Abstract
To further our understanding of powdery mildew biology during infection, we undertook a systematic shotgun proteomics analysis of the obligate biotroph Blumeria graminis f. sp. hordei at different stages of development in the host. Moreover we used a proteogenomics approach to feed information into the annotation of the newly sequenced genome. We analyzed and compared the proteomes from three stages of development representing different functions during the plant-dependent vegetative life cycle of this fungus. We identified 441 proteins in ungerminated spores, 775 proteins in epiphytic sporulating hyphae, and 47 proteins from haustoria inside barley leaf epidermal cells and used the data to aid annotation of the B. graminis f. sp. hordei genome. We also compared the differences in the protein complement of these key stages. Although confirming some of the previously reported findings and models derived from the analysis of transcriptome dynamics, our results also suggest that the intracellular haustoria are subject to stress possibly as a result of the plant defense strategy, including the production of reactive oxygen species. In addition, a number of small haustorial proteins with a predicted N-terminal signal peptide for secretion were identified in infected tissues: these represent candidate effector proteins that may play a role in controlling host metabolism and immunity.
Fungi are the most prevalent cause of disease in plants, and failure to control them results in widespread damage and severe harvest loss. In particular, the powdery mildew Blumeria graminis attacks the cereals wheat and barley, which are two of the main food crops in Western agriculture. The nature of cereal powdery mildew infection and its detrimental effect on crop health require a continual effort aimed in fighting the disease through the use of fungicides and the breeding of novel resistant varieties. Both these approaches are currently effective albeit with very significant economic and ecological impact. However, the gradual withdrawal of some fungicides driven by concerns for their environmental impact, the constant threat of the emergence of fungicide resistance, and new virulence alleles that overcome breed resistance require new tools for disease control. This will entail the development of new classes of fungicides, introgression of novel disease resistance genes, and possibly the engineering of a specific biological control through enhancement of disease tolerance and resistance. Understanding the biology and virulence of plant pathogenic fungi is essential to meet this challenge (1).
Powdery mildews are easily recognizable as white pustules on leaves and stems and are some of the most conspicuous plant diseases affecting a wide range of hosts (2). Powdery mildews are obligate biotrophs, i.e. they have an absolute requirement of a host to grow and complete their life cycle. Blumeria shows a very high degree of host specificity; for example B. graminis f. sp. hordei grows exclusively on barley. In addition to being an economically important pathogen, B. graminis f. sp. hordei is also the best studied powdery mildew and a model for research into these interactions. Most of the life cycle of Blumeria is vegetative and is spent as a rapid succession of asexual cycles in which the airborne conidia germinate on the surfaces of leaves and stems to produce first a primary germ tube and then a secondary germ tube that develops a swollen adhesion structure called an appressorium. Within 8–12 h a thin peg emerges from the underside of the appressorium, breaches the plant cell wall, and penetrates the cell. Inside the plant cell a highly branched feeding structure, the haustorium, develops surrounded by an intact host membrane (3). The haustorium has long been known to actively take up nutrients (4), but like other obligate pathogens, it is now also believed to control host perception and defense (5), enabling the invading pathogen to survive, avoid, and suppress rejection responses. How this is achieved is still unknown, but current thinking postulates that this control is mediated by protein “effectors” secreted by the pathogen into the host cells (6).
To understand the biology and virulence of plant pathogenic fungi, a systematic analysis of the genomes and transcriptomes of plant pathogenic fungi is a top priority, and in recent years it has been actively pursued by a number of institutions and research consortia worldwide (Broad Institute Fungal Genome Initiative and the United States Department of Energy Joint Genome Institute). We are coordinating the sequencing of the barley powdery mildew fungus B. graminis f. sp. hordei genome (Blumeria Genome Sequencing Project) and have extensively researched the transcriptome dynamics of Blumeria during in vivo pathogenic development (7, 8). A comparable effort to analyze the proteome in plant pathogenic fungi is missing despite the fact that it is essential for our understanding of complex biological systems at the post-transcriptional level because transcript levels do not always reflect protein levels and protein activities. This is further complicated by evidence of RNAs acting directly or indirectly with mRNA molecules to influence the expression and, indirectly, the activities of proteins (9). It is also generally accepted that the biological activity of proteins also depends on post-translational modifications and protein-protein interactions. Furthermore a proteogenomics approach can contribute to validating and discovering new ORFs on annotated genomes. In fact, even in well studied and annotated genomes such as the Arabidopsis genome, it has been suggested that around 13% of the Arabidopsis proteome is incomplete because of missing or incorrect gene models or ORFs (10, 11). It is therefore imperative to add a proteomic dimension in an integrated analysis to study gene function, protein expression, and localization.
So far no large scale comparative study of proteomes of phytopathogenic fungi during in planta colonization has been undertaken to unravel key players of the infection, initiation, and establishment of a susceptible interaction with the host, in particular during a biotrophic interaction. There are published reports that analyze the leaf proteome of plants infected by pathogenic fungi, but few of these describe fungal proteins within total extracts from infected leaves, and only a very limited number of fungal proteins have been identified in these tissues (12, 13). In one instance intercellular washing fluids were extracted to detect secreted fungal proteins in the apoplast (14). In the particular case of B. graminis f. sp. hordei, two recent studies have identified a few hundred proteins from conidia (15) and isolated feeding structures (haustoria) (16) of B. graminis. Here we describe the analysis of three different in planta expressed proteomes of Blumeria. We compared proteins extracted from conidia, sporulating epiphytic hyphae growing on the host, and intracellular haustoria embedded in the host epidermis.
Because of the impossibility of obtaining growth of obligate biotrophs in vitro, the analysis of the proteomes of biologically significant life stages must be carried out in the plant host. On the one hand, this analysis is technically challenging as the fungal proteins are embedded in and diluted by the plant proteome; on the other hand, the proteins will be identified in vivo, i.e. under conditions that are biologically relevant to the interaction.
The availability of the recently sequenced B. graminis f. sp. hordei genome now allows the undertaking of a large scale shotgun proteomics approach as the peptides detected by mass spectrometry can directly be matched to the genomic sequence contigs thus avoiding cumbersome and difficult de novo peptide sequencing. At the same time the experimental proof of proteins expressed by Blumeria represents an invaluable proteogenomic resource for the creation and validation of ab initio gene-finding software for the functional annotation of the genome itself.
EXPERIMENTAL PROCEDURES
Barley and B. graminis Cultures
Barley plants (Hordeum vulgaris cultivar Golden Promise) were grown in soil as described previously (7). B. graminis f. sp. hordei strain DH14 was maintained as described previously (7).
Protein Extraction and Separation
Protein extraction in denaturing conditions was performed in a denaturing buffer consisting of 7 m urea, 2 m thiourea, 2% CHAPS, and 20 mm DTT.
Conidia were recovered from infected plants 7 days postinoculation (dpi),1 transferred to a 1.5 ml-polypropylene microcentrifuge tube, and stored at −80 °C until protein extraction. Denaturing extraction buffer (350 μl) was added to 35 mg of conidia. The suspension was transferred with a pipette as small droplets into a small mortar containing liquid N2 and a small amount of quartz sand. After grinding, the powder was transferred to a microcentrifuge tube, left at room temperature to melt, and then incubated for 5 min with regular mixing. Insoluble particulates were removed by two successive 10-min centrifugations at 17,860 × g at room temperature.
Sporulating hyphae were sampled from ∼200 infected primary leaves 5 dpi as described elsewhere (7) and stored at −80 °C. Sporulating hyphae, embedded in the cellulose acetate peels, were transferred to a mortar and pestle and ground with a little sand in liquid N2. The powder was transferred to a centrifuge tube, and 3 ml of denaturing extraction buffer was added. The tissue was left to thaw at room temperature with occasional vortexing. Particulates were removed by two successive 10-min centrifugations at 17,860 × g at 4 °C.
After removing hyphae from primary infected barley leaves (7–10 dpi) as described above, the abaxial epidermis from these primary leaves was stripped using watchmaker forceps. This tissue was defined as the epidermis-haustorial tissue. Epidermal strips were resuspended in 5–10 volumes of precooled denaturing buffer and homogenized in a glass homogenizer on ice. After transfer of the slurry to microcentrifuge tubes, sand was added, and the epidermis was further homogenized with a hand-driven Potter homogenizer. Insoluble particulates were removed as described for the conidia extracts.
Sporulating hyphae (biological replicates HY1–3) and epidermis-haustoria (biological replicates EH1–4) protein extracts were further cleaned up and concentrated by either TCA-acetone (TA) precipitation (samples HY1, HY2, and EH1) or chloroform-methanol (CM) precipitation (samples HY3, EH2, and EH3) or both (sample EH4). For TA precipitation, 19 volumes of cold TA solution (10% (w/v) TCA in cold acetone and 0.07% β-mercaptoethanol) was added to 1 volume of protein extract in a microcentrifuge tube. After 30-min incubation at −20 °C, a protein pellet was recovered following 20-min centrifugation at 17,860 × g at 4 °C. To remove the excess salts the protein pellet was washed three times with cold acetone containing 0.07% β-mercaptoethanol and recovered by 10-min centrifugation at 17,860 × g at 4 °C. For CM precipitation, 400 μl of methanol was added to 100 μl of protein extract and vortexed. Then 100 μl of chloroform was added and vortexed. Finally 300 μl of double distilled H2O was added, and the mixture was vortexed. The two solvent phases were separated by 2-min centrifugation at 17,860 × g. The upper phase was removed, and proteins in the interphase were precipitated by addition of 400 μl of methanol and 5-min centrifugation at 17,860 × g. The pellet was further washed with cold acetone containing 0.07% β-mercaptoethanol, and the pellet was recovered by 3-min centrifugation at 17,860 × g. All liquid was removed, and the pellets were allowed to dry on the bench for 2 min, avoiding complete dryness. Pellets were resuspended in denaturing buffer.
Total leaf (TL) tissue from infected plants (7–10 dpi) was also harvested, flash frozen in liquid N2, and stored at −80 °C until protein extraction. Leaves were ground with some sand in liquid N2 using a mortar and pestle. Soluble proteins were extracted in 3 volumes of ice-cold 50 mm Tris, pH 7.6, 0.33 m sucrose, 1 mm DTT, 1 mm MgCl2, 1% (w/v) poly(vinylpolypyrrolidone), and 0.4% (v/v) protease inhibitor mixture VI (Calbiochem/Merck). Samples were thawed, mixed, and kept on ice for 5 min. Particulates were removed by two successive 10-min centrifugations at 3,300 × g and 17,860 × g at 4 °C. Proteins were selectively precipitated by the addition of ammonium sulfate (520 mg/ml of protein solution). After 20-min incubation on ice, a protein pellet was recovered by 20-min centrifugation at 15,000 × g. Samples were desalted by buffer replacement with 1× PBS buffer (150 mm NaCl, 10 mm NaH2PO4, pH 7.4) in a VivaspinTM 20-ml ultrafiltration device and centrifugation at 3,300 × g (Vivascience, Sartorius, Epsom, UK) with a molecular mass cutoff of 3 kDa. Protein concentrations were estimated with the Bradford method (Bio-Rad) using BSA as protein standard.
For each proteome analyzed, up to 50 μg of protein was separated on a 1-mm-thick SDS-polyacrylamide minigel (Mini-Protean III, Bio-Rad; conidia (“CON”), 30 μg; HY1, 45 μg; EH1, 50 μg). Alternatively bigger gels with a thickness of 1.5 mm were used for scale-up (Protean II, Bio-Rad), and 70–150-μg amounts of protein were loaded (HY2, 70 μg; HY3, 90 μg; EH3, 150 μg; EH4, 75 μg). All samples were separated on 12% acrylamide gels except for HY2 and HY3, which were separated on a 15% acrylamide gel. Gels were stained with colloidal Coomassie (17).
In-gel Digestion and Liquid Chromatography-Tandem Mass Spectrometry
In-gel tryptic digestion was performed as described previously (18). Tryptic digests were then reconstituted in 12–20 μl of 0.2% TFA depending on gel band stain intensity.
Four microliters of sample was loaded on a 10-mm trap column packed with 3.5-μm C18 particles (LC Packings/Dionex, Amsterdam, The Netherlands), washed with 0.2% TFA for 5 min at a flow rate of 30 μl/min, and eluted in 0.2% formic acid for 155 min in a 2–27% ACN gradient and for another 30 min in a steeper 27–50% ACN gradient at a flow rate of 250 nl/min on a 15-cm × 75-μm PepMap C18 reverse phase analytical column (3.5-μm particles; LC Packings/Dionex) using an UltimateTM nano-LC (nLC) system (LC Packings/Dionex). The LC system was coupled to an nESI-MS/MS three-dimensional ion trap mass spectrometer (HCT Esquire, Bruker Daltonics, Bremen, Germany) using a 10-cm-long stainless steel emitter (Proxeon, Odense, Denmark) in the nESI source. The LC system and the ion trap were controlled through HyStar (3.1 build 52.2) and Esquire Control (5.3. build 11) modules in the Compass software suite (Bruker Daltonics, Coventry, UK). Mass spectra were acquired from m/z 300 to 2,000 using parameters optimized at m/z 850 with the trap ion charge control set at 150,000 and a maximum acquisition time of 200 ms averaging three scans per spectrum. The three most abundant 2+ or 3+ ions were selected for MS/MS with a signal threshold of 5,000, the isolation window had a width of m/z 4, and the fragmentation amplitude was 1 V. The selected precursor ions were actively excluded for 45 s after two selections. Samples CON, HY1, HY2, EH1, and EH2 were analyzed under these conditions. Raw LC-MS/MS data were batch-processed in DataAnalysis 3.3.147 (Bruker Daltonics). For each LC run corresponding to one gel band tryptic digest up to 6,000 2+ and 3+ compounds (retention time restriction of 5–210 min) with a signal-to-noise ratio above 5 were extracted and exported as Mascot generic format (mgf) files.
The resuspended digests of the samples HY3, EH3, and EH4 were analyzed by LC-MS/MS using a quaternary LC pump coupled to a hybrid linear ion trap-orbitrap (LTQ-Orbitrap, Thermo Fisher Scientific Bremen, Germany). Depending on the intensity of the Coomassie-stained band, 2–10 μl of sample solution was loaded on a peptide Captrap (Michrom Bioresources, Auburn, CA) with an Accela LC pump (Thermo Fisher Scientific) for 5 min at 10 μl/min using water containing 0.1% formic acid. Peptide separation was performed using a pulled tip column (15 cm × 100-μm inner diameter) containing C18 Reprosil 5-μm particles (Nikkyo Technos, Tokyo, Japan). The flow was passively split from 300 μl/min to ∼250 nl/min before the analytical column. Gradient elution was performed from 0 to 50% ACN containing 0.1% formic acid over 100 min. The eluent was directly sprayed into an LTQ-Orbitrap XL mass spectrometer, and a data-dependent top 5 method was used for data acquisition. For each cycle, one full MS scan in the orbitrap at 60,000 resolution and an automatic gain control target of 100,000 was followed by five MS/MS acquisitions in the LTQ at an automatic gain control target of 5,000 on the five most intense ions. Selected ions were excluded from further selection for 60 s. Singly charged or unassigned ions were also rejected. Maximum ion accumulation times were 500 ms for full MS scans and MS/MS scans. For ion trap MS/MS the normalized collision energy was set to 30, activation Q was set to 0.25, and activation time was set to 30 ms. Raw data generated with the LTQ-Orbitrap XL were converted to mgf files using ProteomeDiscoverer 1.0 software (Thermo Fisher Scientific) excluding singly charged ions and non-deconvoluted MS peaks or peaks with a signal-to-noise ratio below 3. The first line “monoisotopic mass” of the mgf file was removed using Microsoft Wordpad to achieve compatibility for batch processing with Mascot Integra 1.4 (Matrix Science, London, UK).
Protein Identification
For protein identification, the integrated analytical work flow software package Mascot Integra 1.4 (Matrix Science) was used for batch searches with Mascot 2.2.2 on an in-house server and the batch processor Mascot Daemon 2.2.2 (Matrix Science). Data were searched against the Blumeria genome (“Genoscope” draft assembly) and EST databases (Blumeria Genome Sequencing Project). The genomic Blumeria contigs longer than 50,000 bp were fractionated to comply with the Mascot software. The Blumeria DNA database used for the protein identification search consisted of numbered subcontigs rather than annotated and defined ORFs; therefore the percentage of sequence coverage, Mr, and pI of contigs were ignored. Searches were reiterated on a curated database from which repeats were excluded. The EST Blumeria database contains a collection of 17,869 ESTs from different studies. Some barley sequences were filtered out manually from the Mascot data obtained when searching with this database. Infected TL extracts and epidermal strips (epidermis and haustoria (“EH”)) were also searched against the UniProtKB/Swiss-Prot rice database downloaded from the European Molecular Biology Laboratory-European Bioinformatics Institute Web site (version 23214, Oryza sativa Nipponbare, July 22, 2008) and the NCBInr database restricting the taxonomy to higher plants. The updated versions of the publically available databases (NCBInr) were downloaded onto the in-house Mascot server on a weekly basis. The rice genomic database was fused to the Blumeria genomic database for the estimation of the false discovery rate (FDR) of identified proteins in protein samples of infected EH tissues. The rice pseudomolecules database version 5.0 was downloaded from the Michigan State University Rice Genome Annotation Project Database and Resource.
Initially each *.mgf file was searched individually with Mascot. In a second identical search, each mgf file generated from protein extracts from the same tissue (CON, hyphae (“HY”), or EH) and analyzed on the same MS/MS instrument (HCT or LTQ-Orbitrap) were merged on the Integra server to perform the Mascot search resulting in a single Mascot result file. The Mascot search parameters for the HCT Esquire data were the following: 1.2-Da error tolerance in MS mode and 0.4-Da error tolerance in MS/MS mode, allowing up to two tryptic missed cleavages and considering 2+ and 3+ ions. Cysteine carbamidomethylation was set as a fixed modification; methionine and proline oxidation (hydroxyproline) were included as variable modifications. Mascot search parameters for the orbitrap data were identical to the parameters described for the HCT except that 10-ppm error in MS mode and 0.8-Da error in MS/MS mode were tolerated.
Peptide Alignment on Blumeria Genome, Protein Validation, and Annotation
Every peptide identified with a Mascot score above the confidence limit (p < 0.05) was extracted from independent comma-separated value-formatted Mascot result files and aligned on the Blumeria genomic sequence. Genes and their proteolytic peptides were defined through individual exons of an ORF. Protein hits were validated if they had at least two different unique peptides with a score greater than the identity score (p < 0.05). In the case of the haustorial proteome, proteins identified with a single significant peptide (singletons) were also accepted if the same protein was identified in another tissue through multiple peptide identifications and the MS/MS data for the common peptide were found to be similar by manual inspection. These MS/MS spectra were extracted and visualized in the Mascot Applet of the Integra software (Matrix Science). To be accepted, the identification required that most of the intensive peaks from the b- and y-ion series were present in both MS/MS spectra with a similar intensity pattern.
To annotate a group of peptides within a putative ORF within a contig, a homology search against the UniProt database using BlastP (19) was used to find homologous proteins against the concatenated amino acid sequences of the identified peptides of that group, keeping their relative positions. The BlastP search parameters were: -F, F (turn off low complexity filtering); -W, 2 (use a smaller word size); -M, PAM30 (scoring matrix recommended for short queries). The DNA sequence around a group of peptides identified for the haustorial proteins was downloaded from the genome assembly (Genoscope version) as a FASTA format file to predict ORFs. Putative ORFs were determined with the FGENESH prediction program using either the Leptosphaeria maculans or the Sclerotinia sclerotiorum ORF prediction models (SoftBerry, Inc.). Putative classical secretion proteins (with signal peptide) and non-classical secretion proteins were predicted with the SignalP (Hidden Marker Model minimum probability of 0.9) and the SecretomeP (Neural Network-Score cutoff, 0.5) algorithms, respectively. The homology BlastP search was then reiterated with the predicted ORFs using the default settings and the NCBInr or Swiss-Prot databases.
Bioinformatics and Genome Annotation: Mapping of PANTHER Terms to Gene Ontology (GO) Terms
Using the GO terms associated with the UniProt entries homologous to the identified proteins, we applied the “GO to PANTHER mappings” to map the protein functions to the PANTHER ontology. This provided a one-to-one mapping of GO terms to PANTHER terms and indicated whether the PANTHER was an exact mapping or a child term. For example, GO:0006096 (“glycolysis”) maps to PANTHER (glycolysis) in a one-to-one equivalence, GO:0006096 (glycolysis) maps to PANTHER (“monosaccharide metabolism”), and GO:0006096 is a child of PANTHER (monosaccharide metabolism). This mapping was used to produce the PANTHER figures.
RESULTS
Proteomics Analysis of the Obligate Pathogen B. graminis
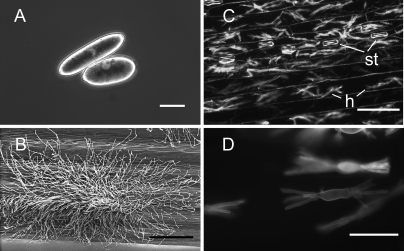
A shotgun proteomics approach was chosen to analyze the proteomes from three tissues of B. graminis f. sp. hordei. The stages analyzed in this study are shown in Fig. 1. The mature asexual reproductive conidia are oblong airborne dispersal structures (Fig. 1A, CON samples). The fungal colonies on the surface of infected leaves (epiphytic hyphae; Fig. 1B) were sampled at a stage when an extensive network of hyphae (runner hyphae) adhering to the leaf surface were visible (7–10 dpi); the conidiophores emerging from the colonies are specialized aerial hyphae that grow at right angles from the leaf surface, they proliferate very extensively, and they produce masses of conidia by a succession of basipetal septation (“sporulating hyphae” or HY samples). The intracellular feeding structures called haustoria (Fig. 1, C and D) are multidigitate cells with a central swollen body found inside the cell wall but outside the cytoplasm, surrounded by invaginated plant-derived plasma membrane of the host epidermal (not visible here). Note that the sample used to analyze the haustorial proteome includes the plant epidermal cells (both infected and non-infected) as well as the non-infected stomata (EH samples).
Fig. 1.
Images of the three different B. graminis f. sp. hordei structures analyzed in this study. A, micrograph of mature ungerminated conidia. The conidia were isolated from sporulating colonies, suspended in water, and photographed using differential interference contrast microscopy (bar, 10 μm). B, scanning electron micrograph of fungal mycelium (sporulating hyphae) growing epiphytically on the barley leaf surface. The “runner hyphae” adhering tightly to the leaf surface are visible at the edge of the colony; the aerial conidiophores emerge from the center of the colony (bar, 100 μm). C, low magnification epifluorescence microscopy image of haustorial cells within epidermal cells of heavily infected primary leaves of barley. Epiphytic structures pictured in B were removed, and the leaf epidermis was dissected. The fungal haustoria (h) and the plant stomata (st) are labeled. The fungal structures were stained by Alexa Fluor 488 bound to wheat germ agglutinin and visualized using a FITC emission/barrier filter (bar, 20 μm). D, close-up micrograph of haustorial structures inside the plant cells. The sample is the same as in C (bar, 10 μm).
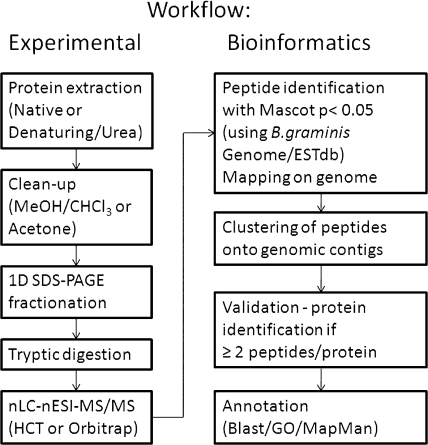
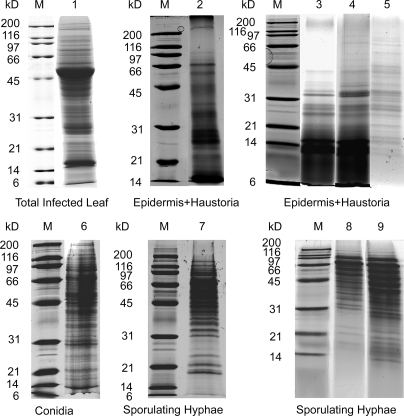
The proteomes of the TL extract and the three fungal tissues were processed as described under “Experimental Procedures” and summarized diagrammatically in Fig. 2. A shotgun gel-LC-MS/MS approach was used to analyze each proteome. Crude protein preparations were first separated by one-dimensional SDS-PAGE (Fig. 3), and the gel lanes were cut into 13–25 gel bands. Digested peptides were then separated by reverse phase nLC. In a preliminary analysis, the soluble proteome of total leaf extract (Fig. 3, lane 1) from barley leaves infected with B. graminis f. sp. hordei was investigated. This led to the identification of 63 Blumeria proteins with two or more significant peptides (supplemental Data 1). These data suggested that the Blumeria biomass in barley leaves at the late stage of infection was sufficient to detect fungal proteins in planta. However, leaf proteomes are characterized by a high dynamic range of protein concentrations that impair the proteome coverage. For instance, it has been estimated that ribulose-bisphosphate carboxylase/oxygenase (Rubisco) accounts for 40% of the total protein content of green leaves (20). In the total leaf extract, this is reflected by the identification of the large subunit of Rubisco as the top hit of the identified plant proteins, and Rubisco is clearly visible as a prominent band of around 50 kDa.
Fig. 2.
Diagram of the experimental and bioinformatics work flow summarizing the main processes used in the analysis of the samples as described in this study.
Fig. 3.
Images of barley/Blumeria protein extracts separated by SDS-PAGE and stained with colloidal Coomassie. Each gel lane was sliced into 2-mm bands for in-gel digestion with trypsin prior to analysis by mass spectrometry. Lane 1, soluble proteins (60 μg) extracted in native conditions from TL tissue of infected plants (10 dpi). Lanes 2–9, proteins extracted under denaturing conditions with a urea-containing buffer following clean-up and concentration by either TA or CM precipitation. Lanes 2–5, barley/Blumeria EH protein extracts prepared from infected barley leaves (10 dpi): lane 2, EH1 (50 μg, TA); lane 3, EH2 (150 μg, CM); lane 4, EH3 (150 μg, CM); lane 5, EH4 (75 μg, TA and CM). Lane 6, CON protein extract (30 μg). Lanes 7–9, sporulating HY protein extracts: lane 7, HY1 (45 μg, TA); lane 8, HY2 (70 μg, TA); lane 9, HY3 (90 μg, CM). All samples were separated on 12% acrylamide gels except for HY2 and HY3, which were separated on a 15% acrylamide gel. Lane M, molecular mass markers.
We then extracted proteins from three different dissected Blumeria tissues and separated them by SDS-PAGE (Fig. 3). These were epidermal strips from infected primary leaves containing Blumeria haustorial cells, conidia, and epiphytic sporulating hyphae.
Although conidial proteins extracted in a urea buffer were abundant enough to be separated directly by SDS-PAGE, the hyphal proteins extracted with urea needed to be concentrated. The epidermis-haustoria protein extracts were even more problematic. Migration on the gel for epidermis-haustoria samples was very poor if the extracts were not first precipitated with an organic solvent. Best results were obtained when they were cleaned with chloroform-methanol, precipitated in methanol, and washed in acetone prior to solubilization in urea denaturing buffer. The epidermis-haustorial extracts contained a higher proportion of low molecular weight peptides in comparison with the conidial and hyphal extracts (Fig. 3, lanes 2–5). This was unlikely to be due to the degradation of the protein sample as precautions were taken to minimize artifacts due to degradation during processing such as flash freezing the dissected epidermis in liquid N2 immediately after dissection and extracting the proteins by macerating the epidermal strips in precooled denaturing buffer. It is not surprising that the epidermis was poor in protein content as most of the cell volume was occupied by the large central vacuole. In addition the cell wall rich in polysaccharides and a cuticle rich in waxes and cutin rendered the protein extraction more challenging.
The peptides from the tryptic digests were first analyzed on a high capacity ion trap mass spectrometer (EH1, EH2, CON, HY1, and HY2; supplemental Data 1) because of its capacity to generate several MS/MS spectra per second, a feature that was ideal for a shotgun approach. We then repeated the analysis using a hybrid linear ion trap-orbitrap on hyphae and epidermis-haustorial samples (EH3, EH4, and HY3; supplemental Data 1); this doubled the number of proteins identified with two or more significant peptides.
To assess the quality of peptide assignment we measured the FDR by submitting the mass spectral data set to a decoy Blumeria database in Mascot. For the conidia and sporulating hyphae samples the FDR was typically below 5% irrespective of the mass spectrometer used (HCT or orbitrap). However, the FDR for the haustorial proteins in the infected epidermis was extremely high at above 30% (EH tissue; Table I). Thus, we analyzed the whole data set against the Blumeria genomic database after the removal of repetitive DNA, which constitutes ∼70% of the total sequences. This, however, only modestly lowered the FDR value. In this context, it is obviously important to consider that the epidermis and haustoria tissue samples contained a combination of two organisms and that the barley biomass was greater than the haustorial biomass. We therefore reasoned that many unassigned peptides were of plant origin. To test this hypothesis, the FDR of analysis using the genomic Blumeria database fused to a rice genomic database was determined. The FDR, calculated for the EH orbitrap data set, dropped significantly from 30.5 to 19.8% and further to 12.6% when the repetitive DNA sequences were removed. Therefore, it can be assumed that most detected peptides were indeed from barley; this was also reflected in a significant increase of the total number of identified peptides in the non-decoy search (Table I). Arguably the FDR could have been further decreased had a barley genomic database been available because only barley peptides that were 100% homologous to rice peptides were identified. Thus, the real number of barley peptides in the epidermis and haustoria samples was well underestimated.
Table I.
Number of identified peptides and proteins in B. graminis f. sp. hordei CON, sporulating HY, and EH
The number of mapped Blumeria peptides, identified proteins, and estimation of the peptide FDR are shown. Estimation of the peptide FDR are shown. ND, not determined; Rep, repetitive; N/A, not applicable.
| Origin of peptides | Identified peptidesa | Unique peptidesa,b | FDR including Rep DNA | FDR excluding Rep DNA | Unique proteinsc |
|---|---|---|---|---|---|
| % | % | ||||
| Total | 11,515 | 3,295 | N/A | N/A | 827 |
| Conidia | 2,890 | 1,464 | 3.4d | 3.2e | 441 |
| Hyphae | 7,547 | 2,341 | 4.0e | 3.9f | 775 |
| Haustoria | 1,078 | 335 | 30.5e | 26.1f | 47 |
| Haustoria-epidermis | N/A | N/A | 19.8e,f | 12.6e,f | ND |
a Peptide numbers correspond to all the significant peptides (0.05% confidence level) identified from HCT and LTQ-Orbitrap and mapped on the Blumeria genome.
b Peptide uniqueness is judged by amino acid sequence (e.g. LVNHFVNEFKR and LVNHFVNEFK count as two unique peptides, although one is a subsequence of the other). Two peptides of the same sequence but differing only in amino acid modification (e.g. ±Met oxidation) did count as a single unique peptide.
c Unique proteins were defined as proteins identified with at least two significant peptides for hyphae and conidia. For haustoria some singleton identifications were accepted provided that the peptide identification was significant, the peptide was also identified in the conidia or hyphae peptide pool, and its MS/MS data matched the MS/MS data of the corresponding peptide in the other tissue(s).
d FDR was estimated for the conidial data set analysed on the HCT using the Blumeria database.
e FDR for LTQ-Orbitrap data only. Similar rates were obtained for HCT data (not shown) using the Blumeria database.
f FDR was calculated as for footnote e but using a hybrid database composed of the Blumeria contigs and the annotated ORFs of a rice genomic database.
Proteogenomics: Genome Annotation through Peptide Mapping
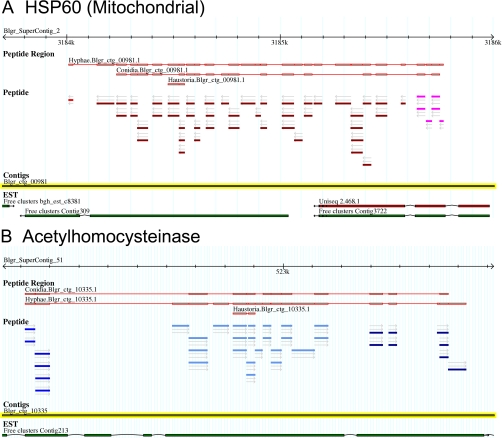
The identified peptides that aligned to the Blumeria genome sequence were added onto GBrowse to visualize their positions relative to each other and to other genome data available. In Fig. 4 we show two representative screen shots of GBrowse for the locus of mitochondrial HSP60 (A) and the cytoplasmic adenosylhomocysteinase (B). The individual peptides are visible in the “Peptide” section. Each peptide pictogram is aligned on the corresponding contig, and the direction of the reading frames is indicated by an arrow; different reading frames are depicted by different colors. The peptides that map to the same locus are clustered into peptide regions. The proteome of origin (conidia, sporulating hyphae, or epidermis/haustoria) and the best “protein” score in Mascot are shown in the “Peptide Region” section. The peptide sequences within this Peptide Region were used for sequence homology blast searches (see “Experimental Procedures” for details). Below these are the genomic DNA contigs and the EST/ cDNAs. The alignments indicate the positions of the introns and exons. The genome sequence information allowed peptide alignment on the genome draft. The aligned peptide groups also helped to identify ORFs. Some of this information highlighted assembly gaps or mistakes such as in the case of the Blumeria catalase with two separated contigs, Blgr_ctg_03976 and Blgr_ctg_03977. In other cases, such as the enolase on contig Blgr_ctg_01683, ORFs were truncated in their N-terminal or C-terminal end at the extremity of the contig as the contig end reflected the presence of a gap in the assembly. Further genomic sequencing with greater sequence coverage will be required to fill these gaps. In a few cases contigs were misorientated, and the peptide alignment highlighted such problems. For instance, an ATP-citrate synthase homolog showed misassembly and misorientation of the subcontigs within the contig Blgr_ctg_05534 (supplemental Data 2).
Fig. 4.
Screen shots of two loci on the B. graminis genome browser GBrowse/Genoscope assembly (Blumeria Genome Sequencing Project). The information visible here is the “supercontig” identifier with the sequence scale (2.4 kb), the assembled predicted protein based on the identified peptides, and the origin of the peptides. The individual color of the peptides identifies their orientation and their reading frames. Note that although the arrows indicate a coherent orientation in these two examples the color of the bars shows frame shifts indicative of separate exons separated by introns. The actual DNA sequence “contigs” and the ESTs available for these loci are shown below. A, the locus of a predicted mitochondrial HSP60. B, the locus of the predicted acetylhomocysteinase enzyme.
Some of the genes in the Blumeria genome are duplicates. hsp30, for instance, is present in 22 copies in the current assembly, 11 of which have clear start and stop codons and are thus likely to represent “full-length” genes. The peptides we identified in this study included representative variants of these genes (Table II). Although peptides 1 and 3 were found in only one form, peptide 2 had two variants with a phenylalanine or an isoleucine in position 5. Likewise peptide 4 had two variants: 4a has an aspartic acid and a serine in positions 10 and 11, whereas 4b has a glutamic acid and an asparagine, respectively. Some polymorphisms, however, were not detected. For example, there is a second version of peptide 3 locus with a glycine and phenylalanine at positions 6 and 7, respectively, and a third version of peptide 4 that is nearly identical to 4a but has an alanine at position 3. In Table III, the 11 hsp30 loci are listed together with the corresponding peptide complements. Note some of the sequence heterogeneity present at the different loci is not covered by the identified peptides. For example, HSP30-1, -2, and -8 shared the same peptides 1 and 2a, but although the DNA sequence encoding peptide 3 is the same for HSP30-1 and -2, the same region in hsp30-8 was encoded by a very different sequence for which no mass spectral data were obtained.
Table II.
HSP30 isoforms and hsp30 loci identified in the Blumeria genome: sequences of the identified peptides corresponding to HSP30 protein isoforms
| Peptide ID | Peptide sequence |
|---|---|
| 1 | AFFHQDQSSSPTIFR |
| 2a | LLDEFDQYSR |
| 2b | LLDEIDQYSR |
| 3 | GTPPAALVEGEADK |
| 4a | SFEFPVPNQDSVK |
| 4b | SFEFPVPNQENVK |
Table III.
HSP30 isoforms and hsp30 loci identified in the Blumeria genome: list of the full-length loci of hsp30 in the Blumeria genome (Blumeria Genome Sequencing Project, Arachne assembly) showing the associated peptide complement identified by mass spectrometry
| hsp30 | Genomic locus | Peptide complement |
|---|---|---|
| 1 | Blgr_ctg_12505 | 1, 2a, 3 |
| 2 | Blgr_ctg_00062 | 1, 2a, 3 |
| 3 | Blgr_ctg_00059 | 1, 2a, 3, 4b |
| 4 | Blgr_ctg_00064 | 1, 2a, 3, 4b |
| 5 | Blgr_ctg_09125 | 1, 2b, 4b |
| 6 | Blgr_ctg_00042 | 1, 2b, 4b |
| 7 | Blgr_ctg_00029 | 1, 2b, 4a |
| 8 | Blgr_ctg_07423 | 1, 2a |
| 9 | Blgr_ctg_00043 | 1, 4b |
| 10 | Blgr_ctg_00047 | 1, 4b |
| 11 | Blgr_ctg_00049 | 1, 4b |
Of the 827 proteins deduced from peptide groups and identified with at least two significant and unique peptides, 803 proteins aligned on the genome; of these, 696 proteins had cognate ESTs. Some proteins (24 proteins) were only represented by ESTs, whereas 107 only had genomic sequences (supplemental Data 3). This reflects the fact that the genome assembly contains gaps in which genes are split across contigs.
Comparative Functional Study of Proteins Identified in Conidia, Hyphae, and Haustoria
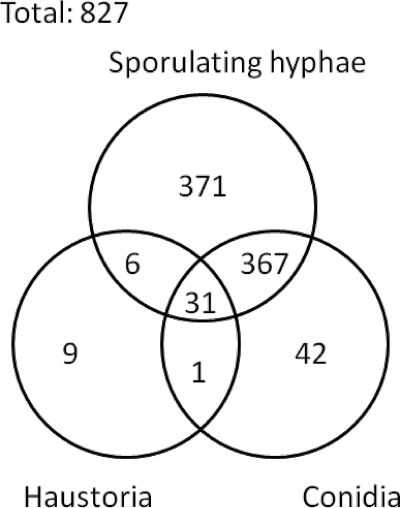
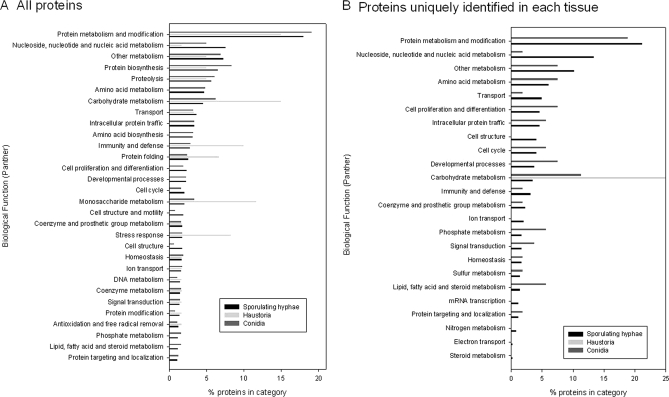
The distribution of the proteins identified in the three different tissues is shown in Fig. 5. In total, 775 were found in sporulating hyphae, 441 were found in conidia, and 47 were found in haustoria. About half (422) of the 827 proteins were found only in one type of tissue. At the time of the study, the genome of Blumeria was represented by a draft assembly (4.2 times coverage, Blumeria Genome Sequencing Project, Genoscope assembly). The number of publicly available Blumeria protein sequences through the NCBI Entrez Web site was limited to 51 entries. Therefore, we first annotated the peptide groups by homology using the BlastP algorithm on the Swiss-Prot database. The GO classification system was used to assign a putative biological function to the Blumeria proteins by selecting the annotation of the protein homolog with the highest similarity score (supplemental Data 3). These “biological function” classifications were then grouped into the PANTHER generic categories, and the distribution of the 30 most represented categories in the three tissues are shown in Fig. 6. Fig. 6A shows the categories of all proteins identified irrespective of whether they are unique to an individual tissue or not. Some categories, such as “protein biosynthesis,” “proteolysis,” and “other metabolism” have a similar proportion of members in all three stages/tissue types. On the whole, the classification distribution in conidia and sporulating hyphae was similar. The haustorial profile, on the other hand, showed some striking differences. The “stress response,” “carbohydrate”, “monosaccharide,” “vitamin metabolism,” and “other intracellular signaling cascade” categories were notably over-represented in the haustoria tissue. A similar analysis was conducted on proteins identified uniquely in one of the life stages (Fig. 6B). This approach highlighted differences in the distribution of proteins into PANTHER categories for proteins identified exclusively in sporulating hyphae or in conidia. In particular, in sporulating hyphae the “nucleoside, nucleotide, and nucleic acid metabolism” category contained 7 times more proteins uniquely identified compared with mature conidia. There are 4% of proteins uniquely identified in sporulating hyphae that are assigned to the “cell structure” PANTHER category, whereas no proteins seen only in conidia could be assigned to this category. In the whole pool of proteins (Fig. 6A), such a difference in this category was less obvious. The proportions of proteins uniquely identified in conidia and that were assigned to “carbohydrate metabolism,” “phosphate metabolism,” and “lipid, fatty acid, and steroid metabolism” PANTHER categories were about 3-fold higher in conidia compared with the proportions of proteins uniquely identified in sporulating hyphae. The only annotated group of proteins we exclusively found in haustoria contained two carbohydrate metabolism enzymes as the other proteins were unknown.
Fig. 5.
Venn diagram representing the distribution of protein identifications in conidia, sporulating hyphae, and haustoria in infected barley epidermis.
Fig. 6.
Histogram illustrating the relative distribution of the identified proteins into categories determined by Gene Ontology and mapped onto the biological function PANTHER categories. The relative distribution values into the top 30 PANTHER categories for each tissue type (black, sporulating hyphae; light gray, haustoria; gray, conidia) are given as the fraction (in percent) of the total identified protein in each tissue. A, all annotated proteins were used in this analysis irrespective of whether they were uniquely identified in one tissue or commonly identified in two or three of the analyzed tissues. B, the relative distribution into the 30 top PANTHER categories of the annotated proteins only identified in one of the three tissues. Note that some categories represent subfamilies of others (e.g. carbohydrate metabolism includes monosaccharide metabolism).
MapMan, a tool used for Arabidopsis microarray analysis (21), was adapted for Blumeria proteomics and allowed us to survey how much of the primary metabolism was represented by the proteins identified in this study. This survey showed that most of the main pathways were active (supplemental Data 4A). Of particular note we observed that although glycolysis was well represented anaerobic fermentation was completely absent in the Blumeria proteome (supplemental Data 4B).
The Haustorial Proteome
Proteins identified in the epidermis and haustoria samples were further investigated. The haustorium is the feeding organ specific to biotroph pathogens and at the interface between the pathogen and the host. It is a key structure for the establishment of the virulence of the fungus. As mentioned previously, 47 unique Blumeria proteins were identified. The putative open reading frames of the 47 haustorial proteins were determined with the FGENESH algorithm (SoftBerry, Inc.). As there was no gene model for the Blumeria ORFs, alternative models of closely related fungi, e.g. L. maculans or S. sclerotiorum, were used. The predicted ORFs were validated by confirming the presence of the identified peptides on each locus. From this analysis it became apparent that the Sclerotinia gene model was more suitable for Blumeria ORF prediction (Tables IV and V and supplemental Data 5).
Table IV.
ORF description of proteins identified in the Blumeria haustorial proteome: proteins identified in the haustoria and other Blumeria tissues
aa, amino acids; pep, peptide; non-class., non-classical; Miss C-term, missing C terminus; Miss N-term, missing N terminus; NA, not applicable; DH, dehydrogenase.
| Contig | ORF homology (BlastP) description | UniProt accession no. | Secretion signal pep/non-class. | ORF length |
|---|---|---|---|---|
| aa | ||||
| Blgr_ctg_00049 | CFEM-containing domain (in membrane fungal proteins) | Q4WLB9 | Yes (21 aa) | 226 |
| Blgr_ctg_00113 | Homolog to M. grisea elicitor with STM1p domain | ABK56833 | No | 365 |
| Blgr_ctg_00198 | HSP90 | O43109.1 | No | 701 |
| Blgr_ctg_00981 | HSP60, mitochondrial, GroEL, chaperone 60 kDa | Q5B041.1 | No | 584 |
| Blgr_ctg_00994 | Mitochondrial ATPase β chain | P23704.1 | Non-class. | Miss C-term |
| Blgr_ctg_01001 | Glyceraldehyde-3-phosphate dehydrogenase | Q00640.1 | Non-class. | 372 |
| Blgr_ctg_01060 | Vacuolar aspartyl protease | Q01294.2 | Yes (17 aa) | 404 |
| Blgr_ctg_01176 | Phosphoglycerate kinase | P41756.1 | No | 423 |
| Blgr_ctg_01522 | Gar2 31-kDa RNA-binding protein | P41891.2 | Non-class. | 307 |
| Blgr_ctg_01605 | HSP70 SSB1 | P87222.1 | No | 598 |
| Blgr_ctg_01682 | Enolase, 2-phosphoglycerate dehydratase | Q7RV85.1 | No | Miss C-term |
| Blgr_ctg_01693 | 14-3-3 protein homolog | Q99002.1 | No | 268 |
| Blgr_ctg_01803 | Ubiquitin (3 copies) | P59263.1 | No | 229 |
| Blgr_ctg_01893 | N-terminal ubiquitin/C-terminal ribosomal protein S27a | P15357.1 | No | 276 |
| Blgr_ctg_02341 | HSP70 | Q5B0C0.1 | No | 511 |
| Blgr_ctg_02392 | Mannitol DH; xylulose/sorbitol reductase | P0C0Y5.1 | Non-class. | 311 |
| Blgr_ctg_02854 | GTP-binding protein like (OBG homolog) | P38219.1 | No | 557 |
| Blgr_ctg_03010 | S-Adenosylmethionine synthase | P48466.1 | No | 395 |
| Blgr_ctg_03652 | Translation elongation factor 3 (EF-3) | Q75EV6.1 | No | 1,054 |
| Blgr_ctg_03869 | Elongation factor 1α (EF-1a) | Q00251.1 | No | 529 |
| Blgr_ctg_03976 | Catalase (Blumeria Q8X1P0); no ESTs | Q8X1P0 | Yes (17 aa) | 718 |
| Blgr_ctg_03977 | Catalase (Blumeria Q8X1P0); no ESTs | Q8X1P0 | Yes (17 aa) | 718 |
| Blgr_ctg_04433 | 60 S acidic ribosomal protein P0 | Q96TJ5.1 | No | 314 |
| Blgr_ctg_04468 | Protein-disulfide isomerase | P55059.1 | Yes (20 aa) | Miss C-term |
| Blgr_ctg_05446 | ADP/ATP translocase or carrier protein, mitochondrial | P02723.1 | NA | N-term wrong |
| Blgr_ctg_05813 | Pyruvate carboxylase, acetyl-CoA carboxylase | O93918.1 | No | 1162 |
| Blgr_ctg_06141 | Peptidyl-prolyl cis-trans isomerase; cyclophilin | P10255.1 | Non-class. | 211 |
| Blgr_ctg_07097 | Pyruvate dehydrogenase E1 component β subunit | P32473.2 | Non-class. | 377 |
| Blgr_ctg_07154 | Actin | O13419.1 | Non-class. | 388 |
| Blgr_ctg_07248 | HSP70 | O13419.1 | No | 637 |
| Blgr_ctg_07813 | Transaldolase (Triose phosphate isomerase phosphate-binding protein) | O42700.2 | No | 320 |
| Blgr_ctg_08720 | Phosphoglycerate mutase | Q898R1.1 | NA | Miss N-term |
| Blgr_ctg_09741 | Plasma membrane ATPase | P07038.1 | Non-class. | 961 |
| Blgr_ctg_10335 | Adenosylhomocysteinase | P39954.1 | Non-class. | 488 |
| Blgr_ctg_10383 | 78-kDa glucose-regulated protein homolog, BiP | P78695.3 | Non-class. | 685 |
| Blgr_ctg_11103 | Bleomycin hydrolase (peptidase C1B, Cys proteinase) | Q13867.1 | No | 502 |
| Blgr_ctg_11163 | Calnexin (calreticulin family) | Q6Q487.2 | Yes (21 aa) | 576 |
| Contig4118 | Ras GTPase Rab11 | P46638.3 | No | 212 |
| 5 out of 34 ORFs with secretion signal peptide (SignalP probability p ≥ 0.9) | Average | 496 | ||
| 10 out of 34 ORFs “non-classical” secretion (SecretomeP NN-score ≥ 0.5) | S.D. | 238.5 | ||
Table V.
ORF description of proteins identified in the Blumeria haustorial proteome: proteins identified in the haustoria only
aa, amino acids; sig, significant; non-class., non-classical; pep, peptide; GPI, glycosylphosphatidylinositol.
| Contig | ORF homology (BlastP) description | UniProt accession no. | Secretion signal pep/non-class. | ORF length |
|---|---|---|---|---|
| aa | ||||
| Blgr_ctg_01259 | No sig homology | Yes (26 aa) | 118 | |
| Blgr_ctg_06599 | No sig homology | Yes (23 aa) | 118 | |
| Blgr_ctg_08144 | No sig homology | Yes (19 aa) | 139 | |
| Blgr_ctg_11206 | No sig homology | Yes (21 aa) | 159 | |
| Blgr_ctg_11305 | Putative β-1,3-glucosidase | P15703.1 | Yes (18 aa) | 281 |
| Blgr_ctg_11334 | GPI-anchored cell wall β-1,3-glucosidase | P15703.1 | Yes | 293 |
| Blgr_ctg_13376 | No sig homology | Yes (24 aa) | 85 | |
| Blgr_ctg_18350 | No sig homology | Yes (24 aa) | 129 | |
| Blgr_ctg_20478 | No sig homology | Yes (19 aa) | 295 | |
| 9 out of 9 ORFs with secretion signal peptide (SignalP p ≥ 0.9) | Average | 180 | ||
| S.D. | 85 | |||
Of the 47 haustorial proteins, 33 proteins were identified with two or more unique and significant peptides (p < 0.05 as determined in Mascot). The remaining 14 proteins were identified with a single significant peptide in haustoria (to be referred to as singletons), but the MS/MS mass spectra of these singletons were validated by similarity comparison with the corresponding peptide of the proteins identified by two or more peptides in conidia or sporulating hyphae extracts (supplemental Data 6). Among the 33 Blumeria proteins detected in haustoria with two or more significant peptides, nine of these were exclusively detected in haustoria and not in conidia or sporulating hyphae samples (Fig. 5).
All of these nine proteins are predicted by the SignalP program to be secreted (Tables IV and V). Another interesting feature of the proteins only detected in haustoria is that the average amino acid length of these nine proteins is 180 (S.D. = 85), which is significantly smaller than the 489 (S.D. = 239) of the remaining haustorial proteins. Many of the 47 proteins detected in the haustoria tissue are common and abundant proteins, such as the various heat-shock proteins, translation elongation factors, and enzymes such as glyceraldehyde-3-phosphate dehydrogenase. It is interesting to mention the presence of two β-1,3-glucosidase homologs only detected in the haustoria. None of the seven other proteins exclusively detected in the haustoria tissue have any homolog to other proteins. The Blumeria catalase Q8X1PO peptides were highly represented in the haustoria samples even if they were not detected as unique for this cell type. In fact, the HCT data showed that 12 (orbitrap:7) were associated to this catalase in the infected epidermis, whereas 0 (orbitrap:3) unique peptides were detected in the hyphase. In addition it must be remembered that the epidermis extracts were a mixture of plant and fungal proteins, whereas the hyphae protein extracts contained only fungal proteins. Taking all this into account it seems that this catalase was present in a higher amount in the haustoria compared with the other cell types.
Proteins Identified in Conidia or Hyphae
In addition to HSP30, heat-shock proteins from other families such HSP70 (six members) and HSP90 (four members) were expressed in all Blumeria tissues. Hydrogen peroxide-detoxifying enzymes such as a catalase (hyphae and haustoria tissues) and a catalase/peroxidase (hyphae tissue) were identified. A single plasma membrane H+-ATPase was expressed in conidia and in haustoria. A protein sharing similarities with the virulence factor SnodProt1 (Blgr_ctg_04801; Stagonospora nodorum protein 1) was expressed in conidia and hyphae. The Blumeria homolog to a protein involved in gene silencing, quelling-deficient 2 (QDE2) protein in Neurospora crassa, was expressed in conidia and hyphae.
Forty-one proteins were detected only in the conidia sample. Among those are a small GTPase sharing similarities with the Colletotrichum trifolii Rac1 (a Ras protein homolog) important for the hyphae morphology and C. trifolii virulence, including the production of reactive oxygen species through a cytosolic phospholipase A2 signaling pathway (22). Another interesting protein only identified in the conidia sample is the neuronal calcium sensor 1 (NCS1) homolog that in yeast regulates sporulation and is required for calcium tolerance (23).
DISCUSSION
The proteogenomics approach described here allowed the identification of 827 proteins of B. graminis f. sp. hordei in conidia, hyphae, and haustoria. Given that Blumeria, like all powdery mildews, is an obligate biotroph, the samples were obtained directly from infected plants. The airborne conidia were collected in pure form by aspirating the sporulating colonies from the surface of infected leaves. A similar sample was analyzed in a recently published study of powdery mildew conidia proteome (15). However, a different work flow was used. There proteins were separated on two-dimensional gels, and single proteins were identified from tryptic digests of gel spots analyzed on MALDI-MS/MS instrumentation. This led to the identification of 186 proteins, some of which were isoforms of the same EST/locus resulting in 129 unique protein identifications (15). In the shotgun approach using one-dimensional SDS-PAGE-nLC-nESI-MS/MS presented here, we identified 441 proteins with at least two unique and significant peptides. This significantly increases and complements the pool of identified proteins in conidia so far. While two-dimensional gel electrophoresis enables the separation of protein isoforms, the mass spectral shotgun information is typically insufficient to distinguish between isoforms. However, the one-dimensional gel-LC approach is more suitable for a larger scale proteomics study and the analysis of membrane proteins.
More than 90% of the proteins identified from the conidia were also identified in the samples from epiphytic hyphae. The epiphytic hyphae were collected from sporulating colonies. This tissue therefore contained surface, “runner” hyphae; secondary appressoria; the conidiophores that emerge perpendicularly from the substrate; and conidia at various stages of maturity up to and including release (sporulation). From such sporulating hyphae, 775 proteins were identified.
Haustoria only develop in the epidermal cells. In this study we opted to manually remove the epiphytic structures (described above), and we then dissected the epidermis from the rest of the leaf. This sample therefore contained proteins from plant cells (infected and non-infected epidermis plus stomata) and proteins from Blumeria haustoria, including proteins secreted from the haustoria into the plant cells. A total of 47 Blumeria proteins were detected in or secreted by the haustoria. We combined bioinformatics and the newly available Blumeria genome to identify bona fide haustorial proteins. In another study, ∼200 haustorial proteins were identified from isolated haustoria purified by homogenization of the infected epidermis followed by a series of density gradient fractionations (16). In both data sets, a substantial number of heat-shock proteins were identified, suggesting a stressed status of the haustoria. The total number of proteins we identified is nominally lower than the number reported by Godfrey et al. (16). The discrepancy can be explained by the different criteria used for protein identification. Godfrey et al. (16) also accepted haustorial proteins identified with a single significant peptide. If we had used similar criteria we would have identified over 120 haustorial proteins. Considering the high FDR in the analysis of haustorial samples, we preferred using more stringent criteria of identification. Moreover our approach yielded additional information because it included proteins that were secreted by Blumeria haustoria in the perihaustorial matrix and potentially taken up by host cells that are not present in the published data set (16). In both cases, the limited number of identified proteins might be explained by the low amount of biomaterial available when proteins were isolated from haustoria, and in our case, the presence of more epidermal tissue in comparison with haustoria might be the main limiting factor.
The availability of large scale genome resources including genomic DNA and EST/cDNA sequences of Blumeria was a significant advantage for peptide identification in this study. We used this resource to aid the first systematic comparison of the proteome diversity during growth in the plant host. As expected and summarized in the Venn diagram of Fig. 5, protein expression is somewhat tissue-specific with half of the proteins represented in only one tissue. What is remarkable is the relatively large proportion of protein hits in the haustoria that are not common to the other proteomes.
The putative role of the proteins was determined by sequence similarity to other proteins that have functional annotations in the relevant databases. The predicted functions were first classified using Gene Ontology, and the categories of biological function were grouped according to the PANTHER classification system (24). Assuming that the percentages of peptide hits in each category are indicative of the activities prevalent in the source tissue at any given time, the comparison of these values gives an insight into the functional status of the tissue. We restricted this analysis to the 30 most frequent categories. The fact that many of these groups share a similar percentage of peptides shows that similar processes are common to all three tissues, such as protein metabolism (biosynthesis, degradation, and traffic). Primary metabolism is also equally well represented as most of the main pathways are active; the notable exception is anaerobic fermentation, a fact also noted in isolated haustoria and commented on by Godfrey et al. (16). The absence of coding capacity and expression of anaerobic fermentation should not be regarded as too surprising given that B. graminis like all powdery mildews live in an exclusively aerobic environment, the host leaf surface.
When all the identified proteins were considered (Fig. 6A), the global functional distribution of proteins in conidia and the sporulating hyphae was very similar, whereas the haustorial proteome showed intriguing differences. The over-representation of carbohydrate metabolism in general and monosaccharide metabolism in particular was not surprising given our understanding that haustoria are the primary feeding structures (25) and that monosaccharides are thought to be the main form of fixed carbon taken up by the fungus (26). This also correlates well with our previous findings based on analysis of transcriptome dynamics where we have shown that genes encoding glycolytic enzymes are coordinately up-regulated in haustoria (8). Although the number of annotated Blumeria proteins from the infected epidermis (i.e. from the haustoria) is low, carbohydrate metabolism proteins are not over-represented in the much more abundant data sets from conidia and sporulating hyphae.
The high relative incidence of proteins involved in the stress response is important. In addition to these, a subset of the proteins involved in “protein folding” are associated with the stress response (e.g. chaperones). Hitherto it was thought that host defense was manifested at penetration, whereas these data indicate that even in compatible interactions, in which haustoria developed to penetrate the host cell, the environment was also stressful possibly as the result of defense responses mounted by the host. Moreover catalase appeared to be particularly highly expressed in the haustoria and probably more so than in the other two tissues. This hints that the fungus needs to detoxify the reactive oxygen species, in particular hydrogen peroxide at the interface with the host plant. In this context it is worth noting that several peroxidases and germin-like proteins, enzymes with a role in the production of hydrogen peroxide in plants, were expressed in the epidermal cells of the infected barley leaves (data not shown).
A comparison of the classification of proteins identified uniquely in the tissues (Fig. 6B) is really only valid for the conidia and sporulating hyphae where the number of identified proteins is substantially higher and allowed us to highlight differences between both tissues. Sporulating hyphae had a higher representation of proteins involved in “nucleic acid metabolism,” reflecting the active proliferation of the fungus and nuclear division required for the formation of the conidia. Conversely the conidia contained a much higher number of proteins involved in lipid, carbohydrate, and phosphate metabolism, i.e. enzymes that are presumably required for the breakdown and processing of storage compounds such as lipids and glycogen following germination. Again these results corroborate our earlier findings based on the analysis of the transcriptome dynamics during development (8) confirming that conidia are primed for a rapid and effective breakdown of nutrient reserves following germination without the need for new transcription to be activated.
A very significant contribution of our study is to provide proteomic evidence for the expression and translation of genes identified either through gene-finding algorithms such as FGENESH or by the existence of transcribed sequences determined as ESTs/cDNAs. This is crucial because the former are known to be error-prone (including failure to determine short ORFs) and at best make predictions with a limited degree of certainty. In the latter case recent advances in high throughput cDNA sequencing programs have revealed that much more of the genome is actually transcribed than translated into protein (27). Therefore demonstrating the existence of predicted proteins is not trivial.
In addition to the proteomics confirmation of gene expression, this study provided data for the determination of the intron/exon gene structure. For example, in Fig. 4A, there are peptides corresponding to DNA sequences that are not represented by cDNA sequences, indicating that this indeed represents a whole exon. Moreover in the case of the existence of gene families of closely related sequences, polymorphism in the sequence enabled us to show that more than one of the gene copies was translated and transcribed. Such fine discrimination between isoform expression is not possible in global transcriptomics analysis using microarrays. In the example of HSP30, there were 22 copies corresponding to this gene in the Blumeria genome of which only 11 appeared to have a complete protein-encoding sequence. The EST/cDNA data suggests that of these at least five are transcribed (not shown), and the proteomics data demonstrate unequivocally that at least three of these must have been translated. The existence of a high level of full expression (i.e. transcription and translation) of multiple copies of genes involved in the stress response was further indication of an organism that needs to adapt and cope with high degree of stress possibly induced by the host plant.
The survey of protein function through homology to known proteins identified a number of proteins that play a role as pathogenicity and virulence factors in other plant pathogenic fungi. We have here identified in conidia a Blumeria homolog of SnodProt1 that is important at the initiation of the infection process in Magnaporthe oryzae (synonym Magnaporthe grisea) (28). A SnodProt1 homolog is secreted in planta by Fusarium graminearum during the infection of wheat ears (14). The Blumeria CAP20-like perilipin protein was expressed in conidia and hyphae and has been reported previously to be required for the appressorium formation and the virulence of Colletotrichum gloeosporioides (29). Similarly the neuronal calcium sensor 1 (NCS1) homolog was detected in conidia only and was shown to be involved in calcium tolerance and the regulation of sporulation (23). We found QDE2 peptides in hyphae and in conidia, whereas others have seen this protein in isolated haustoria (16). Therefore one can assume that this protein is ubiquitous in Blumeria. QDE2 is involved in silencing through specific mRNA degradation (30). It is important to highlight its presence here because one of the reasons that may have led to the massive expansion of the powdery mildew genome is the proliferation of repetitive genomic “parasites” (transposons and retrotransposons). The identification of an active RNA silencing pathway suggests that this in unlikely to be due to the absence of the QDE-dependent pathway.
The β-1,3-glucosidases identified here are predicted to be secreted. The two isoforms detected only in haustoria might be involved in the degradation of the host cell wall and/or remodeling the interfacial matrix of the haustorium.
Although this type of study does not yield truly quantitative data, it is reasonable to assume that the number of peptides identified for a protein partly reflects protein abundance. Thus, the nine Blumeria proteins only detected in epidermis and haustoria samples, but not in conidia or sporulating hyphae, can be assumed to be present at a relatively much higher level in haustoria compared with the other tissues that only contained fungal proteins. A closer analysis of these proteins indicates that all have a putative secretion signal and are smaller than average. Of these, six of the nine are of unknown function. All these criteria are typical of effector proteins that are injected into the plant cells (6). We therefore propose that these proteins are prime candidates for novel effector proteins produced by Blumeria and secreted into the host tissues. It remains to be seen whether functional analyses of these proteins confirm this hypothesis.
In conclusion, the work published here represents a significant advance in our understanding of the biology of powdery mildews by probing the protein profiles present during development in the host. We used these data to complement and validate in silico prediction of gene models and the data sets obtained from EST analysis. We have revealed that the fungus is subject to significant plant-generated stress even in fully functional haustoria and identified a number of candidate effector proteins that are likely to take part in the control of host metabolism and immune response. We have thus demonstrated the significant value of in vivo proteomics studies of pathogens.
Supplementary Material
Footnotes
* This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/E000983/1.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- dpi
- days postinoculation
- CM
- chloroform-methanol
- CON
- conidia
- EH
- epidermis and haustoria
- FDR
- false discovery rate
- HY
- hyphae
- nLC
- nano-LC
- nESI
- nano-ESI
- TA
- TCA-acetone
- TL
- total leaf
- mgf
- Mascot generic format
- LTQ
- linear trap quadrupole
- EST
- expressed sequence tags
- NCBInr
- National Center for Biotechnology Information non-redundant
- HCT
- high capacity trap
- GO
- Gene Ontology
- PANTHER
- Protein Analysis through Evolutionary Relationships
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- QDE
- quelling-deficient.
REFERENCES
- 1.Marris E., Dodds P., Glover J., Hibberd J., Zhang J. H., Sayre R. (2008) Agronomy: five crop researchers who could change the world. Nature 456, 563–568 [DOI] [PubMed] [Google Scholar]
- 2.Glawe D. A. (2008) The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol 46, 27–51 [DOI] [PubMed] [Google Scholar]
- 3.Both M., Spanu P. (2004) Blumeria graminis f. sp. hordei, an obligate pathogen of barley, in Plant Pathogen Interactions (Talbot N. ed) pp. 202–218, Blackwell Publishing, Oxford [Google Scholar]
- 4.Bushnell W. R. (1972) Physiology of fungal haustoria. Annu. Rev. Phytopathol 10, 151–176 [Google Scholar]
- 5.Catanzariti A. M., Dodds P. N., Ellis J. G. (2007) Avirulence proteins from haustoria-forming pathogens. FEMS Microbiol. Lett 269, 181–188 [DOI] [PubMed] [Google Scholar]
- 6.Hogenhout S. A., Van der Hoorn R. A., Terauchi R., Kamoun S. (2009) Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact 22, 115–122 [DOI] [PubMed] [Google Scholar]
- 7.Both M., Eckert S. E., Csukai M., Müller E., Dimopoulos G., Spanu P. D. (2005) Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol. Plant Microbe. Interact 18, 125–133 [DOI] [PubMed] [Google Scholar]
- 8.Both M., Csukai M., Stumpf M. P., Spanu P. D. (2005) Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17, 2107–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 10.Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansong C., Purvine S. O., Adkins J. N., Lipton M. S., Smith R. D. (2008) Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief. Funct. Genomic. Proteomic 7, 50–62 [DOI] [PubMed] [Google Scholar]
- 12.Rampitsch C., Bykova N. V., McCallum B., Beimcik E., Ens W. (2006) Analysis of the wheat and Puccinia triticina (leaf rust) proteomes during a susceptible host-pathogen interaction. Proteomics 6, 1897–1907 [DOI] [PubMed] [Google Scholar]
- 13.Zhou W., Eudes F., Laroche A. (2006) Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 6, 4599–4609 [DOI] [PubMed] [Google Scholar]
- 14.Paper J. M., Scott-Craig J. S., Adhikari N. D., Cuomo C. A., Walton J. D. (2007) Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 7, 3171–3183 [DOI] [PubMed] [Google Scholar]
- 15.Noir S., Colby T., Harzen A., Schmidt J., Panstruga R. (2009) A proteomic analysis of powdery mildew (Blumeria graminis f.sp. hordei) conidiospores. Mol. Plant Pathol 10, 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godfrey D., Zhang Z., Saalbach G., Thordal-Christensen H. (2009) A proteomics study of barley powdery mildew haustoria. Proteomics 9, 3222–3232 [DOI] [PubMed] [Google Scholar]
- 17.Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 18.Bindschedler L. V., Palmblad M., Cramer R. (2008) Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry 69, 1962–1972 [DOI] [PubMed] [Google Scholar]
- 19.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 20.McCabe M. S., Garratt L. C., Schepers F., Jordi W. J., Stoopen G. M., Davelaar E., van Rhijn J. H., Power J. B., Davey M. R. (2001) Effects of P-SAG12-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol 127, 505–516 [PMC free article] [PubMed] [Google Scholar]
- 21.Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 22.Chen C., Dickman M. B. (2004) Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol 51, 1493–1507 [DOI] [PubMed] [Google Scholar]
- 23.Hamasaki-Katagiri N., Molchanova T., Takeda K., Ames J. B. (2004) Fission yeast homolog of neuronal calcium sensor-1 (Ncs1p) regulates sporulation and confers calcium tolerance. J. Biol. Chem 279, 12744–12754 [DOI] [PubMed] [Google Scholar]
- 24.Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., Guo N., Ladunga I., Ulitsky-Lazareva B., Muruganujan A., Rabkin S., Vandergriff J. A., Doremieux O. (2003) PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voegele R. T., Mendgen K. (2003) Rust haustoria: nutrient uptake and beyond. New Phytol 159, 93–100 [DOI] [PubMed] [Google Scholar]
- 26.Fotopoulos V., Gilbert M. J., Pittman J. K., Marvier A. C., Buchanan A. J., Sauer N., Hall J. L., Williams L. E. (2003) The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, At beta fruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol 132, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carninci P. (2009) Molecular biology: the long and short of RNAs. Nature 457, 974–975 [DOI] [PubMed] [Google Scholar]
- 28.Jeong J. S., Mitchell T. K., Dean R. A. (2007) The Magnaporthe grisea snodprot1 homolog, MSPI, is required for virulence. FEMS Microbiol. Lett 273, 157–165 [DOI] [PubMed] [Google Scholar]
- 29.Hwang C. S., Flaishman M. A., Kolattukudy P. E. (1995) Cloning of a gene expressed during appressorium formation by Colletotrichum gloeosporioides and a marked decrease in virulence by disruption of this gene. Plant Cell 7, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catalanotto C., Azzalin G., Macino G., Cogoni C. (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev 16, 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.