Summary
Neuronal morphology plays an essential role in neuronal function. The establishment and maintenance of neuronal morphology is intimately linked to the actin cytoskeleton; however, the molecular mechanisms that regulate changes in neuronal morphology are poorly understood. Here we identify a novel myosin-Va (MyoVa)-interacting protein, RILPL2, which regulates cellular morphology. Overexpression of this protein in young or mature hippocampal neurons results in an increase in the number of spine-like protrusions. By contrast, knockdown of endogenous RILPL2 in neurons by short hairpin RNA (shRNA) interference results in reduced spine-like protrusions, a phenotype rescued by overexpression of an shRNA-insensitive RILPL2 mutant, suggesting a role for RILPL2 in both the establishment and maintenance of dendritic spines. Interestingly, we demonstrate that RILPL2 and the Rho GTPase Rac1 form a complex, and that RILPL2 is able to induce activation of Rac1 and its target, p21-activated kinase (Pak). Notably, both RILPL2-mediated morphological changes and activation of Rac1-Pak signaling were blocked by expression of a truncated tail form of MyoVa or MyoVa shRNA, demonstrating that MyoVa is crucial for proper RILPL2 function. This might represent a novel mechanism linking RILPL2, the motor protein MyoVa and Rac1 with neuronal structure and function.
Keywords: RILPL2, Myosin-V, Rac1, Actin, Dendritic spine
Introduction
Throughout development, cells differentiate and undergo cytoskeletal and membrane rearrangements to establish a distinct morphology, specific for their physiological function. One of the most complex morphologies is exhibited by neurons, which typically extend one long axon housing the molecular machinery for releasing signals for neuronal transmission, as well as numerous, relatively shorter dendrites containing neurotransmitter receptors for receiving signals. These dendrites are the processes from where dendritic filopodia and spines emerge. These dendritic protrusions are thought to serve different purposes. Filopodia, thought to be immature spines, might serve an exploratory role to identify presynaptic partners. Dendritic spines are bulbous protrusions that contain the postsynaptic density (PSD), a structure that has been implicated in the composition, function and plasticity of glutamatergic synapses (Harris and Kater, 1994; Kim and Sheng, 2004; Matus, 2000; Yuste and Bonhoeffer, 2004).
The actin cytoskeleton plays an important role in the formation and maintenance of all of these structures; however, despite intense research, the mechanisms that control actin dynamics in neurons have yet to be fully understood.
Members of the Rho family of small GTPases, including Rac1, Cdc42 and RhoA, are important regulators of the actin cytoskeleton (Hall, 1998; Hall and Nobes, 2000). In neurons, Rho GTPases play a pivotal role in various developmental and maintenance processes, including cell migration, cell polarity, axon growth and guidance, dendrite elaboration and maintenance, as well as in the formation and plasticity of dendritic spines and synapses (Arimura and Kaibuchi, 2007; Linseman and Loucks, 2008; Luo, 2002; Newey et al., 2005; Tada and Sheng, 2006). A growing body of evidence indicates that Rac is particularly crucial for the formation, maintenance and structural plasticity of the dendritic spine (de Curtis, 2008; Penzes et al., 2008). For instance, overexpression of Rac in young dissociated hippocampal neurons is sufficient to induce the formation of spines (Wiens et al., 2005), whereas expression of a dominant-negative (DN) Rac in rat hippocampal slices results in a progressive elimination of dendritic spines (Nakayama et al., 2000). The activity of small GTPases such as Rac1 is directly modulated by guanine-nucleotide exchange factors (GEFs) that promote the formation of active GTP-bound Rac1, and GTPase-activating proteins (GAPs) that promote the hydrolysis of GTP and increase levels of inactive, GDP-bound Rac1 (Van Aelst and Cline, 2004). Following its activation, GTP-bound Rac1 activates downstream targets that are involved in spine morphogenesis, such as p21-activated kinase (Pak), which regulates dendritic-spine density and length by regulating effectors of actin polymerization (Bagrodia and Cerione, 1999; Hayashi et al., 2007; Hayashi et al., 2004).
Besides its crucial role in dendritic-spine morphogenesis and structural plasticity (Carlisle and Kennedy, 2005; Cingolani and Goda, 2008; Luo, 2002; Matus, 2000), the actin cytoskeleton also acts as a track for transport mediated by actin-based motor proteins of the myosin family. Recent studies have implicated myosin-V isoforms in trafficking of α-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA)-type glutamate receptors and in the delivery of mRNA-protein complexes within dendrites and dendritic spines in hippocampal neurons (Correia et al., 2008; Lise et al., 2006; Wang et al., 2008; Yoshimura et al., 2006). These studies suggest a crucial role for myosin-V in trafficking events required for proper neuronal function and plasticity; however, the identity of neuronal cargos transported by myosin-V-family members remains largely unknown. Also unclear is whether myosin-V, in addition to its role in local transport events within the dendritic-spine compartment, plays a role in the regulation of the actin cytoskeleton during neuronal development and maturation.
In this study, we identify RILP-like protein 2 (RILPL2) as a novel myosin-Va (MyoVa)-interacting protein. RILPL2 is related to Rab-interacting lysosomal protein (RILP) (Wang et al., 2004) – a downstream effector of the small GTPase Rab7, which regulates the morphology of lysosomal and late-endosomal compartments (Cantalupo et al., 2001; Jordens et al., 2001; Progida et al., 2007). In contrast to RILP, RILPL2 lacks the Rab7-interacting region, and ectopic expression of RILPL2 failed to alter lysosomal-compartment morphology (Wang et al., 2004). To date, the cellular function of RILPL2 has remained unknown. Here we uncover a novel role for RILPL2 in the regulation of cellular shape, particularly dendritic-spine morphogenesis in developing neurons. We also reveal that RILPL2-mediated morphological changes are correlated with activation of Rac1-Pak signaling. Disruption of RILPL2 interaction with MyoVa, using truncated RILPL2 mutant forms, or loss of MyoVa function abrogated RILPL2-mediated effects and Rac1 activation, suggesting that proper MyoVa-mediated trafficking is crucial for the proper functioning of RILPL2. Overall, we reveal a novel and intriguing mechanism involving RILPL2, the actin-based motor MyoVa and Rac1 for the regulation of neuronal morphogenesis.
Results
Identification of RILPL2 as a novel MyoVa-interacting protein
To identify novel MyoVa binding partners in the brain, we performed a yeast two-hybrid screen of a rat brain cDNA library with MyoVa C-terminal (MyoVa-CT) tail region as bait. This region encompasses the MyoVa proximal, medial and globular tail regions, the latter containing the cargo-binding domain (Fig. 1A). One of the proteins isolated from our screen was a small protein of 197 residues, previously identified as RILPL2 (Wang et al., 2004). RILPL2 is related to RILP and RILPL1, on the basis of the presence of two small regions of significant amino acid similarities, referred to as RILP-homology region-1 and -2 (RH1 and RH2, respectively; Fig. 1B). Overall, RILPL2 shares ∼22% and ∼32% amino acid identity with RILP and RILPL1, respectively (Wang et al., 2004). Similar to RILP and RILPL1, RILPL2 is predicted to adopt two α-helical coiled-coil secondary structures, between amino acids 62 and 96, and 121 and 154 (Fig. 1B) (Lupas et al., 1991). Database searches failed to identify any other conserved structural or functional domains in RILPL2.
Fig. 1.
RILPL2 is a novel MyoVa-interacting protein. (A) Schematic diagram depicting the primary structure and functional domains of the brain-specific isoform of MyoVa, including the predicted coiled-coil (CC) regions, PEST region, the alternatively spliced region and the DIL motif, which is also found in the gene products of Drosophila Canoe and human AF-6 (Ponting, 1995). Asterisk indicates the Ca2+ calmodulin-dependent protein kinase II phosphorylation site of MyoVa (Karcher et al., 2001). (B) Schematic diagram of RILPL2 primary structure with its predicted CC and RILP-homology (RH) regions. The truncated forms of RILPL2 (RILPL2-ΔCT; RILPL2-ΔNT) used in this study are shown below. (C) Compiled results of yeast two-hybrid assays showing interaction of the MyoVa globular tail (MyoVb-GT) with RILPL2-ΔCT, as assayed by HIS3 and β-galactosidase induction. (D) Co-immunoprecipitation assays from COS-7 cells expressing either HA–RILPL2-FL with either GFP–MyoVa-CT or GFP–MyoVb-CT reveal interaction of RILPL2 with MyoVa-CT but not MyoVb-CT. Left panels show expression of the different constructs in starting lysates. Right panels show co-immunoprecipitates detected with anti-HA or anti-GFP antibodies. (E) Pull-down assays (shown in duplicate) from rat brain tissue extracts using purified GST-RILPL2 or GST alone, as a control, reveal the interaction of RILPL2 with MyoVa.
Because our original MyoVa yeast two-hybrid bait contained a large portion of the MyoVa tail, we sought to determine more precisely the regions of interaction between MyoVa and RILPL2. We generated MyoVa medial tail (MyoVa-MT) and globular tail (MyoVa-GT) constructs, as well as N- or C-terminally truncated forms of RILPL2 (RILPL2-ΔNT; RILPL2-ΔCT) (Fig. 1B). Yeast two-hybrid analysis revealed that the N-terminal (NT) portion of RILPL2, encompassing amino acid residues 1 to 113, is sufficient for association with the MyoVa-GT region (Fig. 1C). Co-immunoprecipitation of MyoVa and RILPL2 exogenously expressed in COS-7 cells further confirmed the interaction (Fig. 1D; supplementary material Fig. S1). Notably, we found that RILPL2 associates with the C-terminal (CT) region of MyoVa, but not MyoVb (Fig. 1D), a closely related family member also expressed in the brain (Lise et al., 2006; Zhao et al., 1996). These results reveal a novel and selective interaction of RILPL2 with a member of class-V myosins.
Next, we assessed whether RILPL2 associates with endogenous MyoVa present in the brain. The lack of a commercially available anti-RILPL2 antibody and our unsuccessful attempts to generate specific RILPL2 antiserum prevented us from detecting and immunoprecipitating endogenous RILPL2. As an alternative, to test RILPL2 association with endogenous MyoVa we performed pull-down assays using purified full-length RILPL2 (RILPL2-FL) fused to glutathione-S-transferase (GST). We found that GST–RILPL2-FL, but not the control GST, associates with endogenous MyoVa (Fig. 1E). These results suggest that RILPL2 and MyoVa interact in the brain.
RILPL2 transcript is present in the brain and other tissues
Previous examination of the expression of the RILPL2 transcript in cDNA panels from multiple human tissues showed the presence RILPL2 in several tissues, including lung, placenta, brain, heart, liver, kidney and pancreas (Wang et al., 2004). Our reverse-transcriptase (RT)-PCR analyses confirmed RILPL2 presence in numerous brain regions from embryonic day 18 (E18) and adult rats, as well as in other tissues, including spleen, liver, kidney and thymus (Fig. 2A). In cultured cortical neurons, we found RILPL2 transcript expression to be developmentally regulated, with higher levels of expression achieved between days in vitro (DIV) 8 and DIV14 (Fig. 2B), a period when MyoVa is also expressed (Bridgman and Elkin, 2000; Espindola et al., 1992) and when neurons undergo intense synapse development (Rao et al., 1998).
Fig. 2.
Tissue distribution of RILPL2 mRNA. (A) RT-PCR shows expression of RILPL2 transcript in the brain and other tissues from E18 (left panel) and adult (right panel) rat. (B) RT-PCR shows expression of RILPL2 transcript in cultured primary cortical neurons at DIV1, 3, 8 and 14. (C-G) In situ hybridization for RILPL2 in E18 rat brain. (C) Low-magnification image of RILPL2 expression in the developing cortex. (D) Sense control for cortical expression. (E) Higher-magnification image of cortical layers showing that RILPL2 is highly expressed in the MZ and uppermost CP, as well as in the SPL. (F) Image of the developing hippocampus showing the relative distribution of RILPL2 in nascent DG and CA1/3 fields. Black box is the higher-magnification image shown in G. (G) Higher-magnification image of RILP2 expression in cells in the developing CA1/3 fields. MZ, marginal zone; CP, cortical plate; SPL, subplate layer; IZ, intermediate zone; VZ, ventricular zone; DG, dentate gyrus; CA, cornu ammonis. Scale bars: 50 μm (C,E,G); 200 μm (F).
To further explore the regional distribution of RILPL2 during neuronal development, we performed in situ hybridization on brains of E18 rats. Consistent with our RT-PCR data, RILPL2 transcript was detected in both the hippocampus and cortex at E18 (Fig. 2C-G). RILPL2 transcript was found throughout the various layers of the developing neocortex, strongly localizing within the marginal zone (MZ) and the transient subplate layer (SPL) (Fig. 2C,E). In the hippocampus, RILPL2 was predominantly expressed in the developing cornu ammonis (CA) fields, as well as in regions of the dentate gyrus (Fig. 2F,G). Because the period investigated is one of neurite outgrowth and synaptogenesis, the widespread expression of RILPL2 in these developing regions suggests a possible role in neuronal differentiation and morphogenesis.
RILPL2 expression alters cell morphology
In contrast to the well-documented role of RILP in regulating the morphology of late-endosomal and lysosomal compartments (Cantalupo et al., 2001; Progida et al., 2007; Wang et al., 2004), the cellular function of RILPL2 has not been characterized. To obtain insights into RILPL2 intracellular targeting and putative cellular function, we transiently expressed HA-tagged full-length RILPL2 (HA–RILPL2-FL) in COS-7 cells. Control COS-7 cells, expressing GFP and/or RFP, typically displayed a fibroblast-like morphology featuring relatively smooth edges and a flat surface (Fig. 3A; Fig. 4A). Phalloidin staining showed F-actin in stress fibers and in lamellipodia at the edges of GFP-transfected cells (Fig. 3A). Interestingly, approximately 45% of the cells expressing HA–RILPL2-FL underwent significant morphological changes, showing an elongated cell body extending one or more long processes, as well as regions with increased lamellipodia enriched in F-actin (Fig. 3B,C). To characterize the region of RILPL2 necessary for this morphological rearrangement, we expressed truncated forms of RILPL2 and monitored their effects on cellular morphology. Both HA–RILPL2-FL and HA–RILPL2-ΔCT displayed a diffuse cytoplasmic distribution with some enrichment at membrane ruffles, as shown by immunostaining with an anti-HA antibody (Fig. 3B-D). Similar to HA–RILPL2-FL, the C-terminally truncated form induced the formation of cellular extensions (Fig. 3D). By contrast, HA–RILPL2-ΔNT and HA–RILPL2-ΔRH1 accumulated as intracellular aggregates and membrane patches at the extremities of the cell, and failed to induce the formation of cellular extensions (Fig. 3E,F). Taken together, these observations reveal a novel function for RILPL2 in controlling cellular shape, a function that requires its NT RH1 region and proper intracellular localization.
Fig. 3.
Expression of RILPL2 alters cellular morphology. (A-F) COS-7 cells were transfected with HA–RILPL2-FL or truncated forms of RILPL2 (HA–RILPL2-ΔCT, HA–RILPL2-ΔNT, HA–RILPL2-ΔRH1) and visualized 48 hours later by immunofluorescence using an anti-HA antibody. F-actin was visualized using Alexa-Fluor-568 phalloidin staining. (A) Representative example of a GFP-transfected cell shape, with F-actin present at lamellipodia (arrow) and stress fibers (asterisk). (B-D) Expression of HA–RILPL2-FL (B,C) or HA–RILPL2-ΔCT (D) results in morphological remodeling characterized by the apparition of cellular extensions (arrowheads) and F-actin enrichment at lamellipodia (arrows). (E,F) Expression of HA–RILPL2-ΔNT (E) or HA–RILPL2-ΔRH1 (F) leads to the formation of abnormal inclusions of unknown nature. Scale bars: 10 μm.
Fig. 4.
A DN form of MyoVa, but not of MyoVb, blocks RILPL2-induced morphological changes. (A-E) COS-7 cells were co-transfected with HA–RILPL2-FL and either GFP, GFP–MyoVa-CT or GFP–MyoVb-CT. (A,B) Representative examples of cells expressing control GFP (A) or HA–RILPL2-FL (B) are shown. Arrowheads in B indicate cellular extensions. (C,D) Coexpression with GFP–MyoVa-CT, but not GFP–MyoVb-CT, induces a redistribution of RILPL2 into vesicular structures (black arrowheads) and blocks RILPL2-mediated formation of cellular extensions (white arrowheads). (E) Quantification of the proportion of co-transfected cells displaying a change in morphology. Data represent means ± s.e.m. **P<0.01; ***P<0.001. Scale bar: 10 μm.
We next investigated whether RILPL2-mediated morphological changes could be prevented by expressing truncated forms of MyoVa or MyoVb, lacking the head motor domain, which have been shown to have a DN effect on MyoV-dependent cargo transport in various cell types (Lapierre et al., 2001; Lise et al., 2006; Rodriguez and Cheney, 2002; Volpicelli et al., 2002; Wu et al., 1998). Overexpression of GFP–MyoVa-CT or GFP–MyoVb-CT in COS-7 cells led to the formation of numerous bright vesicular accumulations in the perinuclear region as well as throughout the cell (Fig. 4C,D). Notably, coexpression of GFP–MyoVa-CT with HA–RILPL2-FL resulted in the recruitment and redistribution of RILPL2 to MyoVa-positive vesicular structures (Fig. 4C). This redistribution of RILPL2 with MyoVa-CT strongly supports an association between the two proteins; however, this accumulation most probably represents mistargeted RILPL2. In contrast to MyoVa-CT, MyoVb-CT failed to recruit RILPL2 (Fig. 4D). As described above, a significant percentage of HA–RILPL2-FL-transfected cells (45.3±3.2%) showed an altered morphology, characterized by at least one cellular extension, compared with control GFP- and RFP-transfected cells (19.7±2.3%) (Fig. 4E). Coexpression with GFP–MyoVa-CT blocked the RILPL2-mediated protrusive effect, as shown by a return to control levels (26.6±2.9%) (Fig. 4E). This effect was specific for MyoVa-CT, because MyoVb-CT did not block the RILPL2-induced phenotype (50.4±4.3%) (Fig. 4E). This is consistent with our biochemical analysis showing a specific interaction of RILPL2 with MyoVa, but not MyoVb. These results suggest that MyoVa might be required for proper trafficking and localization of RILPL2 and for RILPL2-mediated changes in cell shape.
RILPL2 alters hippocampal dendritic-spine development
The presence of RILPL2 in the brain combined with the observed effects of RILPL2 expression in non-neuronal cells led us to hypothesize that RILPL2 might play a role in neurons, particularly in neuronal morphogenesis. To test this hypothesis, we first evaluated the long-term effects of RILPL2 expression on dendritic-spine morphogenesis. Cultured hippocampal neurons were co-transfected at DIV7 with HA–RILPL2-FL or GFP, together with RFP as an unbiased marker to visualize dendritic structures. In transfected neurons, HA–RILPL2-FL was distributed throughout the neuron and showed partial colocalization with endogenous MyoVa (supplementary material Fig. S2). Neurons were fixed 12 days after transfection (DIV19) and the effect of RILPL2 expression on spine density was evaluated. At this stage, neurons displayed mostly spines, with very few or no filopodia, as shown in control cells transfected with GFP and RFP (Fig. 5A). Notably, dendrites of neurons expressing HA–RILPL2-FL displayed a significantly greater number of spine-like protrusions per dendritic length (36.4±2.1; Fig. 5B,E) than GFP control cells (26.3±3.4; Fig. 5A,E). By contrast, expression of HA–RILPL2-ΔNT or HA–RILPL2-ΔCT did not increase the number of spine-like structures. Expression of HA–RILPL2-ΔCT resulted in a number of spine-like protrusions similar to control levels (29.8±1.8; Fig. 5C,E), whereas expression of HA–RILPL2-ΔNT reduced the number of spine-like structures below control levels (18.0±2.3; Fig. 5D,E), suggesting a DN effect. It should also be noted that approximately 30-50% of neurons transfected with HA–RILPL2-ΔNT displayed abnormal inclusions in their soma and dendrites similar to those observed in COS-7 cells, whereas the remaining cells showed a diffuse pattern of expression (Fig. 5D; and data not shown). To avoid potential problems associated with the health of neurons with abnormal inclusions, we analyzed only RILPL2-ΔNT-transfected neurons showing a diffuse distribution. Together, these data suggest that RILPL2 plays a role in dendritic-spine morphogenesis.
Fig. 5.
Effect of long-term expression of RILPL2 on dendritic-spine and axonal morphogenesis. (A-I) Dissociated primary hippocampal neurons (DIV7) were transfected with RFP and either (A) GFP, (B) HA–RILPL2-FL, (C) HA–RILPL2-ΔCT or (D) HA–RILPL2-ΔNT. At DIV19, neurons were fixed and recombinant proteins were detected by immunofluorescence using anti-GFP or anti-HA antibodies. (E) Quantification of the effect of overexpression of different recombinant forms of RILPL2 on the number of dendritic-spine-like protrusions per unit of dendritic length. Total numbers of cells analyzed per group are shown. Data represent means ± s.e.m. *P<0.05. (F,G) Expression of HA–RILPL2-FL, but not GFP, results in altered axonal morphology characterized by the apparition of ring-like structures (enlarged in G). (H,I) Staining with the presynaptic marker synaptophysin (Syn) shows that RILPL2-induced axonal varicosities might correspond to enlarged synaptic terminals that are enriched for Syn (arrowheads in H and I). Scale bars: 5 μm (A-D); 10 μm (F-I).
RILPL2 expression results in the formation of axonal varicosities
Besides the presence of RILPL2 in the somatodendritic compartment and its effects on dendritic-spine morphogenesis, ectopic RILPL2 was also detected within the axon. Notably, we observed abnormally large varicosities along the axons of HA–RILPL2-FL-transfected neurons, compared with GFP control neurons (Fig. 5F,G). These axonal structures were enriched for the presynaptic marker synaptophysin (Fig. 5I), indicating a possible HA–RILPL2-FL-mediated enlargement of presynaptic terminals. This observation indicates that RILPL2 might control some aspects of presynaptic morphology or function in addition to its involvement in dendritic morphogenesis.
RILPL2 signals through the small GTPase Rac1
Cellular morphology and protrusive outgrowths in neuronal and non-neuronal cells are known to be controlled by actin cytoskeletal rearrangements (Matus, 2000). Rho-GTPase proteins, including Rac1, are major regulators of the actin cytoskeleton in both cell types. In non-neuronal cells, Rac1 is known to promote the growth of lamellipodia (Ridley et al., 1992) and, in some cases, formation of neurite-like processes (Kozma et al., 1997; Miyashita et al., 2004). In neurons, Rac1 is involved in the formation, maintenance and structural plasticity of dendritic spines, as well as in the outgrowth of axons (de Curtis, 2008; Luo, 2002; Nakayama and Luo, 2000; Penzes et al., 2001a; Penzes et al., 2001b; Penzes and Jones, 2008). Hence, we hypothesized that the effects of RILPL2 on cell morphology might be due, at least in part, to the activation of Rac1. To test this hypothesis, we expressed FL and truncated forms of RILPL2 in COS-7 cells and measured the activation of endogenous Rac1 by affinity isolation of its activated GTP-bound form (Fig. 6A). We found that the expression of HA–RILPL2-FL significantly increased Rac1 activity compared with control untransfected cells (control: 0.3±0.1-fold; HA–RILPL2-FL: 2.3±0.3-fold bound Rac1 relative to total Rac1). Moreover, we found that the expression of HA–RILPL2-ΔCT significantly increased Rac1 activity, whereas HA–RILPL2-ΔNT failed to activate Rac1 (HA–RILPL2-ΔCT: 2.0±0.2-fold; HA–RILPL2-ΔNT: 0.6±0.3-fold). Rac1 activation by HA–RILPL2-FL or HA–RILPL2-ΔCT was blocked by coexpression with GFP–MyoVa-CT, but not with GFP–MyoVb-CT (GFP–MyoVa-CT: 0.9±0.2-fold; GFP–MyoVb-CT: 2.1±0.3-fold). Expression of GFP–MyoV-CT constructs alone had no significant effect on Rac1 activity. On the basis of our observations that GFP–MyoVa-CT alters the subcellular localization of both HA–RILPL2-FL and HA–RILPL2-ΔCT (Fig. 4C; and data not shown), these results indicate that proper trafficking of RILPL2, possibly mediated by MyoVa, is crucial for RILPL2-mediated Rac1 activation. To clarify the link between the RILPL2-induced morphological changes and Rac1 signaling, we assessed the effect of blocking Rac1 function in RILPL2-expressing cells. Coexpression of a DN form of Rac1 (Rac1-T17N) with RILPL2 blocked the formation of cellular elongations (Fig. 6B,C), suggesting that the effects of RILPL2 are mediated, at least in part, through Rac1 signaling.
Fig. 6.
RILPL2 transient expression activates Rac1. (A) COS-7 cells were transfected with HA–RILPL2-FL, HA–RILPL2-ΔCT or HA–RILPL2-ΔNT, with or without GFP–MyoVa-CT or GFP–MyoVb-CT. Cells were harvested 48 hours post-transfection, and activation of endogenous Rac1 was measured by evaluating Rac binding to the binding domain of Pak fused to GST. Total or bound Rac1 was detected using anti-Rac1 antibody (lower panels). Expression of RILPL2 or MyoV constructs was detected using anti-HA or anti-GFP antibodies, respectively (top panels). Graph shows the quantification of relative Rac1 activation (bound Rac/total Rac) upon expression of various recombinant forms of RILPL2 and MyoV. *P<0.05. (B) Co-expression of a DN form of Rac1 (GFP–Rac1-T17N) blocked RILPL2-mediated formation of cellular extensions (arrowheads) in COS-7 cells. (C) Quantification of the proportion of co-transfected COS-7 cells from B displaying a change in morphology. *P<0.05; **P<0.01. (D,E) Cultured hippocampal neurons (DIV10) were transfected with either GFP or HA-tagged RILPL2. At DIV15, neurons were fixed and immunolabeled with anti-GFP or anti-HA antibodies, and anti-phospho-Pak (labeled P-Pak). (D) Increased phospho-Pak intensity was observed in neurons transfected with HA–RILPL2-FL (lower panels), as compared with GFP control (top panels). (E) Graph shows average integrated intensity of phospho-Pak immunostaining in dendrites from neurons expressing GFP or HA–RILPL2-FL. Total numbers of cells analyzed per group are shown. *P<0.05. Data represent means ± s.e.m. (F) Pull-down assays reveal interaction of RILPL2 with endogenous Rac1 from rat brain tissue extract. Both Rac1 and MyoVa were pulled down by GST–RILPL2-FL, but not by the AMPA-type glutamate-receptor subunit GluR2. GST alone was used as a negative control. Scale bars: 10 μm (B); 5 μm (D).
We then investigated whether RILPL2 could activate Rac1 in neurons. Pak is a major downstream effector of Rac1 and has been implicated in spine morphogenesis (Bagrodia and Cerione, 1999; Hayashi et al., 2007; Hayashi et al., 2004). Binding between Rac1 and Pak causes activation and autophosphorylation of Pak; hence, Pak phosphorylation can be used as a measure of Rac1 activation in neurons (Xie et al., 2008). We examined endogenous levels of phosphorylated Pak (Pak-P) in neurons transfected with HA–RILPL2-FL, or GFP as a control. RILPL2 overexpression resulted in a significant increase in dendritic Pak-P immunoreactivity (HA–RILPL2-FL: 150.2±14.3%) compared with control (GFP: 100.0±9.6%), suggesting that RILPL2 activates the Rac1-Pak signaling pathway to mediate changes in cell morphology (Fig. 6D,E). We further tested whether RILPL2 could associate with endogenous Rac1 by conducting pull-down assays from rat brain lysates, and found that Rac1 bound to GST–RILPL2-FL, but not to GST alone (Fig. 6F). MyoVa was also found associated with GST–RILPL2-FL, whereas a negative control, the AMPA-type glutamate receptor subunit GluR2, was absent. This data suggest that RILPL2 can associate with MyoVa and Rac1 in the brain, possibly forming a tripartite complex. Alternatively, it is also possible that RILPL2 forms independent binary complexes with each protein. Indeed, it is worth noting that our co-immunoprecipitation experiments failed to show association of RILPL2, MyoVa or RILPL-MyoVa with endogenous Rac1 present in COS-7 cells (supplementary material Fig. S3). However, we cannot rule out the possibility that RILPL2 or RILPL2-MyoVa association with Rac1 might require active Rac1, or an unknown adaptor protein that might be absent in COS-7 cells. Another possibility is that the interaction might be too transient within the cell to be detected or be labile in the experimental conditions tested.
Knockdown of RILPL2 or MyoVa results in a reduction of dendritic spines
To further examine the effect of RILPL2 on spine development, we used shRNAs to specifically interfere with the expression of rat RILPL2. The efficiency of our shRNA was first tested in COS-7 cells and we found that two of our RILPL2 shRNAs (shRNA-493 and shRNA-496) significantly reduced the expression levels of rat GFP–RILPL2-FL (supplementary material Fig. S4). Expression of the related transcripts, rat RILPL1 or mouse RILPL2 (HA–RILPL2-res), both of which have significant sequence divergence from the rat RILPL2 at the shRNA-recognition sites, was unaltered by the RILPL2 shRNAs (Fig. 7B; supplementary material Fig. S4), indicating the specificity of the RILPLP2 shRNAs for rat RILPL2. The highest knockdown efficiency was demonstrated by shRNA-496, which was used for the subsequent experiments. Lack of anti-RILPL2 antibody prevented us from testing the potency of our shRNAs for depleting endogenous RILPL2 in cultured neurons. As an alternative, we confirmed that ectopic expression of rat GFP–RILPL2-FL was significantly reduced by RILPL2 shRNA in neurons (supplementary material Fig. S4).
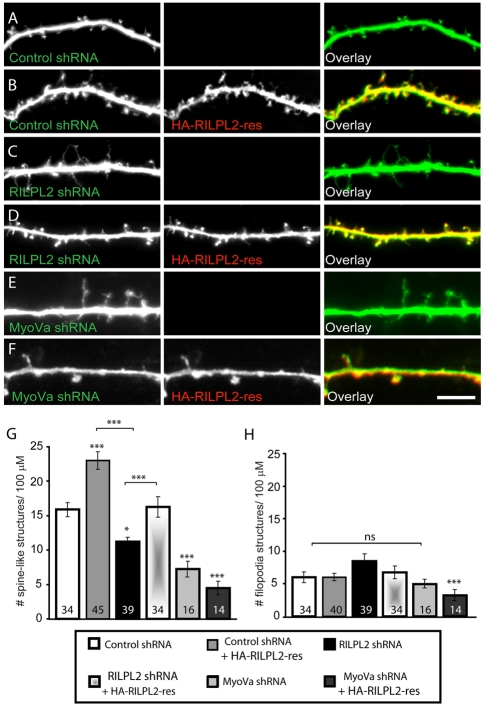
Fig. 7.
RILPL2 or MyoVa knockdown reduces spine number. (A-F) Dissociated primary hippocampal neurons (DIV10) were transfected with control shRNA, RILPL2 shRNA (RILPL2 shRNA-496) or MyoVa shRNA, with or without HA–RILPL2-res. At DIV15, neurons were fixed and exogenous proteins were detected by immunofluorescence with anti-GFP or anti-HA antibodies. Graphs show a summary of changes in the number of spine-like protrusions (G) or filopodia (H) per unit of dendritic length. Expression of either RILPL2 shRNA or MyoVa shRNA reduces the number of spine-like protrusions. Total numbers of cells analyzed per group are shown. Data represent means ± s.e.m. ***P<0.001, *P<0.05. ns, not significant. Scale bar: 5 μm.
We assessed the effects of endogenous RILPL2 knockdown by transfecting young hippocampal neurons (DIV10) and allowing the shRNA constructs to express for 5 days prior to fixation and immunostaining. As expected, these younger neurons (DIV15) displayed a lower basal number of spine-like structures compared with older neurons (DIV19) (Fig. 5). Consistent with our results in DIV19 hippocampal neurons, expression of HA–RILPL2-FL significantly increased the number of spine-like structures by ∼30-40% (Fig. 7B,G). By contrast, loss of RILPL2 resulted in a significant reduction in the number of spine-like structures, compared with control shRNA (HA–RILPL2-FL: 23.6±1.1; RILPL2 shRNA: 12.3±0.6; control shRNA: 16.3±0.7) (Fig. 7B,C,G). We also observed a decrease in Pak activation in RILPL2-shRNA-transfected neurons (supplementary material Fig. S5). A modest increase in the number of filopodia per neuron upon expression of RILPL2 shRNA was observed, suggesting that some filopodia might have failed to transform into spines and/or spines were destabilized under these conditions (Fig. 7C,H). The effect of RILPL2 shRNA was rescued by overexpression of HA–RILPL2-res, as the number of spine-like structures in shRNA neurons coexpressing HA–RILPL2-res was restored to control levels (RILPL2 shRNA + HA–RILPL2-res: 18.1±01.3) (Fig. 7D,G). Overall, these loss-of-function data provide further evidence that RILPL2 plays a role in dendritic-spine formation and maintenance.
As described above, we have shown that RILPL2-induced morphological changes can be abrogated with the expression of a DN form of MyoVa. To examine whether reducing MyoVa activity in neurons would have a similar effect, we used a previously characterized shRNA against rat MyoVa to decrease its expression (Correia et al., 2008). At 5 days after transfection, hippocampal neurons exhibited a loss of spines, similar to the loss observed with a reduction in RILPL2 expression (MyoVa shRNA: 7.2±1.1) (Fig. 7E,G). In addition, MyoVa-shRNA-transfected neurons showed a similar decrease of Pak activation as observed upon reduced RILPL2 expression (supplementary material Fig. S5). Although no obvious change in exogenous RILPL2 distribution (perinuclear accumulation or shaft vs spine localization) was detected in the absence MyoVa, neurons coexpressing MyoVa shRNA and RILPL2 also failed to develop spines (MyoVa shRNA + HA–RILPL2-res: 4.5±1.0) (Fig. 7F,G). These data indicate that proper MyoVa function is required for normal RILPL2 function, Rac1-Pak signaling and spine formation.
Expression of RILPL2 in neurons at early stages of development reduces axonal outgrowth
In addition to its role in dendritic-spine development, Rac1 has been implicated in early events of neuronal development, including dendritic and axonal outgrowth (de Curtis, 2008; Koh, 2006). On the basis of the ability of RILPL2 to activate Rac1 and the presence of RILPL2 transcript as early as E18, we hypothesized that RILPL2 might play a role in neuronal morphogenesis in early developmental stages. To test this hypothesis, GFP or HA–RILPL2-FL cDNA was electroporated into hippocampal neurons immediately after dissociation (DIV0) and neuronal morphology was examined 72 hours later, when neurons had developed easily discernible dendrites and typically one clear axon. Surprisingly, we observed a decrease in the average length of the longest neurite in neurons expressing HA–RILPL2-FL, as compared with GFP control cells (Fig. 8A,B). More detailed quantification of morphological changes using antibodies against the axonal protein Tau or the dendritic microtubule-associate-protein 2 (MAP2), revealed that HA–RILPL2-FL specifically reduced total axonal outgrowth with no effect on overall outgrowth or branching of dendrites, as compared with control (control shRNA: 70.0±4.5; control shRNA + HA–RILPL2-FL: 51.7±4.7) (Fig. 8C,D,F,G). By contrast, knockdown of RILPL2 at this stage had no significant effect on either axonal or dendritic outgrowth (RILPL2 shRNA: 70.5±4.9) (Fig. 8E-G). Combined, these data indicate that, although overexpression of RILPL2 impedes axonal outgrowth in young neurons, RILPL2 is not crucial for neurite morphogenesis early in development.
Fig. 8.
RILPL2 expression in young neurons specifically blocks axonal outgrowth. (A,B) Dissociated primary hippocampal neurons were electroporated at DIV0 with GFP or HA–RILPL2-FL and fixed at DIV3 for immunodetection using anti-GFP or anti-HA antibodies. Representative DIV3 neurons expressing GFP (A) or HA–RILPL2-FL (B) are shown in upper panels. Quantitative analysis of the length of the longest process on a given cell reveals an increase in the percentage of neurons displaying a longest process between 0 and 30 μm upon RILPL2 expression. (C-E) Expression of HA–RILPL2-res blocks axonal outgrowth, without affecting dendritic growth, whereas RILP2 shRNA does not alter process outgrowth. Axons (arrowheads) were labeled using anti-Tau antibody. (F,G) Graphs show quantification of the average length (F) and the average number of primary and secondary Tau-positive and Tau-negative processes (G). Data represent means ± s.e.m. **P<0.01. ns, not significant. Scale bars: 5 μm.
Discussion
In the present work, we identified RILPL2 as a novel interacting partner for the actin-based molecular motor MyoVa, and report a novel role for RILPL2 in controlling neuronal morphogenesis. Using a combination of yeast two-hybrid and biochemical assays, we determined that the RILPL2-MyoVa interaction occurs via the N-terminal region of RILPL2 and the MyoVa globular tail. Expression of RILPL2 in non-neuronal cells resulted in a change in cell shape. In both young and mature neurons, manipulation of RILPL2 expression altered the formation and maintenance of dendritic spines. We also showed that the observed effects of RILPL2 expression correlated with increased activity of the small GTPase Rac1, as well as phosphorylation of the Rac1 downstream substrate Pak. Furthermore, we provide evidence that functional MyoVa is required for RILPL2-mediated effects. These combined results uncover a novel role for RILPL2 in the regulation of cellular shape and dendritic-spine morphogenesis, probably via the Rac1-Pak pathway.
The ability of RILPL2 to trigger changes in cellular morphology and to activate Rac1 is particularly intriguing, because it suggests a different function for RILPL2 compared with its related protein RILP, known to regulate trafficking and morphology of late endosomes and lysosomes through distinct interactions with Rab7 and the microtubule motor dynein (Cantalupo et al., 2001; Jordens et al., 2001; Progida et al., 2007). Consistent with differential cellular functions for RILPL2 and RILP, Wang et al. reported the inability of RILPL2 to affect the morphology and distribution of lysosomes (Wang et al., 2004). Here, we provide several lines of evidence for a novel role for RILPL2 in the control of some aspects of cellular morphogenesis, probably mediated through Rac1 signaling, which regulates the actin cytoskeleton. First, expression of RILPL2 in non-neuronal cells resulted in the formation of cellular extensions and, to a lesser extent, membrane ruffles. Second, this remodeling was accompanied by increased Rac1 activity and is consistent with previously reported phenotypes associated with Rac (Kozma et al., 1997; Miyashita et al., 2004; Ridley et al., 1992). Both RILPL2-mediated morphological changes and Rac1 activation were blocked by the expression of a truncated tail version of MyoVa, which sequesters RILPL2 in vesicular compartments. In addition, expression of a DN Rac1 mutant similarly blocked RILPL2-induced phenotypic changes. Finally, overexpression of RILPL2 in developing neurons increased the levels of the Rac1 effector, Pak-P and the number of dendritic-spine-like structures, whereas RILPL2 knockdown had opposite effects. This is consistent with the reported roles of Rac and Pak in neurons as positive regulators of dendritic-spine development (Bagrodia and Cerione, 1999; de Curtis, 2008; Hayashi et al., 2007; Hayashi et al., 2004; Nakayama and Luo, 2000; Penzes et al., 2008). Together, these data suggest that RILPL2-mediated effects on cell morphology are mediated, at least in part, through the Rac1-Pak signaling pathway.
Expression of RILPL2 at early developmental stages (DIV0-DIV3) resulted in reduced axonal elongation, with no significant effect on dendritic outgrowth or arborization. This result seems counterintuitive, because Rac is generally considered as a positive regulator of neurite outgrowth (Luo, 2000; Luo, 2002; Negishi and Katoh, 2005; Van Aelst and Cline, 2004; Watabe-Uchida et al., 2006). One possibility for the observed phenotype is the sequestering by RILPL2 of Rac11 (or a Rac1 GEF) that would normally be required for elongation of axons, resulting in a reduction in axonal length. However, other studies have reported the involvement of Rac in growth-cone collapse (Jin and Strittmatter, 1997; Vastrik et al., 1999) and in the selective blockade of axonal outgrowth (both initiation and elongation), without affecting dendrite growth, suggesting that Rac can also act as a negative regulator of axon outgrowth (Luo et al., 1996; Luo et al., 1994; Penzes et al., 2001b). Thus, another possibility is that RILPL2 expression during this specific developmental time window locally activated Rac signaling in the growth cone, resulting in a reduction in axonal outgrowth. In contrast to our expectations, shRNA-mediated knockdown of RILPL2 at this developmental stage did not facilitate axonal elongation, suggesting that, although RILPL2 overexpression is deleterious for axon elongation, endogenous RILPL2 is not essential for normal neuritogenesis at early development stages.
Our pull-down data from brain lysates support a novel interaction between RILPL2 and Rac1 that might underlie activation of the Rac1-Pak pathway. However, the mechanism of activation of Rac1 signaling by RILPL2 remains unclear. RILPL2 does not have a Dbl-homology (DH) domain, the common motif that mediates nucleotide exchange that is shared by the classical GEFs for Rho GTPases (Cerione and Zheng, 1996). Therefore, it seems unlikely that RILPL2 is a Rac GEF that directly activates Rac1. One possible scenario is that the protein complex containing RILPL2 and Rac1 might also recruit a Rac GEF or another protein that facilitates activation of the small GTPase, therefore resulting in indirect activation of Rac1 by RILPL2 (see model in supplementary material Fig. S6). Further studies will have to be performed to test this hypothesis.
Our analyses of truncated forms of RILPL2 suggest that the N-terminal portion of RILPL2 plays a crucial role in its function, because deletion of this portion abrogates the ability of RILPL2 to associate with MyoVa, activate Rac1 and stimulate the formation of cellular extensions. However, whereas expression of the RILPL2 N-terminal region alone is sufficient for induction of morphological changes and Rac1 activation in COS-7 cells, it is not sufficient to increase spine formation, suggesting that the RILPL2 C-terminal region is also important for RILPL2 function, at least in neurons. By contrast, expression of the RILP2 C-terminal region alone seems to have a DN effect, resulting in a decrease in the number of dendritic-spine-like structures formed to below control levels. Thus, distinct regions seem to be functionally important for RILPL2 activity, possibly through associations with specific proteins within a complex.
The molecular mechanisms that underlie cellular morphogenesis include both cytoskeletal remodeling and membrane trafficking. The ability of RILPL2 to activate Rac1 suggests a function in cytoskeletal remodeling; however, we cannot rule out the possibility that RILPL2 might be implicated in membrane trafficking events as well. Consistent with this idea is our initial finding that RILPL2 interacts with the actin-based motor protein MyoVa, which is involved in the trafficking of membrane organelles in several cells types (Bridgman, 2004; Desnos et al., 2007). In neurons, there is increasing evidence that MyoVa acts as a Ca2+-sensor to control the activity-dependent delivery of cargos from the dendritic shaft to the spine compartment (Correia et al., 2008; Dekker-Ohno et al., 1996; Petralia et al., 2001; Wang et al., 2008; Yoshimura et al., 2006). The recently reported association of MyoVa with Rab11 (Correia et al., 2008; Roland et al., 2009), a small GTPase associated with recycling endosomes and implicated in neurite formation (Shirane and Nakayama, 2006) and plasticity-dependent dendritic-spine growth (Park et al., 2004; Park et al., 2006; Wang et al., 2008), might provide a mechanistic link between RILPL2, MyoVa-mediated trafficking and membrane remodeling associated with cell shape and spine morphology.
We used an shRNA approach to knock down endogenous MyoVa levels. This resulted in a loss of dendritic-like protrusions. Interestingly, this phenotype was not observed in previous studies that employed RNA interference or DN approaches to reduce the levels of functional MyoVa (Correia et al., 2008; Yoshimura et al., 2006). One possible explanation for the discrepancies between the observations of this study and previous studies is the duration of MyoVa inhibition. In the previous studies, shRNA and DN constructs were expressed for relatively short periods of time, from 14 hours to 2.5 days (Correia et al., 2008; Yoshimura et al., 2006). In the current study, MyoVa shRNA constructs were expressed for 5 days before dendritic-spine-like numbers were assayed. This might have reduced MyoVa levels to a critical threshold required to induce spine loss that might not have been reached previously. It is of note that overexpression of RILPL2 in MyoVa-shRNA-expressing cells was not able to rescue this phenotype, suggesting that MyoVa is crucial for RILPL2-mediated effects on dendritic spines. Furthermore, because MyoVa interacts with other factors that control dendritic-spine morphology, notably the small GTPase Rab11 (Correia et al., 2008; Park et al., 2006; Roland et al., 2009), it is entirely possible that overexpression of RILPL2 alone would not rescue the phenotype induced by MyoVa knockdown.
In this study, we focused our investigation on the function of RILPL2 in neuronal cells; however, the presence of RILPL2 in other tissues, notably in the immune system, suggests a role for RILPL2 in other organs. In this regard, it is worth noting that the human homolog of RILP2 has been suggested to be a binding partner for p40-phox (NCF4) (Futoshi Kuribayashi, Tetsuro Ago and Hideki Sumimoto, Department of Biochemistry, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan, unpublished observations), a protein that is part of the nicotinamide adenine dinucleotide phosphate-oxidase (NADPH) enzymatic complex, which is responsible for the production of superoxide within cells (Wientjes and Segal, 1995). Although this complex has been most studied in macrophages and neutrophils, it has also been detected at the synapse, where it has been suggested to be a source of superoxide required for long-term potentiation and memory function (Kishida et al., 2005; Serrano et al., 2003; Tejada-Simon et al., 2005). Intriguingly, Rac is a subunit of the NADPH oxidase and has been involved in regulating the activity of this enzymatic complex (Hordijk, 2006). It is tempting to speculate that RILPL2 might be involved in regulating the NADPH oxidase activity through Rac1. Further analysis will be required to test this hypothesis.
Materials and Methods
cDNA cloning
The generation of a GFP-tagged version of rat brain MyoVa (MyoVa-CT; aa 1005-1830) and rat MyoVb (MyoVb-CT; aa 1221-1846) has been described elsewhere (Lise et al., 2006). Truncated versions of MyoVa corresponding to the medial (MyoVa-MT; aa 1152-1395) and globular (MyoVa-GT; aa 1396-1830) tails were obtained by PCR with specific primers and subcloned into pEGFPC1 (Clontech). GFP-tagged full-length chicken and mouse MyoVa (brain isoforms) were kindly provided by Enilza Espreafico and Michael D. Ehlers, respectively (Espreafico et al., 1992; Wang et al., 2008). Full-length mouse RILPL2 cDNA (aa 1-197) was obtained from whole brain tissue by RT-PCR using specific primers and subcloned into a N-HA-GW1 vector (Sala et al., 2001). Truncated forms of RILPL2 (HA–RILPL2-ΔCT: aa 1-113; HA–RILPL2-ΔNT: aa 114-197; and HA–RILPL2-ΔRH1: aa 57-197) were obtained by PCR amplification from full-length cDNA and subcloned into N-HA-GW1. For pull-down assays, RILPL2 was amplified by PCR with specific primers and subcloned in frame with GST into pGEX4T-1 (GE Healthcare). For RNA-interference experiments, shRNA vectors were constructed using pSUPER (OligoEngine) or a modified version of pSUPER, pSUPERneo+GFP (OligoEngine), in which a short interfering RNA and GFP are dually expressed under the H1 and the PGK promoter, respectively. To create RILPL2 shRNA-493 and shRNA-496, two complementary 60-bp oligonucleotides containing the sense and antisense sequences for 5′-AGAGAGAAAGACGCTATGG-3′ (19 bp, corresponding to nucleotides 768-786 of rat RILPL2) and for 5′-GAGAAAGACGCTATGGTTA-3′ (corresponding to nucleotides 771-789 of rat RILPL2), were annealed and ligated into pSUPER vector in accordance with OligoEngine's instructions. For the rescue experiments, we used a vector expressing the mouse cDNA version of RILPL2 (HA–RILPL2-res). Construction and validation of MyoVa shRNA was described elsewhere (Correia et al., 2008). All constructs were confirmed by DNA sequencing.
Yeast two-hybrid analysis
Brain MyoVa-CT (aa 1005-1830) was subcloned into pGBKT7 (GAL4 DNA-binding-domain vector) and used to screen an adult rat brain cDNA library subcloned into pGADT7 (GAL4 activation-domain vector) (MATCHMAKER system; Clontech, Palo Alto, CA). Three independent clones corresponding to RILPL2 (aa 7-197) were obtained from our screen. Full-length rat RILPL2 cDNA (aa 1-197) was isolated from whole brain tissue by RT-PCR using specific primers and subcloned into pGBKT7. For assays of specificity and binding domains, desired cDNA fragments were amplified by PCR with specific primers and subcloned into pGBKT7 and pGADT7. These fusion proteins were tested for interaction using HIS3 and lacZ as reporter genes, and induction levels were semi-quantified as previously described (Kim et al., 1995). pGBKT7-HCN and pGADT7-p53, containing the GAL4 activation domain or GAL4 binding domain fused to p53 and HCN, respectively, were used as negative controls.
RT-PCR
RNA from E18 adult rat tissues and cultured cortical neurons was isolated using the RNeasy mini kit (Qiagen) following the manufacturer's protocol. We used 1 μg of RNA for each RT reaction with oligo (dt) primers (Invitrogen). The RT reaction was followed by PCR amplification using specific primers for rat RILPL2 (forward 5′-ATGGAGGAGCCCCCAGTACGG-3′ and reverse 5′-CGGTCAGGGAAGCACACCTAG-3′) and β-actin (forward 5′-AGCCATGTACGTAGCCATCC-3′ and reverse 5′-TTCACCACCACAGCTGAGAG-3′) as a control.
In situ hybridization
In situ hybridization was conducted as previously described (de Lecea et al., 1997), with some modifications. E18 rat brains were perfused with 4% PFA, and cryoprotected in 30% sucrose in 4% PFA. Brains were then embedded in grade IV-V bovine albumin (Fisher Scientific), cut into blocks and further cryoprotected in the sucrose/PFA solution prior to sectioning on a Leica tabletop cryostat microtome. Rat full-length RILPL2 cDNA was subcloned into pSPT-19 between EcoRI and XmaI. Probes were synthesized with the Dig-RNA labeling kit (Roche). Hybridization with 1 μg/ml digoxygenin-labeled RILPL2 probes proceeded at 65°C in a solution of 50% deionized formamide, 10% dextran sulphate, 5× Denhardt's solution, 0.62 M NaCl, 10 mM EDTA, 20 mM PIPES-Na, 0.2% SDS, 250 μg/ml heat-denatured salmon sperm DNA and 250 μg/ml heat-denatured yeast transfer RNA (tRNA).
Cell culture and transfections
Briefly, the brain regions were dissociated by papain enzymatic digestion. Cells were cultured on poly-D-lysine (Sigma)-treated coverslips at a density of 125,000-150,000 cells per coverslip (24-well pates). Cultures were maintained in neurobasal media (Gibco-Invitrogen), supplemented with B27, penicillin, streptomycin and L-glutamine as described elsewhere (Brewer et al., 1993). COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco-Invitrogen) containing 10% fetal bovine serum, penicillin and streptomycin. Transfections were performed using Lipofectamine 2000 (Gibco-Invitrogen) according to the manufacturer's protocol. For experiments in young neurons, freshly dissociated cells (DIV0) were electroporated using the AMAXA system following the manufacturer's protocol. Neurons were then plated at a density of 300,000 cells per coverslip and grown for 3 days prior to fixation and immunostaining.
Immunofluorescence
Cells were fixed at room temperature (RT) with 4% paraformaldehyde/4% sucrose for 10 minutes, then washed three times with phosphate-buffered saline (PBS) containing 0.3% Triton X-100 before and after each antibody incubation. The following primary antibodies were used (source and dilution as indicated): anti-GFP (chicken: 1:1000, AbCam; rabbit 1:1000, Synaptic Systems), anti-HA (mouse: 1:1000, Upstate Biotechnology; rat: 1:500, Roche); anti-phospho-Pak1 (Thr423)/Pak2 (Thr402) (rabbit; 1:1000, Cell Signaling); anti-Tau (mouse; 1:1000, Millipore, Clone PC1C6) and anti-MAP2 (mouse; 1:500, Millipore, Clone AP20; chicken; 1:10,000, AbCam). All antibody incubations were performed in blocking solution containing 2% normal goat serum for 1 hour at RT or overnight at 4°C. Cells were incubated for 1 hour at RT in blocking solution containing the appropriate Alexa-Fluor-conjugated secondary antibody (1:1000, Invitrogen Molecular Probes). Coverslips were then mounted on slides (Frost Plus, Fisher) with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL).
Imaging and evaluation of cellular morphology
Images were taken using a 63× or 20× objective affixed to a Zeiss Axiovert M200 motorized microscope using the AxioVision software. For analysis of cell morphology of COS-7 cells, an experimenter blinded to treatment conditions used the 10× objective to randomly select 10-15 fields for analysis. Morphological changes were quantified in terms of the proportion of transfected cells showing at least one cellular extension that had a length at least twice the diameter of the nucleus. A minimum of 200 cells was analyzed per group from two to four independent experiments.
Analysis of dendritic protrusions was preformed as previously described using the Northern Eclipse software (Empix Imaging, Mississauga, Canada) (Arstikaitis et al., 2008). Briefly, an experimenter blinded to the treatment conditions used the RFP or GFP fluorescent signal to detect transfected cells and manually outline protrusions present on all dendritic processes in the field of view. For the shRNA experiments, pSUPERneo+GFP vector was used as a control and the GFP fluorescent signal was used to detect transfected cells. Any protrusion with a clear head greater than 0.35 μm was classified as spine-like structure, whereas any thin protrusion lacking a head and with a length between 2 and 10 μM was considered as filopodia. The data presented represent the average number of spine-like protrusions or filopodia per dendritic length (100 μm).
For quantitative analysis of changes in Pak phosphorylation levels, cultures that were directly compared were stained simultaneously and imaged with the same acquisition parameters. Using the Northern Eclipse software, an experimenter blinded to the treatment conditions used the GFP or HA fluorescent signal to manually outline approximately 50-100 μm of primary dendrites (including dendritic protrusions). In parallel, regions corresponding to areas without cells were outlined to create a background mask. For summary average gray intensities (total immunofluorescence intensity/pixel area), the average gray values of background areas were subtracted to average gray values of the outlined dendritic segments.
For quantitative analysis of morphology in young neurons, images were captured with the 20× objective based on immunoreactivity against GFP. Dendrites and axons were identified based on standard morphological criteria and immunoreactivity against the axonal marker Tau or the dendritic marker MAP2. The total length of the axon and dendrites, as well as the number of primary and secondary branches, were determined manually using Neuron J 1.4.0 (Meijering et al., 2004), a plug-in software for ImageJ (NIH). At least 50 cells were analyzed per group from two to three independent experiments.
All the experiments were repeated two to four times and analysis was performed by an experimenter blinded to the identity of the transfected constructs. Values for the different experimental groups were compared by two-tailed Student's t-test (to compare two groups) or ANOVA with Tukey B Post-Hoc (to compare ≥ three groups) with P<0.05 considered statistically significant.
Immunoprecipitation, pull-down assays and immunoblotting
For co-immunoprecipitation experiments, transfected COS-7 cells were quickly harvested and lysed in TEEN buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl) containing 1% Triton X-100, 1 mM PMSF (Sigma), and a protease inhibitor cocktail tablet (Roche). After rotation for 1 hour at 4°C, insoluble material was removed by centrifugation at ∼16,000 g for 15 minutes at 4°C. Samples were then incubated for 1 hour at 4°C with 3 μg anti-GFP or 3 μg anti-HA polyclonal antibody (rabbit; custom made by Affinity BioReagents). After addition of 40 μl protein A Sepharose 4 Fast Flow beads (GE Healthcare), samples were incubated on a rotator at 4°C for 1 hour or overnight. Immunoprecipitates were washed three times with TEEN buffer containing 0.1% Triton X-100. Samples were heated at 90-100°C in SDS-PAGE sample buffer containing 5 mM DTT for 5 minutes and analyzed by SDS-PAGE. Western blot signals were detected with the Odyssey system (Li-Cor) or ECL reagents (GE Healthcare). Co-immunoprecipitation experiments were repeated three to four times.
For pull-down assays, whole brains from adult Wistar rats were quickly removed. Brain tissue was homogenized in TEEN buffer supplemented with 10 mM ATP and 10 mM MgCl2, 1 mM PMSF (Sigma), 1 mM benzamidine (Sigma) and a protease inhibitor cocktail tablet (Roche). Cells were lysed by the addition of 1% Triton X-100 followed by rotation for 1 hour at 4°C. Insoluble material was removed by centrifugation at ∼257,000 g for 25 minutes at 4°C. Samples were then incubated on a rotator at 4°C overnight with Sepharose 4B beads (GE Healthcare) coupled to 30 μg purified GST-RILPL2 (aa 1-197) or GST. Beads were washed three times with TEEN buffer containing 0.5% Triton X-100. Sample buffer containing 5 mM DTT was added to the beads and samples were heated at 90-100°C for 5 minutes before analysis by SDS-PAGE. Membranes were blotted with anti-MyoVa [rabbit (Espreafico et al., 1998; Evans et al., 1997)], anti-Rac11 (mouse; Cell Biolabs) and anti-GST (rabbit; custom made by Affinity Bioreagent). Pull-down experiments were repeated three to four times.
Rac1 GTPase activation assay and quantification
COS-7 cells were grown in six-well dishes until 60% confluent. A total of 1-3 μg of DNA was transfected using Lipofectamine 2000 reagent (Gibco-Invitrogen) according to the manufacturer's protocol. At 2 days after transfection, cells were lysed by the addition of ice-cold 400 μl MBL with protease inhibitors provided with the Rac1 assay kit (Millipore). Samples were briefly sonicated, spun down at 16,000 g and the supernatant retained. For input/loading control, 40 μl was removed from the samples, and the rest of the sample was used to perform the Rac1 activation assay as previously described (Xie et al., 2007). Quantification was performed by densitometry as previously described (Srivastava et al., 2005; Xie et al., 2007). Intensities were averaged and a one-way ANOVA with Tukey B Post-Hoc was performed using the SPSS package (SPSS, Chicago, IL). Experiments were repeated three to four times.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/20/3810/DC1
Alaa El-Husseini died in a tragic accident on December 23 2007. As a great friend and mentor, he is deeply missed. We thank Rujun Kang, Esther Yu, Xiao-Yan Jiang and Pascale Fretier for excellent technical assistance. We are thankful to Enilza Espreafico and Paul Bridgman for the generous gift of anti-MyoVa antibodies. We are also grateful to John Mercer for valuable discussions on this project. We finally thank the members of the El-Husseini laboratory for valuable scientific discussions and support. This work was funded by grants for A.E.-H. from the Canadian Institutes for Health Research (CIHR), the Michael Smith Foundation for Health Research (MSFHR) and the EJLB foundation. M.-F.L. is supported by MSFHR. D.P.S. is supported by the American Heart Association (AHA). P.P. is funded by grants from National Alliance for Autism Research (NAAR), the National Alliance for Research on Schizophrenia and Depression (NARSAD) and NIH grant MH 071316. R.L.L. and T.P.O. are funded by CIHR grant MOP-13246. Deposited in PMC for release after 12 months.
References
- Arimura, N. and Kaibuchi, K. (2007). Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8, 194-205. [DOI] [PubMed] [Google Scholar]
- Arstikaitis, P., Gauthier-Campbell, C., Carolina Gutierrez Herrera, R., Huang, K., Levinson, J. N., Murphy, T. H., Kilimann, M. W., Sala, C., Colicos, M. A. and El-Husseini, A. (2008). Paralemmin-1, a modulator of filopodia induction is required for spine maturation. Mol. Biol. Cell 19, 2026-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrodia, S. and Cerione, R. A. (1999). Pak to the future. Trends Cell Biol. 9, 350-355. [DOI] [PubMed] [Google Scholar]
- Brewer, G. J., Torricelli, J. R., Evege, E. K. and Price, P. J. (1993). Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567-576. [DOI] [PubMed] [Google Scholar]
- Bridgman, P. C. (2004). Myosin-dependent transport in neurons. J. Neurobiol. 58, 164-174. [DOI] [PubMed] [Google Scholar]
- Bridgman, P. C. and Elkin, L. L. (2000). Axonal myosins. J. Neurocytol. 29, 831-841. [DOI] [PubMed] [Google Scholar]
- Cantalupo, G., Alifano, P., Roberti, V., Bruni, C. B. and Bucci, C. (2001). Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle, H. J. and Kennedy, M. B. (2005). Spine architecture and synaptic plasticity. Trends Neurosci. 28, 182-187. [DOI] [PubMed] [Google Scholar]
- Cerione, R. A. and Zheng, Y. (1996). The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8, 216-222. [DOI] [PubMed] [Google Scholar]
- Cingolani, L. A. and Goda, Y. (2008). Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9, 344-356. [DOI] [PubMed] [Google Scholar]
- Correia, S. S., Bassani, S., Brown, T. C., Lise, M. F., Backos, D. S., El-Husseini, A., Passafaro, M. and Esteban, J. A. (2008). Motor protein-dependent transport of AMPA receptors into spines during long-term potentiation. Nat. Neurosci. 11, 457-466. [DOI] [PubMed] [Google Scholar]
- de Curtis, I. (2008). Functions of Rac GTPases during neuronal development. Dev. Neurosci. 30, 47-58. [DOI] [PubMed] [Google Scholar]
- de Lecea, L., del Rio, J. A., Criado, J. R., Alcantara, S., Morales, M., Danielson, P. E., Henriksen, S. J., Soriano, E. and Sutcliffe, J. G. (1997). Cortistatin is expressed in a distinct subset of cortical interneurons. J. Neurosci. 17, 5868-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker-Ohno, K., Hayasaka, S., Takagishi, Y., Oda, S., Wakasugi, N., Mikoshiba, K., Inouye, M. and Yamamura, H. (1996). Endoplasmic reticulum is missing in dendritic spines of Purkinje cells of the ataxic mutant rat. Brain Res. 714, 226-230. [DOI] [PubMed] [Google Scholar]
- Desnos, C., Huet, S. and Darchen, F. (2007). `Should I stay or should I go?' myosin V function in organelle trafficking. Biol. Cell 99, 411-423. [DOI] [PubMed] [Google Scholar]
- Espindola, F. S., Espreafico, E. M., Coelho, M. V., Martins, A. R., Costa, F. R., Mooseker, M. S. and Larson, R. E. (1992). Biochemical and immunological characterization of p190-calmodulin complex from vertebrate brain: a novel calmodulin-binding myosin. J. Cell Biol. 118, 359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espreafico, E. M., Cheney, R. E., Matteoli, M., Nascimento, A. A., De Camilli, P. V., Larson, R. E. and Mooseker, M. S. (1992). Primary structure and cellular localization of chicken brain myosin-V (p190), an unconventional myosin with calmodulin light chains. J. Cell Biol. 119, 1541-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espreafico, E. M., Coling, D. E., Tsakraklides, V., Krogh, K., Wolenski, J. S., Kalinec, G. and Kachar, B. (1998). Localization of myosin-V in the centrosome. Proc. Natl. Acad. Sci. USA 95, 8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L. L., Hammer, J. and Bridgman, P. C. (1997). Subcellular localization of myosin V in nerve growth cones and outgrowth from dilute-lethal neurons. J. Cell Sci. 110, 439-449. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Hall, A. and Nobes, C. D. (2000). Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K. M. and Kater, S. B. (1994). Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu. Rev. Neurosci. 17, 341-371. [DOI] [PubMed] [Google Scholar]
- Hayashi, K., Ohshima, T., Hashimoto, M. and Mikoshiba, K. (2007). Pak1 regulates dendritic branching and spine formation. Dev. Neurobiol. 67, 655-669. [DOI] [PubMed] [Google Scholar]
- Hayashi, M. L., Choi, S. Y., Rao, B. S., Jung, H. Y., Lee, H. K., Zhang, D., Chattarji, S., Kirkwood, A. and Tonegawa, S. (2004). Altered cortical synaptic morphology and impaired memory consolidation in forebrain-specific dominant-negative PAK transgenic mice. Neuron 42, 773-787. [DOI] [PubMed] [Google Scholar]
- Hordijk, P. L. (2006). Regulation of NADPH oxidases: the role of Rac proteins. Circ. Res. 98, 453-462. [DOI] [PubMed] [Google Scholar]
- Jin, Z. and Strittmatter, S. M. (1997). Rac1 mediates collapsin-1-induced growth cone collapse. J. Neurosci. 17, 6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens, I., Fernandez-Borja, M., Marsman, M., Dusseljee, S., Janssen, L., Calafat, J., Janssen, H., Wubbolts, R. and Neefjes, J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680-1685. [DOI] [PubMed] [Google Scholar]
- Karcher, R. L., Roland, J. T., Zappacosta, F., Huddleston, M. J., Annan, R. S., Carr, S. A. and Gelfand, V. I. (2001). Cell cycle regulation of myosin-V by calcium/calmodulin-dependent protein kinase II. Science 293, 1317-1320. [DOI] [PubMed] [Google Scholar]
- Kim, E. and Sheng, M. (2004). PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771-781. [DOI] [PubMed] [Google Scholar]
- Kim, E., Niethammer, M., Rothschild, A., Jan, Y. N. and Sheng, M. (1995). Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378, 85-88. [DOI] [PubMed] [Google Scholar]
- Kishida, K. T., Pao, M., Holland, S. M. and Klann, E. (2005). NADPH oxidase is required for NMDA receptor-dependent activation of ERK in hippocampal area CA1. J. Neurochem. 94, 299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, C. G. (2006). Rho GTPases and their regulators in neuronal functions and development. Neurosignals 15, 228-237. [DOI] [PubMed] [Google Scholar]
- Kozma, R., Sarner, S., Ahmed, S. and Lim, L. (1997). Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17, 1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre, L. A., Kumar, R., Hales, C. M., Navarre, J., Bhartur, S. G., Burnette, J. O., Provance, D. W., Jr, Mercer, J. A., Bahler, M. and Goldenring, J. R. (2001). Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman, D. A. and Loucks, F. A. (2008). Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front. Biosci. 13, 657-676. [DOI] [PubMed] [Google Scholar]
- Lise, M. F., Wong, T. P., Trinh, A., Hines, R. M., Liu, L., Kang, R., Hines, D. J., Lu, J., Goldenring, J. R., Wang, Y. T. et al. (2006). Involvement of myosin Vb in glutamate receptor trafficking. J. Biol. Chem. 281, 3669-3678. [DOI] [PubMed] [Google Scholar]
- Luo, L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173-180. [DOI] [PubMed] [Google Scholar]
- Luo, L. (2002). Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18, 601-635. [DOI] [PubMed] [Google Scholar]
- Luo, L., Liao, Y. J., Jan, L. Y. and Jan, Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787-1802. [DOI] [PubMed] [Google Scholar]
- Luo, L., Hensch, T. K., Ackerman, L., Barbel, S., Jan, L. Y. and Jan, Y. N. (1996). Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379, 837-840. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M. and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Matus, A. (2000). Actin-based plasticity in dendritic spines. Science 290, 754-758. [DOI] [PubMed] [Google Scholar]
- Meijering, E., Jacob, M., Sarria, J. C., Steiner, P., Hirling, H. and Unser, M. (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58, 167-176. [DOI] [PubMed] [Google Scholar]
- Miyashita, M., Ohnishi, H., Okazawa, H., Tomonaga, H., Hayashi, A., Fujimoto, T. T., Furuya, N. and Matozaki, T. (2004). Promotion of neurite and filopodium formation by CD47: roles of integrins, Rac, and Cdc42. Mol. Biol. Cell 15, 3950-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, A. Y. and Luo, L. (2000). Intracellular signaling pathways that regulate dendritic spine morphogenesis. Hippocampus 10, 582-586. [DOI] [PubMed] [Google Scholar]
- Nakayama, A. Y., Harms, M. B. and Luo, L. (2000). Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20, 5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi, M. and Katoh, H. (2005). Rho family GTPases and dendrite plasticity. Neuroscientist 11, 187-191. [DOI] [PubMed] [Google Scholar]
- Newey, S. E., Velamoor, V., Govek, E. E. and Van Aelst, L. (2005). Rho GTPases, dendritic structure, and mental retardation. J. Neurobiol. 64, 58-74. [DOI] [PubMed] [Google Scholar]
- Park, M., Penick, E. C., Edwards, J. G., Kauer, J. A. and Ehlers, M. D. (2004). Recycling endosomes supply AMPA receptors for LTP. Science 305, 1972-1975. [DOI] [PubMed] [Google Scholar]
- Park, M., Salgado, J. M., Ostroff, L., Helton, T. D., Robinson, C. G., Harris, K. M. and Ehlers, M. D. (2006). Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron 52, 817-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes, P. and Jones, K. A. (2008). Dendritic spine dynamics – a key role for kalirin-7. Trends Neurosci. 31, 419-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Kambampati, V., Mains, R. E. and Eipper, B. A. (2001a). Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J. Neurosci. 21, 8426-8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Sattler, R., Zhang, X., Huganir, R. L., Kambampati, V., Mains, R. E. and Eipper, B. A. (2001b). The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229-242. [DOI] [PubMed] [Google Scholar]
- Penzes, P., Cahill, M. E., Jones, K. A. and Srivastava, D. P. (2008). Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 18, 405-413. [DOI] [PubMed] [Google Scholar]
- Petralia, R. S., Wang, Y. X., Sans, N., Worley, P. F., Hammer, J. A., 3rd and Wenthold, R. J. (2001). Glutamate receptor targeting in the postsynaptic spine involves mechanisms that are independent of myosin Va. Eur. J. Neurosci. 13, 1722-1732. [DOI] [PubMed] [Google Scholar]
- Ponting, C. P. (1995). AF-6/cno: neither a kinesin nor a myosin, but a bit of both. Trends Biochem. Sci. 20, 265-266. [DOI] [PubMed] [Google Scholar]
- Progida, C., Malerod, L., Stuffers, S., Brech, A., Bucci, C. and Stenmark, H. (2007). RILP is required for the proper morphology and function of late endosomes. J. Cell Sci. 120, 3729-3737. [DOI] [PubMed] [Google Scholar]
- Rao, A., Kim, E., Sheng, M. and Craig, A. M. (1998). Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J. Neurosci. 18, 1217-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J., Paterson, H. F., Johnston, C. L., Diekmann, D. and Hall, A. (1992). The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401-410. [DOI] [PubMed] [Google Scholar]
- Rodriguez, O. C. and Cheney, R. E. (2002). Human myosin-Vc is a novel class V myosin expressed in epithelial cells. J. Cell Sci. 115, 991-1004. [DOI] [PubMed] [Google Scholar]
- Roland, J. T., Lapierre, L. A. and Goldenring, J. R. (2009). Alternative splicing in class v myosins determines association with rab10. J. Biol. Chem. 284, 1213-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala, C., Piech, V., Wilson, N. R., Passafaro, M., Liu, G. and Sheng, M. (2001). Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115-130. [DOI] [PubMed] [Google Scholar]
- Serrano, F., Kolluri, N. S., Wientjes, F. B., Card, J. P. and Klann, E. (2003). NADPH oxidase immunoreactivity in the mouse brain. Brain Res. 988, 193-198. [DOI] [PubMed] [Google Scholar]
- Shirane, M. and Nakayama, K. I. (2006). Protrudin induces neurite formation by directional membrane trafficking. Science 314, 818-821. [DOI] [PubMed] [Google Scholar]
- Srivastava, D. P., Yu, E. J., Kennedy, K., Chatwin, H., Reale, V., Hamon, M., Smith, T. and Evans, P. D. (2005). Rapid, nongenomic responses to ecdysteroids and catecholamines mediated by a novel Drosophila G-protein-coupled receptor. J. Neurosci. 25, 6145-6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, T. and Sheng, M. (2006). Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16, 95-101. [DOI] [PubMed] [Google Scholar]
- Tejada-Simon, M. V., Serrano, F., Villasana, L. E., Kanterewicz, B. I., Wu, G. Y., Quinn, M. T. and Klann, E. (2005). Synaptic localization of a functional NADPH oxidase in the mouse hippocampus. Mol. Cell Neurosci. 29, 97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst, L. and Cline, H. T. (2004). Rho GTPases and activity-dependent dendrite development. Curr. Opin. Neurobiol. 14, 297-304. [DOI] [PubMed] [Google Scholar]
- Vastrik, I., Eickholt, B. J., Walsh, F. S., Ridley, A. and Doherty, P. (1999). Sema3A-induced growth-cone collapse is mediated by Rac1 amino acids 17-32. Curr. Biol. 9, 991-998. [DOI] [PubMed] [Google Scholar]
- Volpicelli, L. A., Lah, J. J., Fang, G., Goldenring, J. R. and Levey, A. I. (2002). Rab11a and myosin Vb regulate recycling of the M4 muscarinic acetylcholine receptor. J. Neurosci. 22, 9776-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Wong, K. K. and Hong, W. (2004). A unique region of RILP distinguishes it from its related proteins in its regulation of lysosomal morphology and interaction with Rab7 and Rab34. Mol. Biol. Cell 15, 815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Edwards, J. G., Riley, N., Provance, D. W., Jr, Karcher, R., Li, X. D., Davison, I. G., Ikebe, M., Mercer, J. A., Kauer, J. A. et al. (2008). Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell 135, 535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida, M., Govek, E. E. and Van Aelst, L. (2006). Regulators of Rho GTPases in neuronal development. J. Neurosci. 26, 10633-10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens, K. M., Lin, H. and Liao, D. (2005). Rac1 induces the clustering of AMPA receptors during spinogenesis. J. Neurosci. 25, 10627-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wientjes, F. B. and Segal, A. W. (1995). NADPH oxidase and the respiratory burst. Semin. Cell Biol. 6, 357-365. [DOI] [PubMed] [Google Scholar]
- Wu, X., Bowers, B., Rao, K., Wei, Q. and Hammer, J. A., 3rd (1998). Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J. Cell Biol. 143, 1899-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Srivastava, D. P., Photowala, H., Kai, L., Cahill, M. E., Woolfrey, K. M., Shum, C. Y., Surmeier, D. J. and Penzes, P. (2007). Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron 56, 640-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Photowala, H., Cahill, M. E., Srivastava, D. P., Woolfrey, K. M., Shum, C. Y., Huganir, R. L. and Penzes, P. (2008). Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J. Neurosci. 28, 6079-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, A., Fujii, R., Watanabe, Y., Okabe, S., Fukui, K. and Takumi, T. (2006). Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 16, 2345-2351. [DOI] [PubMed] [Google Scholar]
- Yuste, R. and Bonhoeffer, T. (2004). Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5, 24-34. [DOI] [PubMed] [Google Scholar]
- Zhao, L. P., Koslovsky, J. S., Reinhard, J., Bahler, M., Witt, A. E., Provance, D. W., Jr and Mercer, J. A. (1996). Cloning and characterization of myr 6, an unconventional myosin of the dilute/myosin-V family. Proc. Natl. Acad. Sci. USA 93, 10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.