. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- Labahn J, Scharer OD, Long A, EzazNikpay K, Verdine GL, Ellenberger TE. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005;13:87–98. doi: 10.1016/j.str.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- Robey-Bond SM, Barrantes-Reynolds R, Bond JP, Wallace SS, Bandaru V. Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry. 2008;47:7626–7636. doi: 10.1021/bi800162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica Section D-Biological Crystallography. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. In: Doublié S, editor. Macromolecular Crystallography Protocols. Vol. 364. Totowa, NJ: Humana Press; 2007. pp. 215–230. [DOI] [PubMed] [Google Scholar]
- Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


