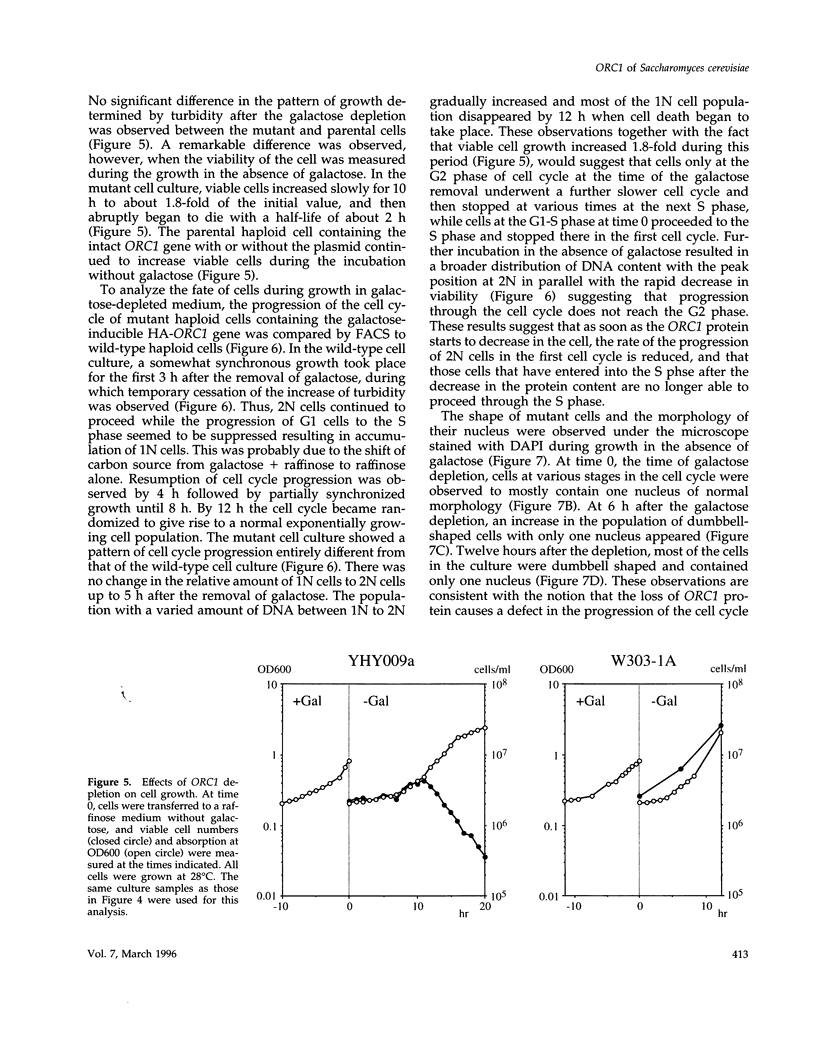
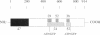Abstract
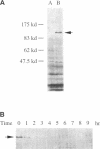
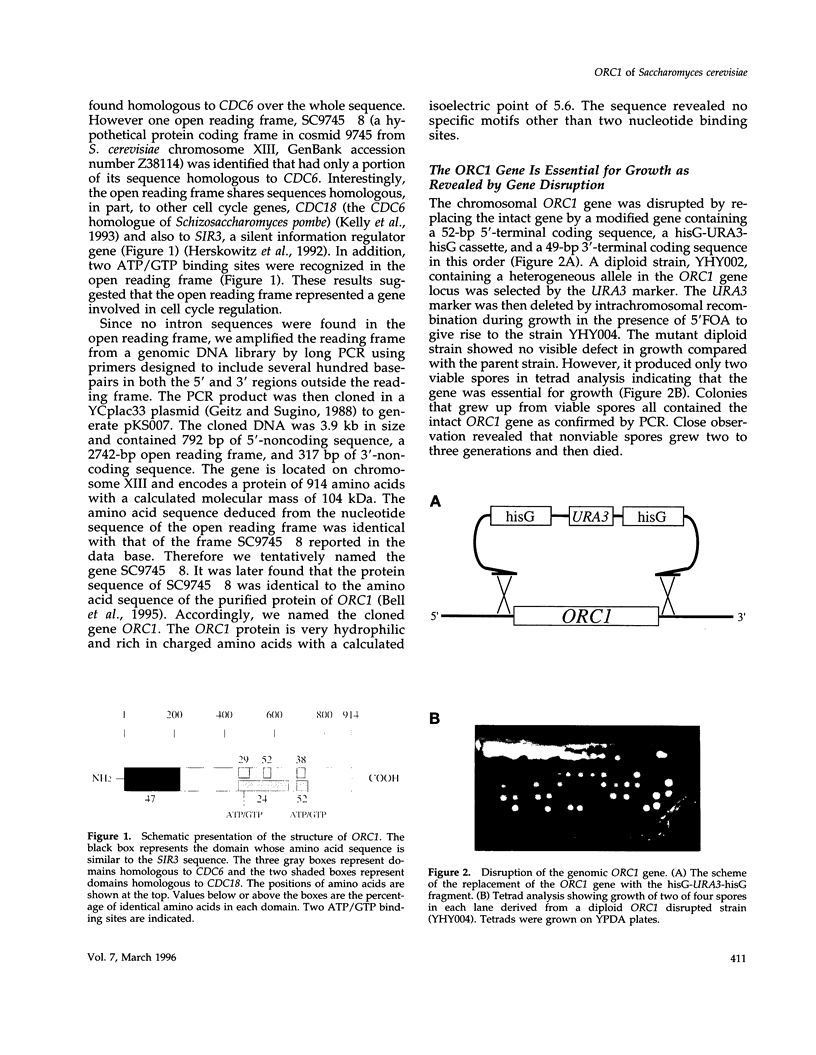
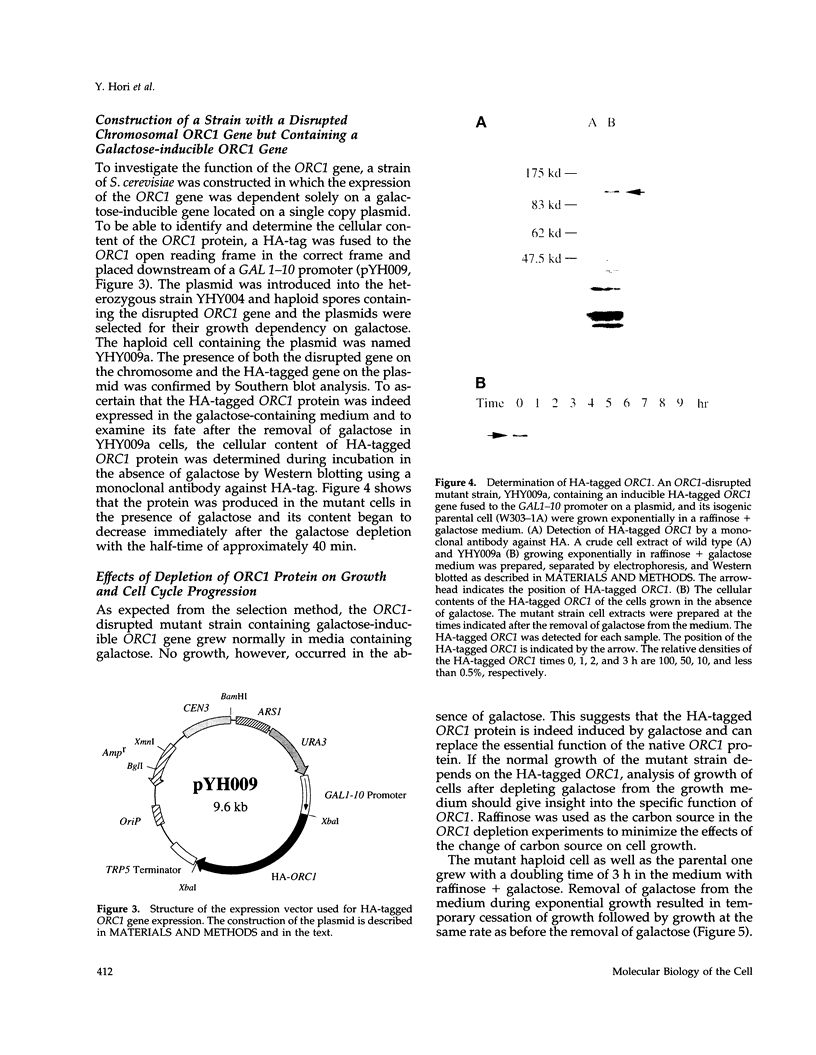
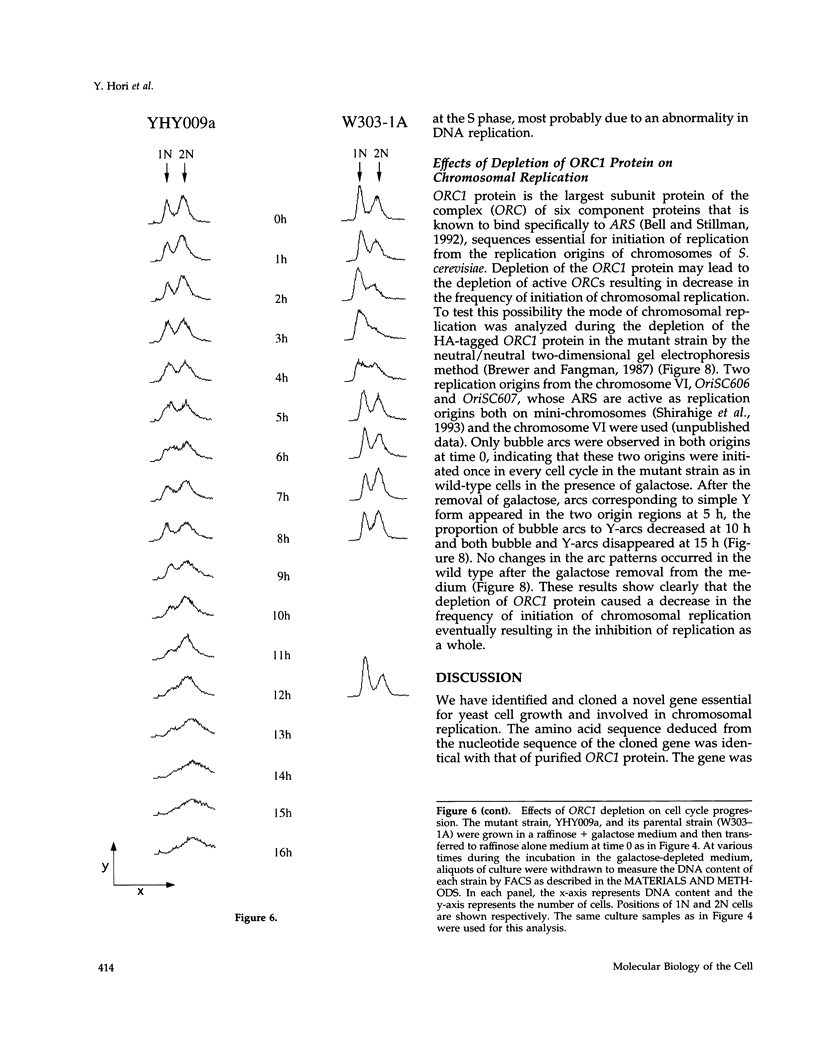
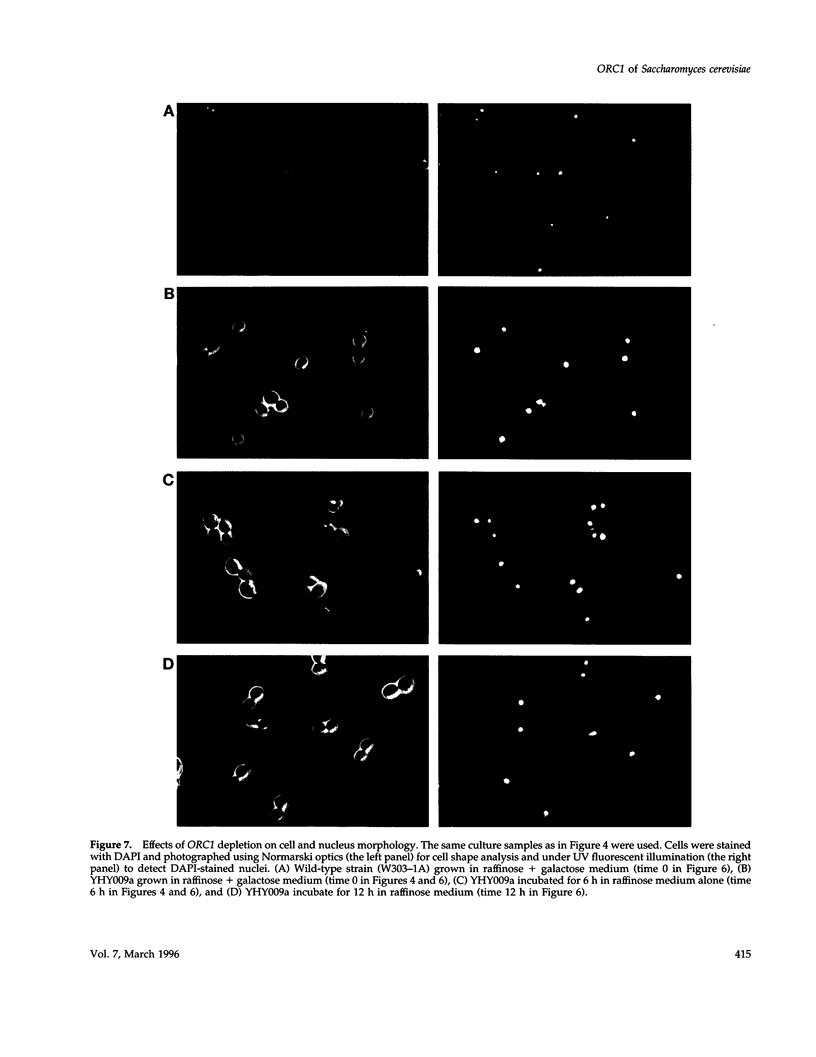
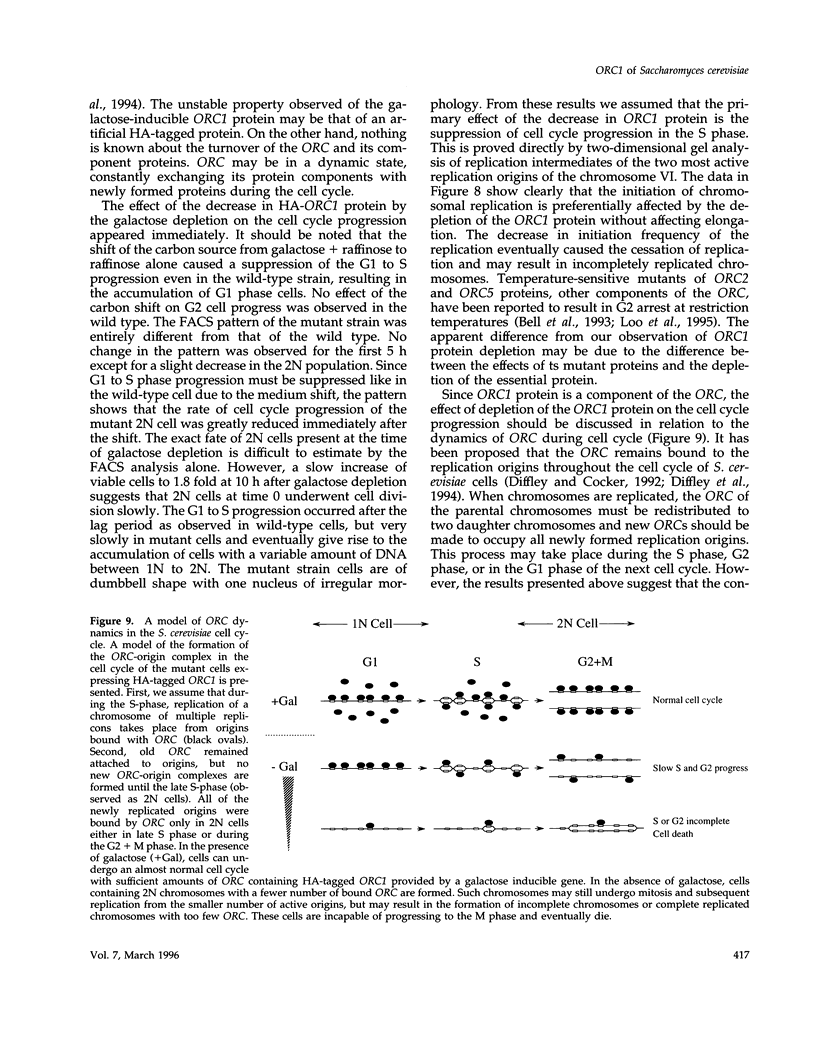
A novel cell cycle gene was identified by a computer search for genes partly homologous to known CDC genes, CDC6 of Saccharomyces cerevisiae and CDC18 of Schizosaccharomyces pombe, using the nucleotide sequence data base for S. cerevisiae produced by the Yeast Sequencing Project. The protein sequence coded by the cloned gene was found to be identical to that of purified ORC1 protein. Disruption of the gene and subsequent tetrad analysis revealed that the gene was essential for growth. The function of the gene product was analyzed by depleting the protein from the cell using a mutant haploid strain containing the disrupted ORC1 gene on the chromosome and a galactose-inducible gene coding for HA-tagged ORC1 protein on a single copy plasmid. The HA-tagged protein was expressed during growth in the presence of galactose but began to decrease rapidly upon depletion of galactose. Analysis of the cell cycle progression of the mutant cells by FACS after the removal of galactose from the medium, and microscope observations of cells and their nuclei revealed that the normal progression of 2N cells was immediately impeded as the ORC1 protein started to decrease. This was blocked completely in the cells that had progressed to the S phase under conditions deficient in ORC1 protein followed by cell death. Two-dimensional gel analysis of the replication intermediates after the galactose removal revealed that the depletion of ORC1 protein caused a decrease in the frequency of initiation of chromosomal replication, eventually resulting in the inhibition of replication as a whole. The function of the ORC1 protein in the cell cycle progression of S. cerevisiae is discussed in light of current information on ORC.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Mitchell J., Leber J., Kobayashi R., Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995 Nov 17;83(4):563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Bueno A., Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992 Jun;11(6):2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Epstein C. B., Cross F. R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992 Sep;6(9):1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Loo S., Dillin A., Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995 Apr 15;9(8):911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Hogan E., Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. A general method for preparing chromatin containing intact DNA. EMBO J. 1985 Apr;4(4):913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Williamson D. H. An alkaline sucrose gradient analysis of the mechanism of nuclear DNA synthesis in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1978 Aug 17;164(2):217–225. doi: 10.1007/BF00267387. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993 Jul 30;74(2):371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Li J. J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993 Dec 17;262(5141):1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Liang C., Weinreich M., Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995 Jun 2;81(5):667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Loo S., Fox C. A., Rine J., Kobayashi R., Stillman B., Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995 Jun;6(6):741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a 'reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995 Aug 1;14(15):3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Dowell S. J., Diffley J. F. Recent developments in the initiation of chromosomal DNA replication: a complex picture emerges. Biochim Biophys Acta. 1994 Apr 6;1217(3):239–256. doi: 10.1016/0167-4781(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Ina S. DNA replication of histone gene repeats in Drosophila melanogaster tissue culture cells: multiple initiation sites and replication pause sites. Mol Cell Biol. 1993 Jul;13(7):4098–4106. doi: 10.1128/mcb.13.7.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T., Ina S. Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: multiple replication origins in a broad zone. Mol Cell Biol. 1994 Nov;14(11):7394–7403. doi: 10.1128/mcb.14.11.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K., Iwasaki T., Rashid M. B., Ogasawara N., Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Aug;13(8):5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]