Abstract
In mammals the retina contains photoactive molecules responsible for both vision and circadian photoresponse systems. Opsins, which are located in rods and cones, are the pigments for vision but it is not known whether they play a role in circadian regulation. A subset of retinal ganglion cells with direct projections to the suprachiasmatic nucleus (SCN) are at the origin of the retinohypothalamic tract that transmits the light signal to the master circadian clock in the SCN. However, the ganglion cells are not known to contain rhodopsin or other opsins that may function as photoreceptors. We have found that the two blue-light photoreceptors, cryptochromes 1 and 2 (CRY1 and CRY2), recently discovered in mammals are specifically expressed in the ganglion cell and inner nuclear layers of the mouse retina. In addition, CRY1 is expressed at high level in the SCN and oscillates in this tissue in a circadian manner. These data, in conjunction with the established role of CRY2 in photoperiodism in plants, lead us to propose that mammals have a vitamin A-based photopigment (opsin) for vision and a vitamin B2-based pigment (cryptochrome) for entrainment of the circadian clock.
Keywords: cryptochromes, retina, suprachiasmatic nucleus
Biological activities are regulated by the daily light–dark cycles (circadian rhythm) in species ranging from cyanobacteria to humans (1–5). The circadian clock that regulates this response to the day–night cycle has at least three components: a photoactive pigment (chromophore) that senses the light and transmits the signal, the clock that oscillates with about 24-hr (circadian) periodicity, and the clock-controlled genes that generate the physiological and behavioral oscillation in response to the changes in the phase and amplitue of the clock (1–5).
In recent years, important progress has been made in understanding the molecular basis of circadian rhythm. In particular, clock genes, which are defined as light-responsive genes with an approximately 24-hr oscillatory transcription period and autoregulatory loops, have been identified in several organisms. The frq gene of Neurospora crassa (6), the per and tim genes of Drosophila melanogaster (7–9) have been isolated and characterized and appear to exhibit the requisite characteristic (self-oscillatory) of circadian clock gene. The mouse clock gene, which does not exhibit an overt oscillatory expression pattern but which is an essential component of the clock mechanism, has also been cloned and characterized (10). All these genes have been shown to encode transcription factors or to have sequence motifs suggestive of a transcription factor. Finally, the recent cloning of the mouse and human homologs of the Drosophila per gene (11–13) strongly suggest the conservation of the basic clock mechanism during evolution. Clearly, these and other related studies have made significant inroads toward molecular description of the clock component of the circadian rhythm. Similarly, several clock-controlled genes for executing the circadian response (output) have been identified in N. crassa (14), Arabidopsis thaliana (15), D. melanogaster (16, 17), and mouse (18–20). In contrast to this wealth of information on the clock and output components of the timekeeping mechanism at the molecular level, the nature of the photosensory molecules that detect the light signal is not known. Because severing the optic nerve abolishes the ability for light entrainment in mammals, it is generally accepted that the eye contains the photopigments for both visual (imaging) and circadian systems (21, 22). However, in mice with a retinal degeneration syndrome (rd) in which all of the rod photoreceptor cells and virtually all of the cone photoreceptors are destroyed light entrainment of the circadian rhythm is normal (23, 24). Similarly, many blind persons with no conscious perception of light exhibit normal photic entrainment of the circadian rhythm (25). Although these photoresponses could be ascribed to residual opsins that may exist in these subjects, the data are also consistent with the presence of a heretofore unknown photoactive pigment in the nonrod noncone types of retinal cells and with the sole role of the entrainment of circadian clock. We have obtained data that indicate that this pigment is the mammalian homolog of the plant blue-light photoreceptor, the cryptochrome.
Plant cryptochromes are 60- to 70-kDa proteins with high degree of sequence homology to the light-activated DNA repair enzymes called DNA photolyases (26–30). Like photolyases, cryptochromes also contain FAD and a pterin as chromophores; however, the cryptochromes have no repair activity (27, 31). Instead, in A. thaliana it has been shown that the two cryptochrome genes (CRY1 and CRY2) that encode highly homologous proteins (29, 30) regulate the plant’s response (inhibition of hypocotyl elongation) to blue light in a partially overlapping manner (32, 33). Of special significance, it was also found that the CRY2 gene is involved in photoperiodism of flowering time in Arabidopsis (34), raising the possibility of a circadian role for this class of proteins, at least in plants.
Recently, two genes with high degree of sequence homology to photolyase/plant blue-light photoreceotor gene family were identified in humans (35–38). Like the plant blue-light photoreceptors, the human cryptochrome homologs were found to contain FAD and a pterin as chromophore/cofactors but exhibit no DNA repair activity (37). Hence, these two human proteins were named cryptochromes 1 and 2 (CRY1 and CRY2) and it was suggested that these pigments may function as photoreceptors for setting the circadian clock in humans and other mammals (37, 39). Herein we present histologic and physiologic evidence that these proteins are most likely the circadian photoreceptors in mammals.
MATERIALS AND METHODS
In Situ Hybridization.
PCR fragments of mouse Cry1 (GenBank accession no. AB000777), Cry2 (accession no. AB003433), and opsin (accession no. M55171), containing positions 1,074–1,793, positions 1,040–1,649, and positions 18 in exon 3 to 12 in exon 5, respectively, were subcloned into pBluescript SK+ plasmid (CLONTECH). The mouse Cry1 and Cry2 genes have 97% and 95% sequence identity to the corresponding human genes. 35S-labeled sense and antisense RNA probes were generated from these plasmids with T7 and T3 RNA polymerase. Animals (male C57BL mice) were maintained on a 12-hr light/12-hr dark cycle. Sample preparation, hybridization, and visualization were carried out as described elsewhere (11). Animals were sacrificed by decapitation. Frozen tissue sections (20 μm thick) were fixed for 20 min in 4% formaldehyde in phosphate buffer. Sections were treated with proteinase K (10 μg/ml) for 10 min, acetylated with acetic anhydride in 0.1 M triethanolamine, and dehydrated. The 35S-labeled sense and antisense RNA probes in hybridization buffer (50% formamide/10% Dextran sulfate/20 mM Tris⋅HCl, pH 8.0/0.3 M NaCl/0.2% sarcosyl/0.02% salmon sperm DNA/1× Denhardt’s solution) were placed on the sections and then incubated at 55°C overnight. The sections were washed at 65°C in 50% formamide/2× SSC/0.1 M DTT for 30 min. Sections were then treated with RNase A (1 μg/ml) for 30 min at 37°C. Subsequently, sections were washed in 50% formamide/2× SSC/0.1 M DTT for 30 min at 65°C. Slides were dipped in nuclear emulsion (Kodak NTB-2) and exposed for 2 weeks at 4°C.
Analysis of RNA Expression Levels.
The Northern blot analysis for mCry1 and mCry2 were performed with multiple mouse tissue filters from CLONTECH. For analysis of the in situ hybridization data, the autoradiograms were scanned with a Molecular Dynamics Conmputing Densitometer Series 300 and analyzed with image quant software (Molecular Dynamics).
RESULTS
Tissue Expression Patterns of mCry1 and mCry2.
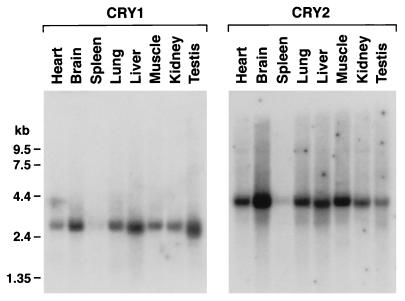
CRY1 is expressed in most human tissues (38). Because we were interested in the expression of cryptochromes in tissues controlling circadian rhythm, we analyzed expression of both mCry1 and mCry2 in various mouse tissues by Northern blot hybridization. Fig. 1 shows that both mCry1 and mCry2 are expressed in most mouse tissues, although in different patterns and at widely different levels. This widespread expression pattern, although not particularly revealing regarding the role of these genes in circadian regulation, is not necessarily inconsistent with CRY1 and CRY2 being circadian photoreceptors because recent studies have revealed that other circadian clock proteins are also expressed throughout the body (40–42). In addition, supporting evidence for CRY1 and CRY2 being circadian photoreceptors were obtained in this study by a detailed analysis in the two key organs associated with circadian rhythm in humans and mice, the eye and the suprachiasmatic nucleus (SCN) (21, 22). These experiments are detailed below.
Figure 1.
Northern blot analysis of mCry1 and mCry2 mRNAs in various tissues. Mouse multiple tissue Northern blots (CLONTECH) were hybridized to random-primed 32P-labeled probes.
Expression of Cryptochromes in the Retina.
Fig. 2 shows the expression of mCry1 and mCry2 in the mouse retina probed by in situ hybridization. As a control, we used opsin, which is highly expressed in the rod photoreceptor cells. In agreement with earlier reports (43), the rhodopsin transcript is localized mostly to the inner segment layer and to a lesser extent to the outer nuclear layer of the retina. Similarly, the cone opsins transcripts are located in the outer nuclear layer (43). The expression patterns of the mouse cryptochromes are in sharp contrast to those of opsins. There is no detectable expression of mCry1 and mCry2 in inner segment or outer nuclear layers. Instead, these genes are expressed in the ganglion cell layer (GCL) and inner nuclear layer (INL) and evenly distributed in central and peripheral retina (Fig. 2). The expression level of mCry2 is particularly high in these layers, indicating that mCry2 plays a more predominant role in the retina compared with mCry1. Genetic and physiologic studies have revealed that a subset of the ganglion cells are at the origin of the retinohypothalamic pathway that transmits the light signal to the master circadian clock in the SCN region of the hypothalmus (21, 22, 44). Hence, the expression patterns of cryptochromes and in particular that of mCry2 are entirely consistent with the pattern expected for the photoreceptor pigment of the circadian clock.
Figure 2.
Expression of mCry1 and mCry2 in mouse retina. In situ hybridization was performed with 35S-labeled antisense RNA of the appropriate genes. (A) Dark-field microgram showing uniform expression of both mCry1 and mCry2 in the GCL and INL from macular (M) region to peripheral retina. Silver grains detected in the outer segment (OS) are due to absorption of light by residual pigment epithelium that remained in these retinal preparations. (Bar = 200 μm.) (B) Bright-field microgram. The mRNA locations of CRY1, CRY2, and opsin are compared. Note that mCry1 and mCry2 are expressed in the GCL and INL only. Arrows indicate clusters of ganglion cells that express Cry1 and Cry2. In contrast, opsin is highly expressed in the inner segment (IS) and the outer nuclear layer (ONL) and is not detected in GCL and INL. The other layers of the retina are also indicated as follows: IPL, inner plexiform layer; OPL, outer plexiform layer. (Bar = 30 μm.)
Circadian Oscillation of mCry1 Expression in the SCN.
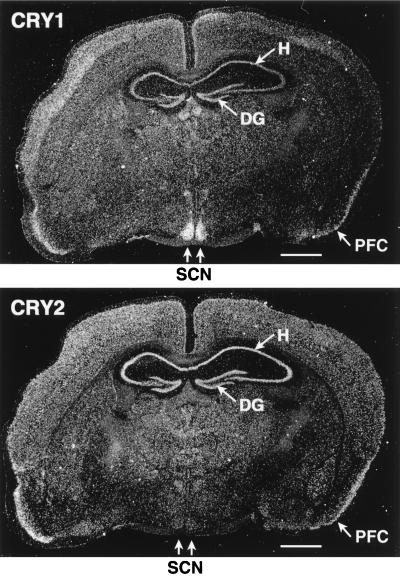
After establishing the specific expression of the cryptochromes within the region of the retina implicated in photoentrainment, we wished to examine their expression in the SCN. As seen in Fig. 1, Northern blot analysis shows that mCry2 is expressed at higher level than mCry1 in the total brain tissue. However, analysis of expression in various regions of the brain by in situ hybridization reveals a striking contrast between the two cryptochromes. Although mCry2 is expressed at relatively high levels throughout the brain, mCry1 is most abundant in the SCN where mCry2 expression is barely detectable (Fig. 3). In fact the expression pattern of mCry1 in the SCN and other areas of the brain is nearly identical to that of the circadian clock protein gene mPer1 (11–13). Thus the cell-specific expressions of mCry1 and mCry2 are entirely consistent with these proteins being part of the circadian clock system. Furthermore, it appears that there is a division of labor between the two proteins, with CRY2 being the main photosensor in the retina and CRY1 playing a separate function in the SCN. The two proteins are 73% homologous and have very similar photochemical properties (37) and at present the molecular basis of this apparent functional division is not known.
Figure 3.
Distribution of mCry1 and mCry2 in the mouse brain. Mouse coronal brain sections (made at zeitgeber time = ZT6) passing through the SCN were probed with mCry1 and mCry2 antisense RNA. Both Cry1 and Cry2 are abundant in all cerebral cortical layers but are particularly abundant in the pyramidal cell layer of the hippocampus (H), the granular cell layer of dentate gyrus (DG), and the pyramidal cell layer of the piriform cortex (PFC). Note that the strongest signal of Cry1 was observed in the SCN where the Cry2 signal is marginal. (Bar = 1 mm.)
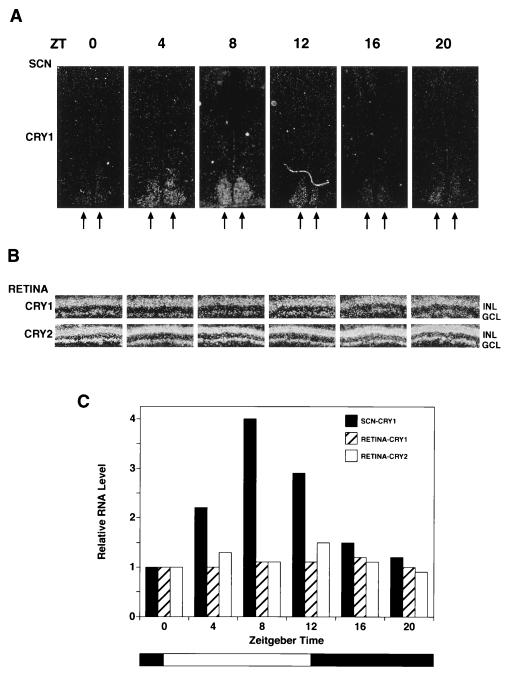
The transcription of the clock genes frq in N. crassa (45), per (46), and tim (47) in D. melanogaster and mPer1 and mPer2 in mice (11–13) exhibit circadian oscillation. Hence, we were interested in finding out whether or not the transcription of mCry1 and mCry2 were regulated in a circadian manner. We performed in situ hybridization on the retinas and the brains of mice kept under 12-hr light/12-hr dark cycles. Fig. 4 shows that transcription of mCry1 and mCry2 in the retina does not oscillate. In contrast, mCry1, which is expressed at high levels in the SCN, shows a clear circadian pattern with maximum expression in the light phase and becoming nearly undetectable after 8 hr in the dark. The same pattern of oscillation was observed (data not shown) when the mice were kept in constant darkness, as is expected of a true circadian regulator (1–5).
Figure 4.
Circadian expression of Cry1 and Cry2 mRNA in the mouse SCN and retina. Mice kept under a 12-hr light/12-he dark cycle were sacrificed every 4 hr, and retinal sections and coronal brain sections encompassing the SCN were prepared for in situ hybridization using 35S-labeled mCry1 and mCry2 RNA probes. Only the results of mCry1 hybridization to SCN are shown because the SCN signal with mCry2 was very weak at all Zeitgeber time (ZT) points and not amenable to quantitative analysis. (A) Expression of mCry1 in SCN. In situ hybridization of mCry1 with coronal brain sections passing through the SCN region are shown. Seven sections (20 μm) were made for each point and the one with the strongest signal for each point are shown in these micrograms. The double arrows indicate the SCN. (B) Expression of mCry1 and mCry2 in the retina. Retinal sections from the central part of the retina were hybridized with mCry1 and mCry2. The GCL and the INL are indicated. (C) Quantitative analyses of Cryptochrome expression in the SCN and the retina as a function of light/dark cycles (34). Grain density from autoradiograms of all section containing SCN were measured and the total grain density for each time point is expressed relative to zeitgeber time (ZT) = 0 value, which represents the values just before turning on the light (at 0800 hr). For retina, for each time point, the grain density was measured in the GCL and INL of the entire retinal region present in the autoradiogram and normalized for the length of the retina present in each section. The intensity for mCry1 and mCry2 are expressed relative to the values of mCry1 and mCry2 at ZT = 0, respectively. In absolute terms, the grain density with mCry2 probe was about 3-fold higher than that obtained with the mCry1 probe at all times.
DISCUSSION
The findings reported in this article suggest that flavin-based blue-light photoreceptors (cryptochromes) are the photoreceptors for circadian clock in mammals. Presently, this conclusion might be considered provisional for lack of a mammalian cryptochrome mutant exhibiting the predicted circadian rhythm defects. However, the following considerations lead us to conclude that CRY1 and CRY2 are circadian photoreceptors even in the absence of supporting genetic data.
(i) The rd/rd mice, which are totally defective in rods and severely depleted in cone photoreceptors, are visually blind but have normal response to photic entrainment of circadian rhythm (23, 24). The GCL and INL regions of the retina where CRY1 and CRY2 are expressed are intact in these animals (23).
(ii) Recently, it has been shown that CRY2 in A. thaliana is involved in photoperiodism of flowering time, thus providing the first evidence for the role of this class of proteins in photic entrainment of an organism (34).
(iii) It has been known for some time that a genetic defect in riboflavin synthesis severely inhibits photoentrainment of circadian rhythm in N. crassa (48), indicating that the photoreceptor in this organism is not an opsin-based pigment but, like cryptochromes, is a flavoprotein.
(iv) Although the expression of mCry2 and mCry1 in the retina is consistent with a photosensor/phototransducer role for these pigments, mCry1 is expressed at high level in the SCN as well. In contrast to the wealth of evidence that the pineal gland is the photoreceptor organ in most nonmammalian vertebrates (21, 22), there are no reports indicating that the SCN may act as the photosensor for circadian entrainment in mammals in an apparent contrast to our proposal that CRY1, which is expressed at high level and in an oscillatory manner in the SCN, is a circadian photoreceptor. However, there are a number of studies that show that light can in fact penetrate into and propagate in sufficient quantities through the human brain and other internal organs (see ref. 49). Hence, it is plausible that light reaches the SCN to excite CRY1 in addition to the ready excitation of CRY1 and CRY2 in the retina. What unique physiologic function such excitation may elicit remains to be elucidated.
(v) Opsins, which are the photoactive pigments responsible for vision (image forming), are expressed almost exclusively in the retina of mammals. Although some opsins have been found in the pineal gland at very low levels, all the evidence suggests that pineal opsins play no role in photoentrainment in mammals (see ref. 5). In contrast, CRY1 and CRY2 are expressed in many tissues even though it is generally accepted that optic input is necessary for circadian entrainment in mammals (1–5, 23, 24). These facts, again superficially, would argue for opsin(s) rather than cryptochromes being the circadian photoreceptor. In fact, closer examination of the relevant data lends further support to the notion that cryptochromes rather than opsins are the photoreceptors for the circadian clock. First, recent reports indicate that in various organisms the entire circadian machinery including the photoreceptor, the oscillator, and the output components are expressed throughout the body and that peripheral tissues and organs contain photoentrainable circadian clocks independent of the master circadian clock located in the brain (40–42). Indeed, the other components of the mammalian circadian clock that have been identified so far, the clock (10) and Per (11–13) genes are, like the CRY1 and CRY2 genes, ubiquitously expressed in most tissues. Second, the widely accepted notion that photic input through the eye is essential for light entrainment of circadian rhythm is mostly based on studies that dealt with physiological and behavioral functions largely controlled by the brain. Even this view has recently been challenged by a report that claimed that the phases and amplitudes of circadian oscillation of human body temperature and blood melatonin concentration could be altered by light exposure behind the knees (50). In fact, we have found significant expression of CRY1 and CRY2 in human skin (data not shown), supporting the view that skin is capable of light entrainment of circadian clock locally. Whether or not it can induce a global change at the organism level is outside the scope of our study.
(vi) Finally, action spectrum data for light entrainment of locomotor activity in mice has been taken as evidence that the photopigment for the circadian clock is rhodopsin (51, 52). These studies found a visible action spectrum peak at around 500 nm and, hence, concluded that an opsin-based photopigment was the light-sensitive material in circadian photoentrainment. However, such experiments do not necessarily exclude cryptochromes (absorption maxima at 420 nm) as the circadian photoreceptors. Disparity between the absorption spectrum of a chromophore responsible for a specific biological phenomenon and the action spectrum of that phenomenon is a common occurrence in photobiology and can be caused by many factors including shielding by other pigments or light scattering (53). In fact in one of these action spectrum studies of circadian locomotor activity, it was found that 357 nm was more effective than 511 nm, which was considered to be the maximum in the visible range (52). Although this latter finding too was ascribed to absorbance by the UV cones present in the retina, the data could equally be taken to be evidence for cryptochromes as the circadian photoreceptors. Thus the whole animal action spectrum data do not contradict but in fact may support our conclusion regarding the function of CRY1 and CRY2 in mammals.
In summary, we propose that the retina of mammals and perhaps of other animals contains two classes of photopigments: vitamin A-based opsins that are used for vision and vitamin B2-based cryptochromes that are the photoreceptors for circadian clock.
Acknowledgments
We thank C. Petit, C. Selby, and R. Thresher for useful discussions and S. Kay, C. Lin, M. Menaker, D. Mu, and J. Takahashi for comments. This work was supported by National Institutes of Health Grant GM31082.
ABBREVIATIONS
- SCN
suprachiasmatic nucleus
- GCL
ganglion cell layer
- INL
inner nuclear layer
References
- 1.Kay S A, Millar A J. Cell. 1995;83:361–364. doi: 10.1016/0092-8674(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi J S. Annu Rev Neurosci. 1995;18:531–553. doi: 10.1146/annurev.ne.18.030195.002531. [DOI] [PubMed] [Google Scholar]
- 3.Hall J C. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 4.Dunlap J C. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 5.Ronnenberg T, Foster R G. Photochem Photobiol. 1997;66:549–561. doi: 10.1111/j.1751-1097.1997.tb03188.x. [DOI] [PubMed] [Google Scholar]
- 6.Aronson B B, Johnson K A, Loros J J, Dunlap J C. Science. 1996;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 7.Bargiello T A, Jackson F R, Young M W. Nature (London) 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 8.Zehring W A, Wheeler D A, Reddy P, Konopka R J, Kyriacou C P, Rosbash M, Hall J C. Cell. 1984;39:369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- 9.Myers M P, Wager-Smith K, Wesley C S, Young M W, Sehgal A. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 10.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M, Steeves T D L, Vitaterna M H, Kornhauser J M, Lowrey P L, et al. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 13.Albrecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 14.Bell-Pedersen D, Shinohara M L, Loros J J, Dunlap J C. Proc Natl Acad Sci USA. 1996;93:13096–13700. doi: 10.1073/pnas.93.23.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar, A. J., Straumer, M., Chory, J., Chua, N. H. & Kay, S. A. Science 267, 1163–1166. [DOI] [PubMed]
- 16.Van Gelder R N, Bae H, Palazzolo M J, Kransnow M A. Curr Biol. 1995;5:1424–1436. doi: 10.1016/s0960-9822(95)00280-6. [DOI] [PubMed] [Google Scholar]
- 17.Rouyer F, Rachidi M, Pikielny C, Rosbash M. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronin N, Sagar S M, Sharp F R, Schwartz W J. Proc Natl Acad Sci USA. 1990;87:5959–5963. doi: 10.1073/pnas.87.15.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornhauser J M, Nelson D E, Mayo K E, Takahashi J S. Science. 1992;255:1581–1584. doi: 10.1126/science.1549784. [DOI] [PubMed] [Google Scholar]
- 20.Kornhauser J M, Mayo K E, Takahashi J S. Behav Genet. 1996;26:221–240. doi: 10.1007/BF02359382. [DOI] [PubMed] [Google Scholar]
- 21.Nelson R J, Zucker I. Comp Biochem Physiol A. 1981;69:145–148. [Google Scholar]
- 22.Foster R G, Provencio I, Hudson D, Fiske S, de Grip W, Menaker M. J Comp Physiol. 1991;169:39–50. doi: 10.1007/BF00198171. [DOI] [PubMed] [Google Scholar]
- 23.Provencio I, Wong S, Lederman A, Argamaso S M, Foster R G. Vision Res. 1994;34:1799–1806. doi: 10.1016/0042-6989(94)90304-2. [DOI] [PubMed] [Google Scholar]
- 24.Foster R G, Froehlich A, Argamaso-Hernan S M, McCall M A. Invest Ophalmol Visual Sci. 1995;36:S422. [Google Scholar]
- 25.Czeisler C A, Shanahan T L, Klerman E B, Martenz H, Brotman D J, Emens A B J, Klein T, Rizzo J F., III N Eng J Med. 1995;332:6–11. doi: 10.1056/NEJM199501053320102. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad M, Cashmore A R. Nature (London) 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 27.Malhorta K, Kim S T, Batschauer A, Dawut L, Sancar A. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 28.Small G D, Min B, Lefebvre P. Plant Mol Biol. 1995;28:443–454. doi: 10.1007/BF00020393. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman P D, Batschauer A, Hays J B. Mol Gen Genet. 1996;253:259–265. doi: 10.1007/s004380050321. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, Ahmad M, Chan J, Cashmore A R. Plant Physiol. 1996;110:1047. [Google Scholar]
- 31.Lin C, Robertson D E, Ahmad M, Raibekas A A, Jorns M S, Dutton P L, Cashmore A R. Science. 1995;269:948–950. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad M, Cashmore A R. Plant Mol Biol. 1996;30:851–861. doi: 10.1007/BF00020798. [DOI] [PubMed] [Google Scholar]
- 33.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore A R. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H, Yang H, Mockler T, Lin C. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 35.Adams M D, Kerlavage A R, Fleischmann R D, Fuldner R A, Bult C J, Lee N H, Kirkness E F, Weinstock K G, Gocayne J D, White O, et al. Nature (London) 1995;377:173–174. [PubMed] [Google Scholar]
- 36.Todo T, Ryo H, Yamamoto K, Inui T, Ayaki H, Nomura T, Ikenaga M. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 37.Hsu D S, Zhao X, Zhao S, Kazantsev A, Wang R P, Todo T, Wei Y F, Sancar A. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 38.van der Spek P J, Kobayashi K, Bootsma D, Takao M, Eker A P M, Yasui A. Genomics. 1996;37:177–182. doi: 10.1006/geno.1996.0539. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S, Sancar A. Photochem Photobiol. 1997;66:727–731. doi: 10.1111/j.1751-1097.1997.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 40.Plautz J D, Kaneko M, Hall J C, Kay S A. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 41.Hege D, Stranewsky R, Hall J C, Giebultowicz J M. J Biol Rhythms. 1997;12:330–338. doi: 10.1177/074873049701200402. [DOI] [PubMed] [Google Scholar]
- 42.Giebultowicz J M, Hege D M. Nature (London) 1997;386:664–665. doi: 10.1038/386664a0. [DOI] [PubMed] [Google Scholar]
- 43.Sun H, Macke J P, Nathans J. Proc Natl Acad Sci USA. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore R Y, Speh J C, Cord J P. J Comp Neurol. 1995;352:351–366. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 45.Crosthwaite S C, Loros J J, Dunlap J C. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 46.Hardin P E, Hall J C, Rosbash M. Nature (London) 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 47.Gekakis N, Saez L, Delahaye-Brown A, Myers M P, Sehgal A, Young M W, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 48.Paietta J, Sargent M L. Proc Natl Acad Sci USA. 1981;78:5573–5577. doi: 10.1073/pnas.78.9.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennisi E. Science. 1997;278:1560–1561. doi: 10.1126/science.278.5343.1560. [DOI] [PubMed] [Google Scholar]
- 50.Campbell S, Murphy P J. Science. 1998;279:396–399. doi: 10.1126/science.279.5349.396. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi J S, DeCoursey P J, Bauman L, Menaker M. Nature (London) 1984;308:186–188. doi: 10.1038/308186a0. [DOI] [PubMed] [Google Scholar]
- 52.Provencio I, Foster R G. Brain Res. 1995;694:183–190. doi: 10.1016/0006-8993(95)00694-l. [DOI] [PubMed] [Google Scholar]
- 53.Coohil T. Photochem Photobiol. 1981;33:941–945. doi: 10.1111/j.1751-1097.1981.tb05518.x. [DOI] [PubMed] [Google Scholar]