Abstract
Background
Streptococcus suis serotype 2 (SS2) is a zoonotic agent that causes death and disease in both humans and swine. A better understanding of SS2-host molecular interactions is crucial for understanding SS2 pathogenesis and immunology. Conventional genetic and biochemical approaches used to study SS2 virulence factors are unable to take into account the complex and dynamic environmental stimuli associated with the infection process. In this study, in vivo-induced antigen technology (IVIAT), an immunoscreening technique, was used to identify the immunogenic bacterial proteins that are induced or upregulated in vivo during SS2 infection.
Results
Convalescent-phase sera from pigs infected with SS2 were pooled, adsorbed against in vitro antigens, and used to screen SS2 genomic expression libraries. Upon analysis of the identified proteins, we were able to assign a putative function to 40 of the 48 proteins. These included proteins implicated in cell envelope structure, regulation, molecule synthesis, substance and energy metabolism, transport, translation, and those with unknown functions. The in vivo-induced changes in the expression of 10 of these 40 genes were measured using real-time reverse transcription (RT)-PCR, revealing that the expression of 6 of the 10 genes was upregulated in the in vivo condition. The strain distribution of these 10 genes was analyzed by PCR, and they were found in the most virulent SS2 strains. In addition, protein sequence alignments of the newly identified proteins demonstrate that three are putative virulence-associated proteins.
Conclusion
Collectively, our results suggest that these in vivo-induced or upregulated genes may contribute to SS2 disease development. We hypothesize that the identification of factors specifically induced or upregulated during SS2 infection will aid in our understanding of SS2 pathogenesis and may contribute to the control SS2 outbreaks. In addition, the proteins identified using IVIAT may be useful potential vaccine candidates or virulence markers.
Background
Streptococcus suis (S. suis) infections have been considered a major problem in the swine industry worldwide, particularly over the past 20 years. S. suis is a gram-positive, facultatively anaerobic coccus, and 35 serotypes (1-34 and 1/2) have been described based on their capsular antigens. Among these, serotype 2 (SS2) is the causative agent of many different syndromes worldwide, including meningitis, septicemia, arthritis, and pneumonia in humans, swine, and other animals [1]. In addition, SS2 is widely recognized as an important zoonotic agent that afflicts people in close contact with infected pigs or pork-derived products [2,3]. Two recent large-scale outbreaks of human streptococcal toxic shock syndrome (STSS) caused by SS2 in China in 1998 and in 2005 have increased public health concerns worldwide. Notably, a major outbreak of SS2 infection emerged in the summer of 2005 in Sichuan Province, China. A total of 215 cases of human S. suis infection were reported, and the outbreak resulted in 38 deaths and massive economic losses [4,5].
Little is known about the virulence factors of SS2. To date, only a few SS2 virulence associated factors have been identified and characterized; these include the capsular polysaccharide (CPS) [1], suilysin (SLY) [6], muramidase-released protein (MRP) [7], extracellular protein factor (EF) [8], adhesin [9], cell wall-associated and extracellular proteins [10], fibronectin- and fibrinogen-binding protein (FBP) [11], a serum opacity factor [12], and the arginine deiminase system [13,14].
An understanding of SS2-host molecular interactions is crucial for understanding SS2 pathogenesis and immunology. Conventional genetic and biochemical approaches used to study SS2 virulence factors are unable to take into account in the complex and dynamic environmental stimuli associated with the infection process. Recently, several technologies, including in vivo expression technology (IVET), differential fluorescence induction (DFI), signature-tagged mutagenesis (STM), transcriptional and proteomic profiling, and in vivo-induced antigen technology (IVIAT) have been developed to identify the pathogen genes expressed during the infection process [15,16].
IVIAT is a method that allows for the direct identification of microbial proteins expressed at sufficient levels during host infection to be immunogenic. A schematic of the IVIAT procedure was described by Rollins et al [16]. The advantage of IVIAT is that it enables the identification of antigens expressed specifically during infection, but not during growth in standard laboratory media. It was speculated that the genes and gene pathways identified by IVIAT may play a role in virulence or pathogenesis during bacterial infection [17,18]. IVIAT has been successfully used to identify arrays of in vivo induced proteins in Salmonella enterica serovar Typhi [19], Escherichia coli O157 [18], Group A Streptococcus (GAS) [17], Vibrio cholerae [20], and others, and these proteins have been shown to contribute to the pathogenesis or virulence of the infecting organisms. When IVIAT was applied to E. coli O157, it identified 223 O157 proteins expressed during human infection. Among these, four proteins--intimin-γ (an adhesin), QseA (a quorum-sensing transcriptional regulator), TagA (a lipoprotein), and MsbB2 (an acyltransferase)--had been previously identified as virulence-related proteins [18].
To identify SS2 proteins that are immunogenic and expressed uniquely during SS2 infection, we applied the newly developed and modified IVIAT method. Briefly, we screened a library of SS2 proteins expressed in E. coli to identify clones that were immunoreactive with convalescent-phase sera, which had been previously fully adsorbed against in vitro-grown SS2 and E. coli organisms. The adsorption process eliminates antibodies reactive with in vitro-expressed antigens and allows for the identification of clones expressing protein antigens that are upregulated during swine infection. Specifically, we hypothesized that by using IVIAT, we could identify proteins that play a role in the SS2-specific host-bacterium interactions unique to SS2 infection in pigs. In this study, we identified 48 putative in vivo-induced (IVI) proteins, which included proteins associated with bacterial cell wall structure, metabolism, regulation, molecule synthesis, substance transport and others. Of these, 10 genes were selected for analysis by real-time PCR to confirm their in vivo upregulation. Six genes were shown to be upregulated in vivo. These results suggest that these newly identified genes may contribute to SS2 pathogenesis.
Results
Sera selection and adsorption
IVIAT depends on the presence of antibodies directed against pathogen antigens expressed in vivo, so the selection of convalescent sera for use in IVIAT must be carefully considered. In this study, sera were selected that had an antibody titer of at least 10,000. All eight convalescent-phase sera, which were collected from recovered pigs as described in the materials and methods, had antibody titers above 12,800. These eight pooled convalescent-phase sera were mixed at equal volumes to create a sera cocktail for IVIAT, in order to best balance individual immune variability with the effects of dilution.
The adsorption efficiency was determined by examining the immunoreactivity of the serum aliquots from the pooled swine convalescent-phase sera after each adsorption step with whole cells and cell lysates of in vitro-grown ZY05719. As shown in Figure 1, the immunoreactivity of the pooled sera with in vitro-grown SS2 progressively decreased with each round of adsorption; the decrease in immunoreactivity was particularly noticeable after the first adsorption step.
Figure 1.
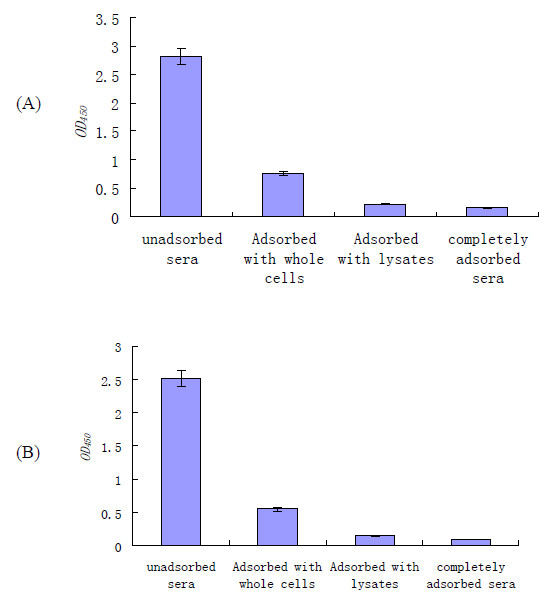
Enzyme immunoassay reactivities of sera with lysates of an in vitro-grown SS2 strain after each step in sequential adsorption. Optical density values (OD450) were corrected for background and for dilution during adsorption. Swine convalescent sera cocktail sets were sequentially adsorbed with SS2 whole cells, cell lysates, and E. coli whole cells and cell lysates. Following sufficient adsorption with all these antigens, sera were considered to have been completely adsorbed. (A) ELISA plates coated with whole SS2 cells. (B) ELISA plates coated with SS2 cell lysates. The results are expressed as means of absorbance values, and error bars represent the standard errors of the means.
The immunoreactivity of the adsorbed pooled convalescent sera against in vitro-derived SS2 proteins was further assessed with dot-ELISA using the individually purified proteins MRP, EF, and GAPDH, which are reportedly expressed on the cell surface (Figure 2). Dot-ELISA results showed that unadsorbed sera strongly reacted with MRP, EF, and GAPDH (Figure 2A). However, when the sera had been completely adsorbed with in vitro antigens, there were no spots on the NC membrane (Figure 2B). These results further validated that antibodies specific to in vitro antigens had been fully removed from the convalescent-phase sera set.
Figure 2.

Dot-ELISA results of reactivities of pooled unadsorbed (A) and adsorbed (B) swine convalescent sera against the three previously reported SS2 virulence-associated proteins MRP, EF, and GAPDH. BSA was used as a negative control.
Use of adsorbed convalescent-phase sera to probe a genomic DNA expression library of the SS2 isolate ZY05719
To provide tenfold coverage of a SS2 genome (2 × 106 bp), a plasmid library containing inserts whose average size is 2 kb would contain about 5.7 × 104 independent recombinants. The SS2 genomic library, prepared from strain ZY05719 isolated from a Sichuan SS2 outbreak (Table 1), in E. coli DH5α consisted of approximately 6 × 104 clones for each expression vector (pET30 a, b, and c). These three libraries were used for IVIAT selection with the adsorbed convalescent sera. During the primary screening, 300 of the most intensely immunoreactive clones were selected. Following rigorous selection, 60 clones that continuously showed a strong positive reaction with the adsorbed convalescent-phase sera antibodies were identified. Their immunoreactivity was confirmed by an additional screening, in which these clones were compared with clones bearing the vectors alone without any inserts present. The positive clones were picked out and then cultured in broth. The presence of a cloned DNA insert in all 60 clones was confirmed by PCR analysis and sequencing.
Table 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Serotype, Genotype and/or phenotype | Reference/source |
|---|---|---|
| Strains | ||
| HA9801 | serotype 2;cps2J+, mrp+, ef+, sly+, gapdh+, gdh+, orf2+ | Jiangsu outbreak SS2 isolate, 1998, China |
| ZY05719 | serotype 2; cps2J+, mrp+, ef+, sly+, gapdh+, gdh+, orf2+ | Sichuan outbreak SS2 isolate, 2005, China |
| T15 | serotype 2; cps2J+, mrp-, ef- | The Netherlands |
| Plasmids | ||
| pET30(abc) | Expression vectors allowing cloning of fragments in each of three reading frames; Kanr | Novagen |
| pMRP | pET30(a) with partial mrp gene amplified from strain ZY05719, and cloned into EcoRI and XhoI sites, in vector, Kanr | This work |
| pEF | pET30(a) with partial ef gene amplified from strain ZY05719, and cloned into EcoRI and XhoI sites, in vector, Kanr | This work |
| pGAPDH | pET32(a) with partial gapdh gene amplified from strain ZY05719, and cloned into BamHI and SalI sites, in vector, Ampr | Previous work |
Kanr, kanamycin-resistant; Ampr, ampicillin-resistant.
Categorization of the IVI proteins according to the actual or putative functions of the genes identified by IVIAT
The sequencing results showed that most of the immunoreactive clones contained only a portion of the coding sequence of the relevant protein, and that these 60 clones encoded 48 different proteins. The difference in the number of positive clones and proteins is due to several clones encoding the same protein. For instance, clones 6, 34, and 73 encoded the protein ysirk1. The nucleotide/protein sequence data of the 48 in vivo-induced (IVI) proteins were deposited in the NCBI GenBank database http://www.ncbi.nlm.nih.gov, and the accession numbers that were assigned to these sequences are listed in Table 2.
Table 2.
Proteins encoded by IVIAT selected clones
| Category | Gene name | Function and/or feature† | Bacterial cell sub-localization‡ | GenBank Accession number |
|---|---|---|---|---|
| Cell envelope structure |
ysirk1 | YSIRK Gram-positive signal peptide | cell wall | EF127690 |
| ysirk2 | YSIRK Gram-positive signal peptide | cell wall | EF127720 | |
| ss-1616 | hypothetical protein SsuiDRAFT 0718 | cell wall | EF127691 | |
| cwh | Cell wall hydrolase/autolysin, peptidoglycan catabolism | cell wall | EF127695 | |
| trag | TRAG protein, essential for DNA transfer in bacterial conjugation | Membrane | EF127701 | |
| srt | sortase-like protein/Sortase family | Cytoplasmic | EF127708 | |
| nlpa | NLPA lipoprotein | Cytoplasmic | EF127718 | |
| Regulation | lac | LacI:Periplasmic binding protein/LacI transcriptional Regulator | EF127711 | |
| hprk | HPr(Ser) kinase, two-component signal transduction system/two-component sensor activity/regulates carbohydrate | Cytoplasmic | EF127733 | |
| flps | FlpS, regulation in arginine deiminase system | Cytoplasmic | EF127749 | |
| Molecule synthesis | ss-896 | Tagatose-6-phosphate kinase/pfkB family carbohydrate kinase | Cytoplasmic | EF127693 |
| ss-1611 | Aspartate kinase/Amino acid kinase family | Cytoplasmic | EF127726 | |
| ss-1759 | Amino acid biosynthesis/glutamate-cysteine ligase activity | Cytoplasmic | ||
| pol1 | DNA polymerase I | Cytoplasmic | EF127735 | |
| cpn60 | 60 kDa chaperonin (Protein Cpn60) (groEL protein)/TCP-1/cpn60 chaperonin family | Cytoplasmic | EF127737 | |
| prpn | Primosomal protein n | Cytoplasmic | EF127740 | |
| pol3 | DNA polymerase III, epsilon subunit:DNA polymerase III, alpha subunit, Gram-positive type | Cytoplasmic |
EF127741 EF127751 |
|
| Substance and energy metabolism | gh3 | Glycoside hydrolase, family 3, N-terminal/Tim barrel glycosyl hydrolase superfamily. carbohydrate metabolism/hydrolase activity, hydrolyzing O-glycosyl compounds | Cytoplasmic | EF127713 |
| ss-999 | hypothetical protein, carbohydrate metabolism/hydrolase activity | Cytoplasmic | EF127746 | |
| ss-1647 | tRNA (guanine-N(7)-)-methyltransferase/Methyltransferase superfamily, Central intermediary metabolism | unknown | EF127722 | |
| ss-862 | Uracil DNA glycosylase/Uracil DNA glycosylase superfamily | Cytoplasmic | EF127729 | |
| ss-802 | ATPase | Cytoplasmic Membrane | EF127747 | |
| ss-1766 | protein metabolism/ATP-dependent Clp protease, ATP-binding subunit | Cytoplasmic | EF127719 | |
| ss-607 | ABC transporter | Cytoplasmic Membrane | EF127697 | |
| sdh | L-serine dehydratase, alpha subunit | Cytoplasmic Membrane | EF127704 | |
| ss-1617 | Fructokinase/Actin-like ATPase Superfamily | unknown | EF127707 | |
| ppc | Phosphoenolpyruvate carboxylase | EF127727 | ||
| ss-1305 | ABC transporter | Cytoplasmic Membrane | EF127736 | |
| Transport | ss-1955 | conserved hypothetical protein, transport and binding | unknown | EF127692 |
| ss-273 | ABC transporter, transport and binding/coupled to transmembrane movement of substances | Cytoplasmic Membrane | EF127694 | |
| ss-1298 | ABC transporter | Cytoplasmic Membrane | EF127714 | |
| ss-1829 | peptidase, S54 (rhomboid) family protein/Rhomboid family | Cytoplasmic Membrane | EF127742 | |
| ss-403 | PTS system sorbose subfamily IIB component | cytoplasmic | EF127754 | |
| Translation | exc-b | Excinuclease ABC, B subunit | Cytoplasmic |
EF127698 EF127700 |
| ss-887 | O-acetylhomoserine sulfhydrylase/PLP dependent aminotransferase superfamily | Cytoplasmic | EF127706 | |
| ss-349 | possible product: hypothetical protein SMU_684 | Cytoplasmic | EF127738 | |
| ss-1935 | Ribosomal protein S4, bacterial and organelle form | Cytoplasmic | EF127739 | |
| Others | ss-154 | 3-phosphoglycerate kinase, phosphoglycerate kinase activity/glycolysis | Cytoplasmic | EF127702 |
| ss-485 | Metal-dependent phosphohydrolase, HD subdomain/HD/PDEase superfamily | Cytoplasmic | EF127730 | |
| ss-1478 | Inorganic diphosphatase, | Cytoplasmic | EF127734 | |
| smc | SMC protein, N-terminal: Structural maintenance of chromosome protein SMC, C-terminal: SMCs flexible hinge | Cytoplasmic | EF127743 | |
| Function unknown | g19 | unknown | EF127696 | |
| g59 | Protein of unknown function DUF150 | Cytoplasmic | EF127717 | |
| g77 | unknown | EF127724 | ||
| g78 | unknown | EF127725 | ||
| g98 | hypothetical protein Franean1DRAFT_0529 | EF127732 | ||
| g130 | unknown | EF127744 | ||
| g132 | unknown | EF127745 |
† Putative functions of hypothetical proteins were determined from NCBI database and CBS Prediction Servers when available.
‡ Predicted by the PSORTb v.2.0 program.
By evaluating previously published studies of Streptococcus species and performing sequence alignments, we were able to assign a functional classification to 40 of the 48 proteins identified by IVIAT. Since the SS2 genomes have not been completely annotated, the genes identified by IVIAT were named after their homolog in other bacteria, when possible. When an IVI gene was unnamed, it was assigned an annotation number http://www.sanger.ac.uk/Projects/Microbes. These unnamed genes did not demonstrate good homology with the SS2 P1/7 genome determined by the Sanger Institute http://www.sanger.ac.uk/Projects/Microbes. Therefore, these genes were annotated http://www.ncbi.nlm.nih.gov. Table 2 shows the 48 IVI proteins encoded by these 60 clones that were categorized according to the actual or putative functions of the open reading frames (ORFs) identified by IVIAT. The IVI proteins were grouped into eight categories: cell envelope, regulatory, molecule synthesis, substance and energy metabolism, transport, translation, others, and those with unknown functions.
Among these 48 newly identified IVI proteins, we found three proteins previously reported to have putative roles in Streptococcus virulence. (i) Protein YSIRK1, when performing protein-protein BLAST with the newly identified IVI proteins against the NCBI database, we found that the protein encoded by ysirk1 is a potential virulence protein. Its sequence indicates that it is a subtilisin-like serine protease homologous to surface-associated subtilisin-like serine protease PrtA of Streptococcus pneumoniae. Interestingly, PrtA was also identified by screening a genomic expression library from Streptococcus pneumoniae using a convalescent-phase serum. Bethe and colleagues reported that PrtA is a highly conserved virulence factor of Streptococcus pneumoniae, and might be a promising candidate for a protein-based vaccine [21]. (ii) Autolysin, the autolysin encoded by cwh is also a reported virulence-associated factor in SS2 [22]. Most bacteria possess several autolysins that are able to degrade their cell walls, and are implicated in various biological functions including cell separation, cell wall turnover, restructuring of cell walls, and bacterial autolysis. In addition, certain autolysins have also been reported to contribute to the pathogenicity of gram-positive bacteria. For example, an intact autolytic function is required for the full virulence of Streptococcus pneumoniae [23]. (iii) protein TRAG, TRAG is a component of the type IV secretion system (T4SS), a virulence-associated pathway of SS2 [22]. The bacterial T4SS, which is widely distributed among the gram-negative and -positive bacteria and is ancestrally related to bacterial conjugation machines (which mediate protein and gene transfer), contributes to pathogenicity [24].
Analysis of the in vivo gene expression profiles
Strain ZY05719 was selected for real-time PCR analysis because it is one of the strains isolated from the 2005 SS2 outbreak in China; ZY05719 was also used for constructing the genomic library. Of the 48 putative IVI genes, 10 (ss-1616, trag, nlpa, srt, cwh, hprk, ysirk, ss-1955, sdh, ss-1298) were selected for further analysis of gene expression by real-time PCR. We selected these genes based on their putative functions, such as involvement in cell structure, metabolism, regulation, and transport, in order to maximize the variety of genes chosen for further analysis. The in vitro expression of these 10 putative IVI genes was observed in early lag phase, log phase, late log phase, and stationary phase of growth, with the highest level of expression occurring at late log phase (data not shown). Before comparing the expression of these 10 putative IVI genes under the in vitro condition, they were first tested under in vivo conditions (expression after challenge with bacterial cells via intravenous inoculation measured at 12, 24, and 36 h pi). All of the putative IVI genes were expressed in vivo under the conditions tested (data not shown). With the exception of ysirk and ss-1955, which were expressed at 12 h pi but not at 24 and 36 h pi, and ss-1298, which was expressed until 36 h, the remaining 7 IVI genes were expressed at 12, 24 and 36 h post-inoculation in vivo. The aim of this study was to identify the genes whose expressions are upregulated in vivo; therefore, we determined the in vivo gene expression relative to the highest level of expression in vitro. Of the 10 analyzed genes, six genes, namely, ss-1616, trag, nlpa, hprk, sdh, and ss-1298, were upregulated in vivo relative to the highest level of expression of the corresponding gene in vitro (Figure 3).
Figure 3.
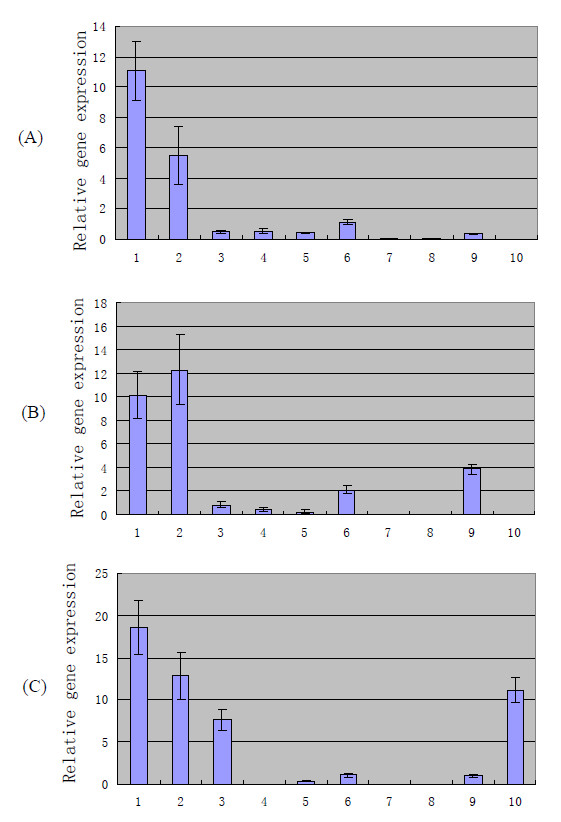
In vivo gene expression at 12 h (A), 24 h (B), and 36 h (C) relative to the highest level of expression in vitro by real-time PCR analysis. Total bacterial RNA extracted from strain ZY05719 grown in LB broth media was used as the template to assay the in vitro expression levels of the 10 newly identified genes. SPF minipigs were employed as model to study the in vivo expression levels. Pigs were inoculated intravenously with strain ZY05719, and bacterial cells recovered from blood at 12 h, 24 h, and 36 h post-inoculation were considered as in vivo growth bacteria. Total bacterial RNAs extracted from in vivo growth bacterial cells were further analyzed by real-time PCR. To determine whether RNA expression level is induced or upregulated under in vivo conditions, we compared in vivo gene expression with the highest level of expression in vitro. The standard deviations are presented from three pigs each, blood collected at 12, 24 and 48 h. 1, ss-1616; 2, trag; 3, nlpa; 4, srt; 5, cwh; 6, hprk; 7, ysirk; 8, ss-1955; 9, sdh; 10, ss-1298; gapdh was used as reference gene.
Location of the IVI genes on the SS2 chromosome
To learn about location of the 48 IVI genes on the SS2 chromosome, we used BLAST to identify them in the S. suis strain P1/7 genomic sequence (genomic sequence data were generated by the S. suis strain P1/7 Sequencing Group at the Sanger Institute, and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/ss/. Thirty-eight IVI genes were located (data not shown). Four genes (trag, exc-b, lac, and ppc) did not have high homology with P1/7, but demonstrated homology with strains S. suis 89/1591, 98HAH33, and 05ZYH33. The remaining six genes could not be located because their sequences were short and did not show high homology with any other sequence in the database.
Pathogenicity islands (PAIs) are clusters of genes that may contribute to virulence in pathogens, sometimes by responding to environmental signals [25,26]. Wei et al. (2006) predicted eight possible SS2 pathogenicity islands based on a systematic analysis of the SS2 strain P1/7 genomic sequence [27]. In this study, five IVI genes (sdh, srt, ss-1955, ss-1829, and ss-802) were found to be distributed in four pathogenicity islands (Figure 4) when located on the SS2 chromosome.
Figure 4.
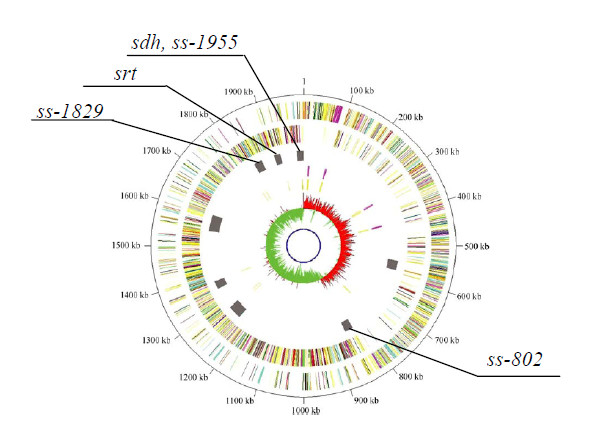
Graphical representation of the locations of five IVI genes on the pathogenicity islands of S. suis serotype 2 strain P1/7. Based on a complete analysis of the SS2 reference strain P1/7 genomic sequence, W. Wei et al. predicted eight putative pathogenicity islands (PAIs). When we determined the locations of the 48 IVI genes identified by IVIAT, we found five IVI genes (sdh, ss-1955, srt, ss-1829, and ss-802) located in four pathogenicity islands in SS2 reference strain P1/7. The genomic map was published by W. Wei et al., 2006 (gray bars the third ring represent eight possible pathogenicity islands).
Strain distribution of IVI genes
To determine whether the newly identified IVI genes are common among SS2 isolates, we examined their distribution in SS2 isolates with different backgrounds, including those isolated at different times and from different regions and sources. PCR was employed to analyze the distribution of 10 IVI genes in Chinese strains (N = 23). Twenty-three SS2 strains isolated from different regions of China in different years were analyzed, and PCR results showed that the distribution ratio of these IVI genes were as follows: ss-1616 (22/23, 95.7%), trag (23/23, 100%), nlpa (22/23, 95.7%), srt (22/23, 95.7%), cwh (23/23, 100%), hprk (23/23, 100%), ysirk (23/23, 100%), ss-1955 (23/23, 100%), sdh (23/23, 100%), ss-1298 (20/23, 87%) (details not shown). The genomic sequences of SS2 strains P1/7, 89/1591, 98HAH33, 05ZYH33 were collected from Sanger or the NCBI data library. The distribution of the 10 IVI genes in these strains was determined by nucleotide sequence alignment (Table 3). With the exception of gene trag, which was not found in strain P1/7, the nine remaining IVI genes were found in all four of the above strains (P1/7, 89/1591, 98HAH33, and 05ZYH33).
Table 3.
Distributions of 10 IVI genes in SS2 strains
| strain | serotype | host | region | year | Gene Name※ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||||
| HA9801* | 2 | Pig | China | 1998 | + | + | + | + | + | + | + | + | + | + |
| ZY05719* | 2 | Pig | China | 2005 | + | + | + | + | + | + | + | + | + | + |
| 89/1591‡ | 2 | N | Canada | N | + | + | + | + | + | + | + | + | + | + |
| P1/7‡ | 2 | N | N | N | + | + | + | + | + | - | + | + | + | + |
| 05ZYH33‡ | 2 | human | China | 2005 | + | + | + | + | + | + | + | + | + | + |
| 98HAH33‡ | 2 | human | China | 1998 | + | + | + | + | + | + | + | + | + | + |
*, The distribution of the 10 IVI genes in strains was analyzed by colony PCR.
‡, The distribution of the 10 IVI genes in strains was performed through alignment the IVI genes with corresponding genomic sequence.
※, 1, cwh; 2, hprk; 3, ysirk; 4, ss-1616; 5, ss-1955; 6, trag; 7, sdh; 8, srt; 9, ss-1298; 10, nlpa.
N, Background not reported in related publication.
+, positive or found in the related genome sequence.
-, negative or not found in the related genome sequence.
Discussion
S. suis infection is a major cause of sudden death of pigs, and is also increasingly becoming a human health concern due to its zoonotic transmission capabilities. Attempts to control the infection have been hampered by our lack of knowledge about SS2 pathogenicity. The identification and characterization of putative virulence factors and other infection-related proteins will aid in the prevention and control of SS2 disease. IVIAT provides a "snapshot" of protein expression during infection, allowing us a glimpse into the possible mechanisms by which this pathogen might counter host defenses and adapt and establish itself within the host to cause disease [18]. In the present study, we used the newly developed IVIAT method to select in vivo-induced proteins.
Convalescent-phase sera collected from pigs naturally infected with SS2 are ideal for IVIAT [16]. However, for multiple reasons, it is not easy to acquire convalescent-phase sera from SS2 natural infection. First, epidemiological studies have found that SS2 outbreaks are usually infrequent and only affect a small number of pigs, which can lead to underdiagnosis or misdiagnosis. Second, pigs infected with SS2 do not always show obvious clinical symptoms, and may become carriers without showing clinical signs. Finally, based on its polysaccharide capsular antigens, at least 35 serotypes of S. suis exist. Isolates belonging to other serotypes (such as 1, 1/2, 3, 4, 5, 7, 8 and 9) have also been associated with disease in pigs [28,29]. Common antigens had been found to be shared between SS2 and these other serotypes (unpublished data from our lab). To reduce these possible interferences, we used pigs with clear backgrounds as animal models, and convalescent sera were prepared following artificial infection.
Until recently, the exact mechanism of SS2 transmission (from pig to human or between pigs) was still poorly understood, but was thought to involve aerosol transmission or other pathways [28-30]. However, some hypotheses about the critical stages of the infection, such as bacterial invasion from the mucosal surfaces to the bloodstream, survival of the bacteria in blood, and invasion from blood into the central nervous system have been presented [28]. Regardless of the mechanism of SS2 invasion, circulation in the blood plays an important role during SS2 disease development. In addition, S. suis is an agent of zoonosis, afflicting people in close contact with infected pigs or pork-derived products. The organisms probably gain entry via small wounds or through inhalation [4,10,29]. Furthermore, transmission between pigs in herds through cutaneous wounds has been suggested [29]. In light of these considerations, intravenous and intramuscular inoculations were employed to assay the expression of SS2 in vivo, and to try to mimic natural infection (such as the middle or late stage of the infection).
In this study, we used real-time PCR to analyze the induction of the expression of IVI genes under different environmental conditions. Real-time PCR results demonstrated that the expression of six of the 10 selected genes was upregulated under in vivo conditions. The upregulation time points for these six genes were 12, 24, and 36 h for ss-1616 and trag, 24 h for hprk and sdh, and 36 h for nlpa and ss-1298. This upregulated expression suggests that these genes may play a significant role during the course of SS2 infection (middle, late, or whole stage of infection). The expression profiles of the other four genes (ysirk, srt, cwh, and ss-1955) showed that they were not obviously upregulated under the in vivo condition (Figure 3). There are two possible explanations for this result. First, since we measured the in vivo gene expression at 12, 24, and 36 h pi, it is possible that we missed the time when the levels of expression of these genes were high relative to the expression of the same gene in vitro. For instance, they may play roles in the rapid replication of SS2 during an early stage of infection. Second, the mRNA concentration might not reflect the amount of protein or antigen produced if these antigens are regulated post-transcriptionally.
The protein TRAG, which is a component of a type IV secretion system (T4SS, virulence associated pathway of SS2), was identified. The T4SS mediates horizontal gene transfer, thus contributing to genome plasticity, the evolution of infectious pathogens, and the dissemination of antibiotic resistance and other virulence traits [31]. The gene trag was found in 98HAH33, 05HAH33, Canada strain 89/1591, and all the tested Chinese SS2 virulent strains, but not in European strain P1/7 or the non-virulent strain T15 (data not shown). Brucella species require a T4SS to reach their proper niche and to replicate within host cells [32]. Whether DNA transfer through a T4SS occurs between SS2 isolates and results in an increase in virulence is unknown, and will only be answered by future studies.
Lipoproteins that are upregulated in vivo in other pathogenic organisms have been identified, and have been shown to be likely important for pathogenesis [33,34]. For instance, in a previous study of Vibrio vulnificus using IVIAT, a putative lipoprotein was also found to be induced in vivo when convalescent-phase sera from patients who survived V. vulnificus septicemia were used to screen a genomic library of this organism [35]. As for nlpa, almost nothing is known about the homolog of this gene in the NCBI database. The partial NLPA protein sequence was identified as a lipoprotein in the 89/1591 genome database, and shares 100% identity with a putative NLPA.
HPr kinase/P-Ser-HPr phosphatase (HprK/P) is a phosphocarrier protein of the phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS). It is also a sensor of two-component signal transduction systems (TCSs) [36]. Thus, HprK/P provides a link between carbon metabolism and the development of virulence [37]. For example, the expression of several virulence genes, such as the hemolysin-encoding hly and the phospholipase-encoding plcA, is repressed when Listeria monocytogenes is grown on cellobiose, glucose, fructose, or other rapidly metabolizable carbon sources [38].
L-Serine dehydratase, an iron-sulfur protein [39], was identified, and this gene was also found in the Canadian strain 89/1591. During the course of the infection, alternative carbon sources (such as amino acids) are utilized by bacteria for growth due to competition for nutrients. The results of Velayudhan et al. showed that the catabolism of L-serine by serine dehydratase was crucial for the growth of Campylobacter jejuni under in vivo conditions [40]. In addition, the fermentation of amino acids produces ammonia that neutralizes the surrounding pH; this neutralization is beneficial since it protects group A Streptococcus (GAS) from acid-induced damage [41].
The protein encoded by ss-1298 is an ABC-type transporter, but its function is still unknown. Gene ss-1616 is a conserved hypothetical outer membrane protein in SS2 genome database, and almost nothing is known about this gene. It was found in all tested strains in this study, and in Canada strain 89/1591 and European strain P1/7.
Many surface proteins of pathogenic gram-positive bacteria are linked to the cell wall envelope by a sorting mechanism that recognizes an LPXTG motif, but surface proteins of Streptococcus pneumoniae harbor another motif, YSIRK-G/S [42]. About 20 surface proteins of Staphylococcus aureus carry the YSIRK-G/S motif, whereas those of Listeria monocytogenes and Bacillus anthracis do not [43,44]. While the function of the YSIRK motif has not been completely elucidated, it may contribute to the efficient secretion of a protein [43]. In the present study, four clones encoded two proteins containing this motif. Although the gene ysirk was only detected 12 h after SS2 infection and then disappeared, and was not strongly upregulated in vivo, the mature protein encoded by ysirk1 showed homology to the surface-associated subtilisin-like serine protease PrtA (a virulence factor) of S. pneumoniae[21]. However, the role of this protein during SS2 infection remains to be determined.
IVIAT enables the identification both of proteins expressed specifically during host infection but not during growth under standard laboratory conditions, and of proteins expressed at significantly higher levels in vivo than in vitro. But IVIAT has its own limitations. IVIAT will not identify all virulence-associated genes. Genes that are expressed both in vivo and in vitro and genes that are not expressed effectively in the E. coli host expression system will not be identified. For instance, some previously reported SS2 virulence factors, such as MRP, EF, FBPs, CPS, and SLY, could not be screened out by IVIAT in this study. We speculate that they are expressed in both in vivo and in vitro growth conditions, and therefore antibodies specific to these antigens had been eliminated during the convalescent sera adsorption steps.
Unexpectedly, some of the genes identified are likely expressed during in vitro growth conditions, such as DNA polymerase I and III, Primosomal protein n, protein Cpn60, and SMC protein (essential for bacterial cell division and cell wall biosynthesis). We speculate that perhaps their expression level was higher during in vivo growth than in vitro growth, and therefore they were detected by the IVIAT.
Conclusion
Taken together, our results suggest that during the course of infection, bacterial metabolism, envelope composition, and virulence will be adjusted for bacteria to survive in the hostile environment. Bacterial pathogens sense their environment, and in response, genes are induced or repressed through spatial and temporal regulation. Genes and gene pathways identified by IVIAT may play a role in virulence or pathogenesis during host infection, and may be appropriate for inclusion in therapeutic, vaccine, or diagnostic applications. The results of this study yielded a set of potentially valuable proteins of a manageable number for future studies on SS2 pathogenicity and for the development of specific diagnostics and vaccines.
Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 1. The S. suis strains were grown in Todd-Hewitt broth (THB) (Oxoid) or Todd-Hewitt agar (THA) (Oxoid) plates supplemented with 2% inactivated calf serum. Strain ZY05719 was originally isolated from the 2005 Sichuan SS2 infection outbreak in China. E. coli DH5α was used as the host strain for cloning, and E. coli BL21 (DE3) was used as the host strain for the recombinant proteins. The E. coli strains were grown in Luria-Bertani (LB) media and stored at -40°C in LB broth containing 20% glycerol. Plasmid-transformed E. coli cells were grown in LB medium supplemented with 30 μg/mL kanamycin (kan).
DNA manipulation and strain construction
DNA manipulations were performed according to standard procedures [45]. All restriction enzymes, DNA polymerases, ligase, and oligonucleotide primers were purchased from TaKaRa. The mrp, ef, and gapdh genes were amplified by PCR, and each gene was separately ligated into pET expression vectors to construct 3 recombinant expression plasmids (Table 1). These recombinant expression plasmids were separately introduced into E. coli BL21 (DE3) and induced to overexpress recombinant proteins.
Indirect ELISA and dot-ELISA
An indirect enzyme-linked immunosorbent assay (ELISA) was used for screening the swine sera with the in vitro-derived SS2 antigens. In brief, microtiter plates (Costar) were coated with SS2 antigen (whole cells and cell lysates). Following incubation and blocking, 100-μL dilutions (1:200-1:51,200, V/V) of sera were added to the wells. The subsequent ELISA protocol was performed as previously described [46]. 3,3',5,5'-tetramethylbenzidine (TMB, Amresco) was used as the substrate, and the optical density at 450 nm (OD450) was determined with an ELISA reader (BIO-RAD550). The antibody titer was defined as the highest serial dilution of serum for which the OD450 value was two standard deviations above the mean OD450 of the negative controls (without primary antibody).
To assay for antibodies specific to MRP, EF, and GAPDH, successively diluted nickel affinity-purified recombinant-expressed MRP, EF, and GAPDH proteins were spotted on a nitrocellulose (NC) membrane (Millipore). Dot-ELISA was performed according to the standard procedure with minor modifications [46]. The reactions were developed with 3,3'-diaminobenzidine (DAB, Amresco) solution with 0.1% H2O2.
Swine convalescent sera and control sera
Recently, a specific pathogen-free (SPF) piglet has been developed as an animal model for studying S. suis [47,47]. Animal experiments were performed as previously reported with minor modifications [48]. All animal experiments were conducted in accordance with national guidelines and were approved by the office of laboratory animal, Shanghai Jiao Tong University and Shanghai administration committee of laboratory animals.
Convalescent sera
To prevent any contact with infectious agents, SPF Bama minipigs and healthy piglets were housed in independent units with absolute filters. Prior to challenge, all the pigs were negative for SS2-specific antibodies, as determined by an ELISA test. SPF minipigs (n = 8, Guizhou line, 7 weeks old) were randomly grouped into 2 units (4/unit, named as group 1 and 2) and piglets (n = 12, 8 weeks old) into 2 units (6/unit, named as group 3 and 4). Bacterial suspensions in THB with 10% inactivated bovine serum were prepared and adjusted to a concentration of 1 × 108 colony forming units (CFU)/mL of S. suis. These pigs were challenged with 2 mL of strain ZY05719 (1 × 108CFU/mL), intramuscularly (i.v.) for group 1 and 3, and intravenously (i.m.) for group 2 and 4, respectively. The pigs were monitored daily post-inoculation (pi) for clinical signs, notably fever and central nervous system dysfunctions such as opisthotonos, tremors, and nystagmus. The rectal temperature was recorded daily. No inflammation was observed at the injection sites. Intramuscularly challenged pigs died naturally between 4 and 8 days after experimental infection, while intravenously challenged pigs died between 2 and 7 days. The pigs, 3 minipigs (1 for i.v. group and 2 for i.m.) and 5 piglets (2 for i.v. group and 3 for i.m.), that recovered after being challenged were used in the subsequent experiments performed in this study. The antibody titer against a homologous strain was determined by indirect ELISA every week after challenge. At week 4, the animals were sacrificed and bled. The sera were collected and kept frozen at -40°C. The flowchart of piglet infections was as shown in Additional File 1: Figure S1. Convalescent sera collected from the recovered pigs were used for IVIAT selection.
Positive control sera
SS2-positive sera were prepared from 3 SPF minipigs immunized with inactivated ZY05719 whole cell bacteria (2 mL of 1 × 108 CFU each) 4 times at 2-week intervals. Ten days after the last injection, the antisera were pooled and used as the positive control in ELISA tests.
Negative control sera
To reduce variability animal to animal, serum samples were obtained from healthy SPF minipigs prior to SS2 infection, negative in ELISA test, used as the negative control for IVIAT or ELISA.
Adsorption of swine convalescent-phase and control sera
To compensate for variations in the immune responses of individual pigs, equal volumes of convalescent sera from 3 minipigs and 5 piglets were pooled and extensively adsorbed with in vitro-derived SS2 antigens to completely remove all antibodies that recognize the antigens that are expressed under the in vitro condition.
The adsorption protocol has been described previously [20]. Briefly, a Protease Inhibitor Cocktail Set II (Merck) formulated for bacterial cells and containing 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF; 20 mM), EDTA (85 mM), bestatin (1.7 mM), pepstatin A (2 mM), and E-64 (0.2 mM) was prepared per the manufacturer's instructions and then added to intact cells and cell lysates at a dilution of 1:10 (V/V). The successive adsorption steps were performed six times with whole bacterial cells, three with native cell lysates, and one with heat-denatured ZY05719 cell lysates and E. coli BL21(DE3) that contain unmodified pET-30abc expression plasmids (Novagen), as described[15,20]. Cell lysates were prepared by sonication, and the protein concentration determined by using spectrophotometer (Smartspec™, BIO-RAD). The cell lysates were first coated onto nitrocellulose membranes and the corresponding antibodies adsorbed to remove antigen-antibody complexes. The resultant adsorbed serum was divided into aliquots that were stored at -40°C. To check the efficacy of each adsorption step, a 10-μL serum aliquot was removed after each adsorption and analyzed with ELISA against either whole SS2 cells or SS2 cell lysates.
Construction of a genomic expression library of the SS2 strain ZY05719
An expression library was constructed with the pET-30abc series of expression vectors, which permit the cloning of inserts into each of the three reading frames under the transcriptional control of the T7 phage promoter. Each vector DNA was digested with BamHI, subjected to agarose gel electrophoresis, purified by using a gel extraction kit (TaKaRa), and treated with shrimp alkaline phosphatase (TaKaRa). Genomic DNA from strain ZY05719 was extracted and partially digested with Sau3AI. Next, we ligated each of the three vectors separately with genomic DNA fragments to create three expression libraries. These libraries were electroporated into competent E. coli DH5α as described previously [18,20]. We assessed the resulting libraries by subjecting a random sample to PCR in order to determine the frequency and size of the inserts. More than 98% of transformants contained inserts of sizes ranging from 0.1 to 4 kbp.
Screening the antigens identified by IVIAT against swine convalescent-phase sera
Immunoscreening was performed according to the procedure described by Sambrook et al. [45]. In short, an aliquot of the library with E. coli BL21 (DE3) as the expression host was diluted and spread on sterile NC membranes that were overlaid on kan/LB plates. After overnight incubation at 37°C, the colonies were lifted onto new sterile NC membranes, and after a 5-h incubation at 37°C, these membranes with the lifted colonies (colony side up) were overlaid on fresh kan/LB plates containing 1 mM isopropyl-D-thiogalactopyranoside (IPTG, Amresco) and incubated overnight at 37°C to induce gene expression of the cloned inserts. The plates were exposed to chloroform vapors for 15 min for partial bacterial lysis and for the release of the induced proteins. Each membrane was removed, blocked with 5% skim milk, and reacted with a 1:100 (V/V) dilution of the pooled adsorbed convalescent-phase sera at room temperature. The clones that reacted with the antibodies in the adsorbed sera were detected by using peroxidase-conjugated staphylococcal protein A (SPA) and visualized with an Enhanced chemiluminescence (ECL) kit (Pierce). The immunoreactive clones were identified by their position on the master membrane. Each positive clone was purified at least two additional times and confirmed as immunoreactive to the adsorbed sera [18,20]. Plasmids from individual positive reactive clones were purified, and the DNA inserts were sequenced in both directions by using pET30-specific primers.
Bioinformatic analysis
Analysis of sequence homologies, protein families, and conserved domains was performed using NCBI BLAST http://blast.ncbi.nlm.nih.gov, information from the Sanger Genome Centre http://www.sanger.ac.uk/Projects/S_suis, and PFAM http://pfam.sanger.ac.uk. The putative functions of the newly discovered proteins were assigned using the CBS Prediction Servers http://www.cbs.dtu.dk/services/ProtFun. The cellular localizations of these proteins were predicted using PSORTb v2.0 http://www.psort.org/psortb/.
Real-time PCR analysis
Gene expression was tested by subjecting the RNA of the bacteria grown under standard laboratory conditions to real-time PCR, and the results were compared to those obtained for bacteria recovered from infected pigs.
In vitro culture
Duplicate cultures of ZY05719 grown under in vitro conditions were harvested at OD600s of 0.1, 0.2, 0.4, 0.6, and 0.8. OD600s in the ranges of 0.1-0.2, 0.2-0.6, and 0.6-0.8 correspond to the lag phase, log phase, and stationary phase, respectively. The bacterial pellet was snap frozen in liquid nitrogen and stored at -80°C.
In vivo gene expression
Three SPF Bama minipigs were inoculated intravenously with ZY05719 for analyzing gene expression under in vivo conditions. The bacterial cells were separated from blood by centrifuging at different speeds. Blood samples were pooled at 12, 24, and 36 h pi, centrifuged at 2,000 rpm to remove blood cells, and repelleted at 12,000 rpm to collect bacterial cell pellets that were subsequently snap frozen in liquid nitrogen and stored at -80°C.
Real-time PCR
Bacterial total RNA was extracted using RNAprep Bacteria Kit (TIANGEN, China), and residual genomic DNA was removed by using a QIAGEN RNase-Free DNase Set (Qiagen) according to the manufacturer's instructions. DNase-treated RNA samples were reverse transcribed by using a first-strand cDNA synthesis kit (TaKaRa) according to the manufacturer's recommendations. The controls for cDNA synthesis and DNase treatment included two negative controls: one with no RNA template and one without reverse transcriptase.
Quantitative real-time PCR (qPCR) assays were performed by using a Chromo4 system (BIO-RAD) and a SYBR-Green PCR kit (Takara). All qPCR reactions were performed in a final volume of 25 μL containing 12.5 μL Premix Ex Taq mix (2×), 0.5 μL (10 μM) of each primer, 2 μL cDNA, and 9.5 μL double-distilled water (ddH2O). The protocol followed for each qPCR was as follows: hot start at 95°C for 10 s, followed by 45 cycles at 95°C for 5 s, 60°C for 20 s. Data were collected and analyzed using Opticon Monitor software V3.1 (BIO-RAD). To normalize the data, primer pairs were designed to amplify the gene glyceraldehyde-3-phosphate dehydrogenase (gapdh) as housekeeping control. Based on the gene classification, 10 genes were selected for the PCR amplification and the specific primer sets that were used are listed in Table 4. The specificity of each resulting amplicon was validated with its corresponding melting curve. The relative level of expression was calculated by comparing the difference in the threshold cycle number of the gene of interest gene with that of the reference gene.
Table 4.
Primers used for real-time PCR in this study
| gene | Sequences of primers (5' to 3') | Amplicon size (bp) |
|---|---|---|
| cwh | TGGTAAATGCCCCATCTAGTC | 137 |
| GGCTGTAACACCAATAATTTCC | ||
| hprk | GAAACCCCTGTTGTCATAGTGG | 126 |
| CAATTCTCCCGATAGACGACTG | ||
| ss-1616 | ACAGGGAATAAGCATCAGCG | 119 |
| ATGTAGTTACGCTCCGCCTT | ||
| ysirk | GCACTTTTATTGCCACGGATT | 160 |
| CAGCACCTTGTTGTCTCGGA | ||
| gapdh | TTGGAAGCTACAGGTTTCTTTG | 98 |
| TTACCACCAGGAGCAGTGACA | ||
| ss-1955 | ATCAGGTTCTAACATTGTTGCG | 122 |
| TAACGCCCCCCTCTAACAAG | ||
| srt | GGTCGACGAAGTGTCATTGC | 123 |
| ATACGTCAGCGTCCTCCCAC | ||
| nlpa | CTGCAACCTGGTCACCAAATAC | 129 |
| ACCCCGGAAAAGTTACGTATGA | ||
| sdh | TAGAAGTCCCTTGTGTCAGACG | 134 |
| AGATCCCACTTGGTACATAGCG | ||
| ss-1298 | TGGATATCGACAGCAAGGAG | 156 |
| CATAGTCGCCCAAATAGAGC | ||
| trag | TCGTGACTTGATGACGGCTG | 167 |
| GATAATGCCACCAGCGTTCA |
Colony PCR analysis
To learn about gene distribution in diverse SS2 isolates with different backgrounds, colony PCR was used. The primers used to detect the 10 IVI genes were same as the oligonucleotides for qPCR (Table 4). Single SS2 colonies were picked from THA plates, suspended in 50 μL of ddH2O and boiled for 10 min to make DNA lysates. Each was assayed using the appropriate primer sets by PCR. PCR reactions were carried out using Taq polymerase according to the manufacturer's recommendation (TaKaRa).
Authors' contributions
HWG carried out the IVIAT selection, participated in the sequence alignment, performed real-time RT-PCR and drafted the manuscript. HDZ carried out the animal experiments and participated in the PCR amplification. CPL conceived of the study, participated in its design and coordination. and critically revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Swine convalescent sera preparation. The data provided represent the preparation of swine convalescent sera. * Time-point of antibody check. ‡ Sacrificed and serum collection. † Number of recovered pigs, and antisera were used as convalescent sera for IVIAT selection. § sera were collected and used as negative control.
Contributor Information
Hongwei Gu, Email: hongweigu999@yahoo.com.cn.
Haodan Zhu, Email: zhuhaodan0311@yahoo.com.cn.
Chengping Lu, Email: lucp@njau.edu.cn.
Acknowledgements
This work was supported by the National Basic Research Program (No. 2006CB504400) from Ministry of Science and Technology of the People's Republic of China.
We appreciate the thoughtful comments of Drs. Huochun Yao, Hongjie Fan, Yongjie Liu, Rongmei Fei, Jianhe Sun, Yaxian Yan, Jianluan Ren, and Yong Yu. We thank Miss Kaicheng Wang for kindly providing rGAPDH for this study, and Dr. Yuling Ma and Mr. Piren Chen for their assistance in sera collection. We also thank Dr. H.E. Smith for providing the SS2 T15 Strain. We are extremely grateful to Dr. Xiuguo Hua for providing SPF minipiglets.
References
- Smith HE, Damman M, van d V, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–1756. doi: 10.1128/iai.67.4.1750-1756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum T, Adam R, Eggelnpohler I, Matalon D, Seibt A, GE KN, Galla HJ, Schroten H. Strain-dependent disruption of blood-cerebrospinal fluid barrier by Streptoccocus suis in vitro. FEMS Immunol Med Microbiol. 2005;44:25–34. doi: 10.1016/j.femsim.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Smith TC, Capuano AW, Boese B, Myers KP, Gray GC. Exposure to Streptococcus suis among US swine workers. Emerg Infect Dis. 2008;14:1925–1927. doi: 10.3201/eid1412.080162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Wang Y, Xu J, Yang W. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12:914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE. 2007;2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AA, Loeffen PL, Berg AJ van den, Storm PK. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect Immun. 1994;62:1742–1748. doi: 10.1093/benz/9780199773787.article.b00034458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot-Herault F, Morvan H, Keribin AM, Gottschalk M, Kobisch M. Production of muraminidase-released protein (MRP), extracellular factor (EF) and suilysin by field isolates of Streptococcus suis capsular types 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet Res. 2000;31:473–479. doi: 10.1051/vetres:2000133. [DOI] [PubMed] [Google Scholar]
- Segura M, Gottschalk M. Extracellular virulence factors of streptococci associated with animal diseases. Front Biosci. 2004;9:1157–1188. doi: 10.2741/1287. [DOI] [PubMed] [Google Scholar]
- Tikkanen K, Haataja S, Finne J. The galactosyl-(alpha 1-4)-galactose-binding adhesin of Streptococcus suis: occurrence in strains of different hemagglutination activities and induction of opsonic antibodies. Infect Immun. 1996;64:3659–3665. doi: 10.1128/iai.64.9.3659-3665.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Punaro MC, Segura M, Plante MM, Lacouture S, Rivest S, Gottschalk M. Streptococcus suis serotype 2, an important swine and human pathogen, induces strong systemic and cerebral inflammatory responses in a mouse model of infection. J Immunol. 2007;179:1842–1854. doi: 10.4049/jimmunol.179.3.1842. [DOI] [PubMed] [Google Scholar]
- de GA, Buys H, Verhaar R, Dijkstra J, van AL, Smith HE. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect Immun. 2002;70:1319–1325. doi: 10.1128/IAI.70.3.1319-1325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baums CG, Kaim U, Fulde M, Ramachandran G, Goethe R, Valentin-Weigand P. Identification of a novel virulence determinant with serum opacification activity in Streptococcus suis. Infect Immun. 2006;74:6154–6162. doi: 10.1128/IAI.00359-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruening P, Fulde M, Valentin-Weigand P, Goethe R. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J Bacteriol. 2006;188:361–369. doi: 10.1128/JB.188.2.361-369.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterhoff N, Goethe R, Gruening P, Rohde M, Kalisz H, Smith HE, Valentin-Weigand P. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the Arginine Deiminase system of Streptococcus pyogenes. J Bacteriol. 2002;184:6768–6776. doi: 10.1128/JB.184.24.6768-6776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M, Brady LJ, Progulske-Fox A, Hillman JD. IVIAT: a novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 2000;8:336–339. doi: 10.1016/S0966-842X(00)01775-3. [DOI] [PubMed] [Google Scholar]
- Rollins SM, Peppercorn A, Hang L, Hillman JD, Calderwood SB, Handfield M, Ryan ET. In vivo induced antigen technology (IVIAT) Cell Microbiol. 2005;7:1–9. doi: 10.1111/j.1462-5822.2004.00477.x. [DOI] [PubMed] [Google Scholar]
- Salim KY, Cvitkovitch DG, Chang P, Bast DJ, Handfield M, Hillman JD, de Azavedo JC. Identification of group A Streptococcus antigenic determinants upregulated in vivo. Infect Immun. 2005;73:6026–6038. doi: 10.1128/IAI.73.9.6026-6038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M, Kudva IT, Griffin RW, Dodson AW, McManus B, Krastins B, Sarracino D, Progulske-Fox A, Hillman JD, Handfield M, Tarr PI, Calderwood SB. Use of in vivo-induced antigen technology for identification of Escherichia coli O157:H7 proteins expressed during human infection. Infect Immun. 2005;73:2665–2679. doi: 10.1128/IAI.73.5.2665-2679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JB, Baresch-Bernal A, Rollins SM, Alam A, LaRocque RC, Bikowski M, Peppercorn AF, Handfield M, Hillman JD, Qadri F, Calderwood SB, Hohmann E, Breiman RF, Brooks WA, Ryan ET. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect Immun. 2006;74:5161–5168. doi: 10.1128/IAI.00488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang L, John M, Asaduzzaman M, Bridges EA, Vanderspurt C, Kirn TJ, Taylor RK, Hillman JD, Progulske-Fox A, Handfield M, Ryan ET, Calderwood SB. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc Natl Acad Sci USA. 2003;100:8508–8513. doi: 10.1073/pnas.1431769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethe G, Nau R, Wellmer A, Hakenbeck R, Reinert RR, Heinz HP, Zysk G. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol Lett. 2001;205:99–104. doi: 10.1111/j.1574-6968.2001.tb10931.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE. 2007;2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry AM, Lock RA, Hansman D, Paton JC. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- Wei W, Ding GH, Wang XJ, Sun JC, Tu K, Hao P, Wang C, Cao ZW, Shi TL, Li YX. A comparative genome analysis of streptococcus suis. Chinese Science Bulletin. 2006;51:808–818. [Google Scholar]
- Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol. 2000;76:259–272. doi: 10.1016/S0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/A:1005870317757. [DOI] [PubMed] [Google Scholar]
- Madsen LW, Bak H, Nielsen B, Jensen HE, Aalbaek B, Riising HJ. Bacterial colonization and invasion in pigs experimentally exposed to Streptococcus suis serotype 2 in aerosol. J Vet Med B Infect Dis Vet Public Health. 2002;49:211–215. doi: 10.1046/j.1439-0450.2002.00491.x. [DOI] [PubMed] [Google Scholar]
- Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol Microbiol. 2003;49:105–116. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- Zygmunt MS, Hagius SD, Walker JV, Elzer PH. Identification of Brucella melitensis 16 M genes required for bacterial survival in the caprine host. Microbes Infect. 2006;8:2849–2854. doi: 10.1016/j.micinf.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Zhang H, Niesel DW, Peterson JW, Klimpel GR. Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect Immun. 1998;66:5196–5201. doi: 10.1128/iai.66.11.5196-5201.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DJ, Smith A, VanWagoner TM, Seale TW, Whitby PW, Stull TL. Lipoprotein e (P4) of Haemophilus influenzae: role in heme utilization and pathogenesis. Microbes Infect. 2007;9:932–939. doi: 10.1016/j.micinf.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YR, Lee SE, Kim CM, Kim SY, Shin EK, Shin DH, Chung SS, Choy HE, Progulske-Fox A, Hillman JD, Handfield M, Rhee JH. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun. 2003;71:5461–5471. doi: 10.1128/IAI.71.10.5461-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini FC, Groisman EA. Two-component regulatory systems can interact to process multiple environmental signals. J Bacteriol. 1996;178:6796–6801. doi: 10.1128/jb.178.23.6796-6801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr--a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Milenbachs AA, Brown DP, Moors M, Youngman P. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol Microbiol. 1997;23:1075–1085. doi: 10.1046/j.1365-2958.1997.2711634.x. [DOI] [PubMed] [Google Scholar]
- Hofmeister AE, Textor S, Buckel W. Cloning and expression of the two genes coding for L-serine dehydratase from Peptostreptococcus asaccharolyticus: relationship of the iron-sulfur protein to both L-serine dehydratases from Escherichia coli. J Bacteriol. 1997;179:4937–4941. doi: 10.1128/jb.179.15.4937-4941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Gardner DJ, Long RD, Welty DM, Barry WT, Johnson CA, Parkins LD, Wright FA, Musser JM. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. 3. ColdSpring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Charland N, Jacques M, Lacouture S, Gottschalk M. Characterization and protective activity of a monoclonal antibody against a capsular epitope shared by Streptococcus suis serotypes 1, 2 and 1/2. Microbiology. 1997;143(Pt 11):3607–3614. doi: 10.1099/00221287-143-11-3607. [DOI] [PubMed] [Google Scholar]
- Berthelot-Herault F, Cariolet R, Labbe A, Gottschalk M, Cardinal JY, Kobisch M. Experimental infection of specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can J Vet Res. 2001;65:196–200. [PMC free article] [PubMed] [Google Scholar]
- Berthelot-Herault F, Gottschalk M, Morvan H, Kobisch M. Dilemma of virulence of Streptococcus suis: Canadian isolate 89-1591 characterized as a virulent strain using a standardized experimental model in pigs. Can J Vet Res. 2005;69:236–240. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Swine convalescent sera preparation. The data provided represent the preparation of swine convalescent sera. * Time-point of antibody check. ‡ Sacrificed and serum collection. † Number of recovered pigs, and antisera were used as convalescent sera for IVIAT selection. § sera were collected and used as negative control.


