Abstract
Background and Aims
Understanding the genetic basis underlying domestication-related traits (DRTs) is important in order to use wild germplasm efficiently for improving yield, stress tolerance and quality of crops. This study was conducted to characterize the genetic basis of DRTs in soybean (Glycine max) using quantitative trait locus (QTL) mapping.
Methods
A population of 96 recombinant inbred lines derived from a cultivated (ssp. max) × wild (ssp. soja) cross was used for mapping and QTL analysis. Nine DRTs were examined in 2004 and 2005. A linkage map was constructed with 282 markers by the Kosambi function, and the QTL was detected by composite interval mapping.
Key Results
The early flowering and determinate habit derived from the max parent were each controlled by one major QTL, corresponding to the major genes for maturity (e1) and determinate habit (dt1), respectively. There were only one or two significant QTLs for twinning habit, pod dehiscence, seed weight and hard seededness, which each accounted for approx. 20–50 % of the total variance. A comparison with the QTLs detected previously indicated that in pod dehiscence and hard seededness, at least one major QTL was common across different crosses, whereas no such consistent QTL existed for seed weight.
Conclusions
Most of the DRTs in soybeans were conditioned by one or two major QTLs and a number of genotype-dependent minor QTLs. The common major QTLs identified in pod dehiscence and hard seededness may have been key loci in the domestication of soybean. The evolutionary changes toward larger seed may have occurred through the accumulation of minor changes at many QTLs. Since the major QTLs for DRTs were scattered across only six of the 20 linkage groups, and since the QTLs were not clustered, introgression of useful genes from wild to cultivated soybeans can be carried out without large obstacles.
Key words: Soybean, Glycine max, domestication related traits, QTL, hard seededness, seed size, pod dehiscence, twinning habit
INTRODUCTION
The morphological and physiological changes associated with domestication can be delimitated into adaptation syndromes resulting from natural or deliberate human selection (Harlan, 1992). Selection pressures associated with harvesting and planting have resulted in non-shattering characteristics, increases in seedling vigour and more rapid germination via loss or reduction of germination inhibitors, whereas human-selected changes have included larger inflorescences, larger seeds, thicker stems, more upright plants, etc., by intentionally selecting for higher yield. The genetic bases underlying distinctive differences between cultivated crops and their wild relatives have long been the subject of evolutionary interest to not only agronomists but also evolutionary geneticists because they have resulted from the different modes of natural and artificial selection (Lande, 1983; Gottlieb, 1984). The development of easily usable and polymorphic DNA markers has facilitated a mapping-based dissection of the genetic basis for domestication-related traits (DRTs) in many plant species such as aubergine (Doganlar et al., 2002), maize (Doebley et al., 1990; Doebley and Stec, 1993; Lauter and Doebley, 2002), pearl millet (Poncet et al., 2000, 2002), rice (Xiong et al., 1999; Cai and Morishima, 2002), sunflower (Burke et al., 2002), tomato (Grandillo and Tanksley, 1996), common bean (Koinange et al., 1996) and wheat (Peng et al., 2003).
By reviewing recent reports on quantitative trait locus (QTL) mapping of DRTs, Ross-Ibarra (2005) was able to identify three major patterns. First, the QTLs are not randomly or even uniformly distributed throughout the genome, but rather occur in apparently linked clusters in certain regions of the chromosome. Secondly, relatively few QTLs of large effect are involved in most DRTs across a variety of taxa. Thirdly, an extensive synteny exists among the QTLs of major effect among species which belong to Gramineae (Paterson et al., 1995) and Solanaceae (Doganlar et al., 2002).
Cultivated and wild soybeans belong to the same biological species, and are constituents of the primary gene pool of Glycine (Hymowitz, 2004). Ohashi (1982) classified cultivated and wild soybeans into Glycine max (L.) Merr. subsp. max Ohashi and G. max subsp. soja (Sieb. & Zucc.) Ohashi. Crosses between them generally produce viable and fertile progeny, although in some soja accessions there are chromosome interchanges which result in bridges and multivalent formation at meiosis that reduce the pollen and seed fertility in hybrids (Palmer et al., 1987, 2000; Singh and Hymowitz, 1988). Cultivated and wild soybeans differ in a set of various morphological and physiological characteristics collectively designated as the domestication syndrome (Broich and Palmer, 1980, 1981). The typical cultivated phenotype displays a bush-type growth habit with a stout primary stem and sparse branches, bearing large seeds with variable seed coat colours, while the wild phenotype is a procumbent or climbing vine with a slender, many branched stem bearing small, coarse black seeds. The wild soybean also differs in the extent of hard seededness and pod dehiscence from the cultivated soybean, although genetic variations also exist in the latter for these DRTs (Bailey et al., 1997; Chachalis and Smith, 2000; Mullin and Xu, 2001; Funatsuki et al., 2006).
Marker-assisted studies have detected many QTLs for various quantitative characteristics including physiological, morphological, yield-related traits and quality-related traits, such as the protein and oil contents in soybean. Currently, >900 QTLs are reported in SoyBase (http://www.SoyBase.org) (Hyten et al., 2004). However, few genetic studies have been carried out for DRTs in soybean. Although the wild soybean is useful as a genetic resource for cultivar improvements (Sebolt et al., 2000; Wang et al., 2001; Concibido et al., 2003; Fukuda et al., 2005; Luo et al., 2005; Chen et al. 2006; Kanamaru et al., 2006), our understanding of the genetic basis of DRTs still remains insufficient. The objective of this study was to gain a better understanding of the genetic basis of morphological and physiological traits that differentiate cultivated and wild soybeans, by means of molecular mapping of recombinant inbred lines (RILs) resulting from a cross between a cultivated and a wild soybean.
MATERIALS AND METHODS
Plant materials and character evaluation
A population of 96 RILs was developed by a single-seed descendent (SSD) method from an F2 population of the cross between a max line Tokei 780 and a soja accession Hidaka 4. Tokei 780 is an early-maturing, determinate, and yellow-seeded breeding line with grey pubescence, bred at Tokachi Agricultural Experimental Station, Memuro, Hokkaido, Japan. The wild accession was collected in Biratori town along the Saru River of the Hidaka region, Hokkaido. The seed (F2:8) for each RIL and the seed of the parental lines were sown in paper pots (#2, Nippon Beet Sugar Mfg, Co., Ltd, Sapporo, Japan) in the Research Faculty of Agriculture greenhouse at Hokkaido University, Sapporo, Japan, in late May 2004. Two weeks later, four seedlings for each RIL and the parents were transplanted into a pot of 22 cm in diameter and 20 cm in depth. A plastic support was then placed close to each individual plant. Four traits affecting plant architecture, namely plant height (PH), number of nodes (NN), maximum internode length (MIL) and twinning habit (TH), were scored 6 weeks after transplantation. TH was evaluated by the number of times that a main stem of an individual plant wound autonomously around the support. Flowering date, the date of the first flower appearance (R1: Fehr et al., 1971), was individually recorded. Determinate habit (DH) was evaluated by scoring an increment of nodes in a main stem from R1 to maturity, according to Thseng and Hosokawa (1972). Mature but undehisced pods were sampled for each of the RILs and parental lines, and preserved as bulk in a refrigerator at 5 °C and >80 % relative humidity until analysis to avoid further desiccation. Pod dehiscence (PD) was evaluated by two replications and expressed as a ratio of dehiscent pods of the 20 pods that were put in a humidity-controlled chamber of 20 °C and 10 % relative humidity for 24 h. After evaluation, seeds were threshed by hand, and used for scoring the 100 seed weight (SW) and hard seededness (HS). HS was evaluated by two replications as a percentage of impermeable seeds out of 40 seeds after 24 h of immersion in 50 mL of distilled H2O in a 200 mL beaker. The experiment was also carried out in 2005 using the seed (F2:9) obtained from a single F8 plant for each RIL. A total of eight traits were examined in the 2005 test, omitting DH. The seeds were directly sown in pots in the greenhouse in early June 2005, and thinned to four plants 2 weeks later. Character evaluation followed the method in the 2004 test except for the following two points: in the 2005 test, morphological evaluation was carried out 10 weeks after seeding (approx. 2 weeks later than in the 2004 test) to enlarge the differences among the RILs, and PD was evaluated with four replications.
Isozyme analysis
Genotypes at five isozyme loci were determined for each of the 96 RILs, according to the method described by Abe et al. (1992). The loci tested were an acid phosphatase locus (Ap), isocitrate dehydrogenase locus (Idh2), mannose phosphoisomerase locus (Mpi) and two phosphoglucomutase loci (Pgm1 and Pgm2).
DNA isolation and SSR analysis
The parents, Tokei 780 and Hidaka 4, were first surveyed for 317 simple sequence repeat (SSR) markers selected from an integrated soybean genetic linkage map (Cregan et al., 1999; Song et al., 2004). The primer sequences were obtained from the SoyBase Web site of the USDA, ARS Soybean Genome Database (http://soybase.agron.iastate.edu/). DNA was extracted from young leaves sampled from eight F8 plants for each RIL following the method described by Doyle and Doyle (1990). SSR analysis was carried out with either 6 % denatured polyacrylamide gel electorophoresis (PAGE) with fluorescent-labelled primers or 15 % non-denatured PAGE with ethidium bromide staining. The polymerase chain reaction (PCR) mixture contained 30 ng of total genomic DNA, 0·25 µm of 5′ and 3′ end primers, 200 µm of each dNTP, 0·5 U of Taq polymerase (TaKaRa, Otsu, Japan) and 1× PCR buffer (10 mm Tris–HCl, pH 8·3; 50 mm KCl; 1·5 mm MgCl2) for a total volume of 20 µL. The PCRs were performed with a GeneAmp PCR System 9700 (Perkin Elmer/Applied Biosystems, Foster City, CA, USA) using the following program: 32 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. Following the amplifications, in the PAGE method with fluorescent-labelled primers, up to six PCR products were combined and brought to a total volume of 20 µL by adding distilled water. An aliquot (1·5 µL) of the mixed PCR products combined with a loading buffer (1·5 µL) was denatured at 95 °C for 5 min and then loaded and separated using an ABI 377 sequencer (Perkin Elmer/Applied Biosystems). GeneScan software (version 3·1) was used to score the observed polymorphisms. In the PAGE method with ethidium bromide staining, the PCR products were mixed with 5 µL of loading buffer (25 % glycerol, 50 mm Tris, 5 mm EDTA, 0·2 % bromophenol blue) and then were loaded on the gel. Electrophoresis was performed at a constant voltage of 250 V for 4 h. After electrophoresis, the gel was stained with 10 µg mL–1 of ethidium bromide for 5 min and band profiles were recoded with a Typhoon 9410 imaging system (Amersham Bioscences, Piscataway, NJ, USA).
Amplified fragment length polymorphism (AFLP) analysis
About 100 ng of total genomic DNA was digested with 5 U of EcoRI and 1 U of MseI, and simultaneously 5 pmol of EcoRI adaptor and 50 pmol of MseI adaptor were ligated with 1 Weiss unit of T4 DNA ligase (New England Biolabs, Beverly, MA, USA) in a reaction buffer containing 50 mm Tris–HCl (pH 7·5), 10 mm MgCl2, 10 mm dithiothreitol (DTT), 50 mm NaCl, 1 mm ATP and 25 µg mL–1 bovine serum albumin (BSA) in a total volume of 20 µL for 10 h at room temperature. The sequences of the EcoRI and MseI adaptors were the same as described by Vos et al. (1995).
Pre-amplification was performed in a total volume of 20 µL containing 3 µL of 10× diluted digestion–ligation DNA template, 0·25 µm EcoRI + A and MseI + C primers, 0·2 mm of each dNTP, 0·5 U of Taq polymerase (TOYOBO, Osaka, Japan), 1·5 mm MgCl2 and 1× PCR buffer. The pre-amplification was performed in a thermocycler GeneAmp PCR system 9700 using the following program: 25 cycles of 94 °C for 20 s, 56 °C for 30 s and 72 °C for 2 min, followed by one cycle of 60 °C for 30 min. The PCR products were diluted 10-fold and used as templates for selective amplification. Selective PCRs were carried out in a volume of 20 µL containing 3 µL of 10× diluted pre-selective PCR product, 0·25 µm EcoRI + ANN primer and MseI + CNN primers, 0·5 U of Taq polymerase, 0·2 mm of each dNTP, 1·5 mm MgCl2 and 1× PCR buffer. PCR was performed on the thermocycler with the following touchdown program: one cycle of 94 °C for 20 s, 65 °C for 30 s and 72 °C for 2 min, followed by eight cycles of 1 °C decreasing annealing temperature per cycle, and 23 cycles of 94 °C for 20 s, 65 °C for 30 s and 72 °C for 2 min, with a final extension at 60 °C for 30 min. The PCR products were separated on 15 % non-denatured PAGE as for the SSR analysis.
Map construction
Of the SSR markers tested, 204 polymorphic and informative markers were chosen as anchors to construct the linkage groups covering all of the 20 linkage groups. A total of 282 markers, including five isozyme and 73 AFLP markers, were mapped in the RIL population. Marker order and distance were determined by the Map Manager program QTXb17 (http://mapmgr.roswellpark.org/mapmgr.html) using the Kosambi function and a criterion of 0·001 probability (d.f. = 1), equivalent to a LOD score of 2·4. Most of the markers were assigned to the 20 linkage groups as expected from the integrated map (Cregan et al., 1999; Song et al., 2004). A lower criterion of 0·05 probability was applied to a few unlinked distal SSR markers and the markers that belonged to their respective linkage groups.
Statistical analysis and QTL mapping
All traits were tested for deviations from normality using the Shapiro–Wilk test (SPSS, version 14·0). Where necessary, traits were transformed using the Box–Cox transformation (Box and Cox, 1964). Each trait was then analysed by one-way analysis of variance (ANOVA) for the 2004 and 2005 data individually, and by two-way ANOVA for the combined data. Tests of significance and partitions of variance components for the RIL, year and RIL × year effects were carried out based on a Model II ANOVA, where the RIL and year effects were treated as random variables (Sokal and Rohlf, 1981).
Marker order and distance inferred by QTXb17 were used to find the candidate QTL by composite interval mapping (CIM) implemented by MapQTL 5 (Van Ooijen, 2004). A total of 1000 permutations were performed on all traits to establish the empirical LOD thresholds at 0·05 probability (Churchill and Doerge, 1994). QTLs were considered to exist only at positions where a LOD score exceeded the corresponding significance threshold. CIM was performed by using the markers nearest to significant QTLs detected by interval mapping in advance as cofactors, and was repeated by adding the markers nearest to the QTLs newly detected by CIM to cofactors until additional QTLs were not detected. The chromosomal location, magnitude and direction of the additive effect and the proportion of the phenotypic variation explained (PVE) for each detected QTL were obtained from the CIM output. One-LOD support limits for the position of each QTL were calculated from the CIM results. The pair-wise interaction between QTLs was analysed as a two-way ANOVA with tagging markers as factors.
RESULTS
Segregation of markers and map construction
Most of the markers used segregated in an expected 1 : 1 ratio in the RIL population. Twenty-seven markers (9·6 % of the 282 markers) on 12 linkage groups produced a significantly (P = 0·05) distorted segregation. Of these, the distortions observed in Satt038, ATG/CAC340 and Satt505 on linkage group G, and Satt551 on linkage group M, were highly significant (P = 0·01). Two or more neighbouring markers that significantly deviated from the expected 1 : 1 ratio existed in six linkage groups, i.e. C1, C2, E, G, L and M. Particularly in the six distorted markers observed in linkage group G, three closely linked distal markers (Satt038, ATG/CAC340 and Satt570) yielded an allele from the soja parent in surplus, while the other closely linked proximal markers (ATG/CAT100, Satt138 and Satt505) yielded an allele from the max parent in surplus.
A linkage map of 282 markers and covering 2383 cM was constructed using the Kosambi function. The average distance between markers was 8·5 cM. There were still gaps of >25 cM in four linkage groups, i.e. B2, C1, C2 and D1a, because of a lack of polymorphic SSR markers. The map length approximately corresponded to the currently known recombination distance of 2524 cM of the integrated soybean linkage map (Cregan et al., 1999; Song et al., 2004). The marker order constructed in this study was in good agreement with that of the integrated map except for a few SSR markers. Most of the discordant marker orders occurred within a region of <5 cM. However, Sat_024 on linkage group E, which is located between two distal SSR markers, Satt411 and Satt384, in the integrated map (Cregan et al., 1999; Song et al., 2004), was positioned near a proximal marker (Satt606) on the same linkage group in the RIL population tested. The linkage map obtained in this study is available on request.
Parental phenotypes and segregation of domestication-related traits
Table 1 shows averages and standard deviations of parents and variance components (%) obtained from two-way ANOVA for nine traits in the RIL population. Of the nine traits tested, TH, PD, SW and HS are representative DRTs in soybean that characterize the difference between the modern cultivars and the wild soybean, although some of the Asian landraces possess the soja-like phenotypes, such as stone (impermeable) seed, twinning tendency and very small seed size. The parents of the RIL population also differed in flowering time (FT), DH and some morphological traits influencing plant architecture such as PH, NN and MIL. The max parent possessed an early and determinate habit with shorter PH, fewer NN and shorter MIL, whereas the soja parent possessed a late-flowering and indeterminate habit with taller PH, more NN and longer MIL (Table 1).
Table 1.
Averages and standard deviations of parents and variance components (%) obtained from two-way ANOVA for nine traits in a RIL population derived from a cross between G. max ssp. max and ssp.soja
| FT | DH | PH | NN | MIL | TH | PD | SW | HS | |
|---|---|---|---|---|---|---|---|---|---|
| 2004 | |||||||||
| Tokei 780 (max) | 45·7 (1·2) | 2·0 (0·0) | 18·3 (2·2) | 6·3 (0·5) | 4·3 (0·8) | 0·0 (0·0) | 0·12 (0·17) | 26·2 (1·3) | 0·0 (0·0) |
| Hidaka 4 (soja) | 77·5 (2·9) | 11·8 (1·3) | 30·5 (7·1) | 6·9 (0·4) | 7·1 (1·0) | 5·4 (1·1) | 0·78 (0·27) | 3·0 (0·05) | 100·0 (0·0) |
| 2005 | |||||||||
| Tokei 780 (max) | 44·5 (1·3) | – | 27·2 (2·0) | 9·3 (0·4) | 5·1 (0·4) | 0·0 (0·0) | 0·19 (0·18) | 26·3 (1·2) | 0·0 (0·0) |
| Hidaka 4 (soja) | 73·0 (2·2) | – | 62·2 (5·3) | 14·0 (0·8) | 9·3 (0·6) | 9·9 (0·4) | 0·96 (0·05) | 3·6 (0·2) | 98·8 (1·8) |
| Percentage of variance components estimated by two-way ANOVA | |||||||||
| RIL | 90·1*** | – | 15·8*** | 1·8* | 36·9*** | 48·6*** | 45·2*** | 80·0*** | 85·7*** |
| Year | 0·0 | – | 61·7*** | 87·8*** | 24·6 *** | 0·0 | 2·3 | 0·2*** | 0·1 |
| RIL × year | 0·8 | – | 17·2*** | 6·7*** | 19·2 *** | 21·0*** | 26·3*** | 3·8* | 6·2*** |
| Error | 9·1 | – | 5·3 | 3·7 | 19·3 | 30·4 | 26·2 | 16·0 | 8·0 |
Values in parentheses are standard deviations. F-tests and partitions of variance components were carried out based on a Model II ANOVA, where the RIL and year effects were treated as random variables (Sokal and Rohlf, 1981). * and ***, significance at the 5 and 0·5 % levels, respectively.
FT, flowering time (number of days after seeding); DH, determinate habit (number of nodes increased after flowering); PH, plant height (cm); NN, number of nodes; MIL, maximum internode length (cm); TH, twinning habit (number of times that a main stem winds around a support); PD, pod dehiscence (rate of dehiscent pods); SW, 100 seed weight (g); HS, hard seededness (percent of hard seed).
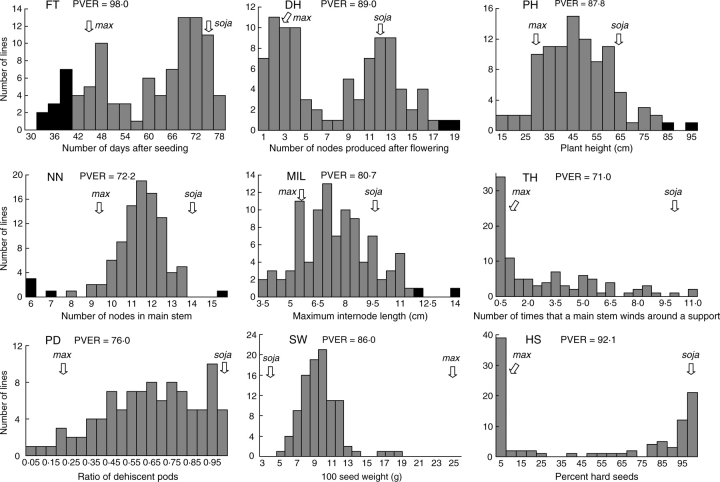
Frequency distributions of phenotypes in the RIL population in the 2005 test (for DH in the 2004 test) are presented in Fig. 1. FT and DH produced a bimodal distribution, suggesting an involvement of one or two major genetic factors. HS also had two modes: one in a class of <5 % (max type) and the other in a class of >95 % (soja type). On the other hand, PH, NN, MIL and SW were characterized by normal distribution. Transgressive segregations were observed in FT, DH, PH, NN and MIL. In the 2005 test, two (PH and MIL, and DH in the 2004 test) to 12 (FT) RILs significantly exceeded the parental values at 0·001 probability (Fig. 1). The frequency distribution of SW was normal, but ranged from the soja parent (3·6 g) to the mid-parent (14·9 g), with only three RILs being distributed over the mid-parent value. Both PD and TH yielded phenotype frequency distributions which were shown to deviate significantly from normality by the Shapiro–Wilk test.
Fig. 1.
Frequency distributions of phenotypes for nine domestication-related traits in a RIL population derived from a cross between G. max ssp. max and ssp. soja. Black bars indicate RILs which significantly exceeded the parental values at 0·001 probability. PVER (the proportion of the phenotypic variation explained by RILs) was calculated based on a Model II ANOVA (Sokal and Rohlf, 1981). FT, flowering time; DH, determinate habit; PH, plant height; NN, number of nodes; MIL, maximum internode length; TH, twinning habit; PD, pod dehiscence; SW, 100 seed weight; HS, hard seededness.
Results of one-way ANOVA revealed that 70 % or more of the total variance observed in each trait was attributable to the variance explained among RILs, suggesting that the variation observed in the RIL population was mostly genetic. Results of two-way ANOVA for the combined data of the two years further indicated that 80 % or more of the total variance was ascribed to the RIL effect in FT, SW and HS (Table 1), suggesting that these three traits were phenotypically stable over the two years tested. TH, MIL and PD showed moderate values of 36·9–48·6 % for the RIL effect and slightly low values of 19·2–26·3 % for the RIL × year effect. On the other hand, the year effect occupied large parts of the total variance in PH (61·7 %) and NN (87·8 %). The high year effects for PH and NN were due to the fact that morphological evaluation was carried out in different growth stages in the 2004 and 2005 tests.
QTL analyses of domestication-related traits
CIM was carried out separately for each of the 2004 and 2005 tests and, except for PH and NN which showed large year effects, the average of the two years. Results of the QTL analysis are presented in Table 2 and graphically in Fig. 2. To compare the results with the QTLs detected in previous studies by the CIM of a lower criterion (lower LOD scores) or by ANOVA, non-significant QTLs with a LOD score of >2·5 are also included in Table 2. CIM revealed a total of 14 significant (P = 0·05) QTLs affecting DRTs by the genome-wide analyses with permutation tests. These QTLs were distributed in six of the 20 linkage groups, and individual QTLs accounted for 13·4–72·5 % of the phenotypic variance observed. In all except a QTL for FT, the direction of additive effects for alleles derived from the soja parent (Table 2) was consistent with that expected from the parental phenotypes (Table 1).
Table 2.
QTLs for domestication-related traits detected in a RIL population derived from a cross between G. max ssp. max and ssp. soja
| Traits | QTL-LG | Position (cM) | Nearest marker | LOD | PVE (%) | Additive effect |
|---|---|---|---|---|---|---|
| FT04 | qFT-C2 | 100·7 | AGG/CGC380 | 14·3 | 55·4 | 11·4 |
| qFT-J | 44·3 | Satt686 | 4·0 | 13·4 | –5·7 | |
| FT05 | qFT-C2 | 101·7 | AGG/CGC380 | 10·7 | 45·4 | 10·8 |
| FT-C | qFT-C2 | 100·7 | AGG/CGC380 | 12·3 | 49·4 | 10·4 |
| qFT-J | 44·3 | Satt686 | 2·9 ns | 8·8 | −4·5 | |
| DH04 | qDH-L | 81·9 | Sat_099 | 23·9 | 72·5 | 4·6 |
| PH04 | qPH-L | 71·0 | ATC/CCG315 | 3·0 ns | 13·4 | 2·5 |
| PH05 | qPH-G | 28·0 | Satt235 | 4·6 | 20·7 | 6·5 |
| NN05 | qNN-A1 | 25·3 | Satt042 | 2·5 ns | 9·9 | 0·5 |
| qNN-G | 22·4 | Satt235 | 3·2 | 14·0 | 0·6 | |
| MIL04 | qMIL-C2 | 5·0 | Sat_130 | 3·3 | 16·8 | 0·8 |
| MIL05 | qMIL-G | 30·0 | Satt235 | 3·8 | 15·2 | 0·8 |
| qMIL-L | 71·0 | ATC/CCG315 | 2·9 ns | 10·6 | 0·7 | |
| MIL-C | qMIL-G | 28·0 | Satt235 | 3·9 | 17·0 | 0·7 |
| qMIL-L | 71·0 | ATC/CCG315 | 3·6 | 16·0 | 0·7 | |
| TH04 | qTH-D1b | 95·0 | Satt546 | 4·2 | 20·5 | 0·8 |
| qTH-G | 25·4 | Satt235 | 2·6 ns | 10·5 | 0·6 | |
| qTH-L | 13·1 | Satt182 | 2·6 ns | 10·8 | 0·6 | |
| TH05 | qTH-G | 28·0 | Satt235 | 7·6 | 34·3 | 1·3 |
| TH-C | qTH-D1a | 0·0 | Satt408 | 2·6 ns | 9·2 | 0·6 |
| qTH-D1b | 95·0 | Satt546 | 2·7 ns | 10·1 | 0·7 | |
| qTH-G | 27·4 | Satt235 | 5·7 | 26·4 | 1·1 | |
| PD04 | qPD-J | 18·4 | AGT/CCA170 | 3·3 ns | 16·3 | 0·10 |
| PD05 | qPD-J | 54·9 | Satt215 | 4·2 | 19·4 | 0·10 |
| PD-C | qPD-E | 47·8 | Sat_124 | 2·7 ns | 9·6 | 0·07 |
| qPD-J | 56·9 | ATG/CCG270 | 4·9 | 21·8 | 0·11 | |
| SW04 | qSW-D2 | 51·9 | Satt154 | 4·6 | 24·2 | −1·1 |
| qSW-H | 60·6 | Satt442 | 2·7 ns | 12·0 | −0·7 | |
| SW05 | qSW-D2 | 48·9 | Satt154 | 3·9 | 18·5 | −1·1 |
| qSW-M | 69·3 | AGA/CAG540/560 | 2·9 ns | 11·5 | −0·9 | |
| qSW-O | 87·9 | Sat_274 | 2·8 ns | 10·8 | −0·4 | |
| SW-C | qSW-D2 | 51·9 | Satt154 | 5·2 | 19·3 | −1·1 |
| qSW-M | 58·6 | AGA/CAG540/560 | 2·7 ns | 10·8 | −0·8 | |
| qSW-O | 87·9 | Sat_274 | 2·8 ns | 11·3 | −0·8 | |
| HS04 | qHS-C2 | 107·3 | ACG/CCG45 | 4·8 | 13·7 | 21·1 |
| qHS-D1b | 124·3 | Satt459 | 13·7 | 47·8 | 42·8 | |
| HS05 | qHS-C2 | 101·7 | AGG/CGC380 | 5·9 | 18·5 | 25·7 |
| qHS-D1b | 124·3 | Satt459 | 12·5 | 42·5 | 39·1 | |
| HS-C | qHS-C2 | 107·3 | ACG/CGC380 | 5·7 | 16·5 | 23·0 |
| qHS-D1b | 124·3 | Satt459 | 13·5 | 46·3 | 39·9 |
QTL analyses were conducted for each of the 2004 and 2005 tests (04 and 05) and the average of the two years (-C). ns, non-significance at 0·05 probability by 1000 permutation tests.
The additive effect of each QTL is shown as a trait unit contribution of the soja allele.
FT, flowering time (number of days after seeding); DH, determinate habit (number of nodes increased after flowering); PH, plant height (cm); NN, number of nodes; MIL, maximum internode length (cm); TH, twinning habit (number of times that a main stem winds around a support); PD, pod dehiscence (rate of dehiscent pods); SW, 100 seed weight (g); HS, hard seededness (percent of hard seed).
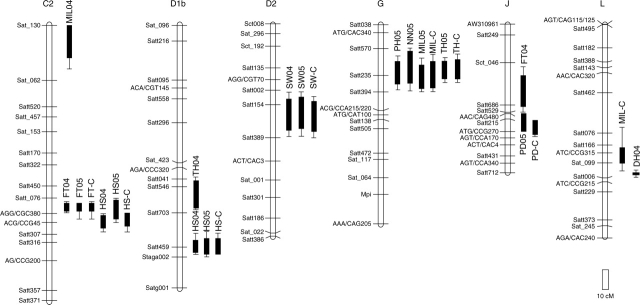
Fig. 2.
Six linkage groups harbouring significant QTLs for domestication-related traits in a RIL population derived from a cross between G. max ssp. max and ssp. soja. QTL analyses were conducted for each of the 2004 and 2005 tests (04 and 05) and the average of the two years (-C). The box delineates the one-LOD support interval, and the whiskers of each box delineate the two-LOD support interval. FT, flowering time; DH, determinate habit; PH, plant height; NN, number of nodes; MIL, maximum internode length; TH, twinning habit; PD, pod dehiscence; SW, 100 seed weight; HS, hard seededness.
A major QTL for flowering time (qFT-C2) with a PVE of >45 % was consistently detected in linkage group C2 in the 2004 and 2005 data. qFT-C2 was flanked with SSR markers Sat_076 and Satt307, which are located in a region harbouring the major gene E1 for maturity (Molnar et al., 2003). Another QTL (qFT-J) on linkage group J was significant in the 2004 test, and had a LOD score of 2·9 in the combined data of the two years. qFT-J had an opposite effect compared with the parental phenotypes (Table 1), which is a plausible factor for causing the transgressive segregation of FT observed in the RIL population (Fig. 1). A single major QTL (qDH-L) for DH with a PVE of 72·5 % was detected on linkage group L. qDH-L was flanked with SSR markers Sat_099 and Satt006, which are located in a region harbouring the major gene dt1 for DH (Cregan et al., 1999).
A significant QTL influencing all of the four morphological traits was detected on linkage group G in the 2005 data and the combined data of the two years. The portion of phenotypic variance explained by the QTL was highest in TH: it accounted for 34·3 and 26·4 % of the total variance for the 2005 data and the combined data of the two years, respectively, suggesting that the QTL mainly controlled the TH. Another significant QTL for TH (qTH-D1b) was detected near Satt546 on linkage group D1b in the 2004 data. In contrast to qTH-G, qTH-D1b had no effect on PH, NN and MIL. Significant QTLs for MIL were detected on linkage group C2 in the 2004 data and on linkage group L in the combined data of the two years.
A significant QTL (qPD-J) for PD was detected on linkage group J in the 2005 data and the combined data of the two years. In the 2004 data, this QTL was not significant, but had a LOD score of 3·3. A non-significant QTL with a LOD score of 2·7 was detected on linkage group E only for the combined data of the two years. qPD-J accounted for 19·4 and 21·8 % of the total variance observed in the 2005 data and the combined data of the two years, respectively.
A significant QTL (qSW-D2) for SW was consistently detected on linkage group D2 in the 2004 and 2005 data. It accounted for 24·2 and 18·5 % of the total variance observed in the two years. In addition, there were three non-significant QTLs for SW, one (qSW-H) on linkage group H in the 2004 data and two (qSW-M and qSW-O) on linkage groups M and O in the 2005 and combined data of the two years. These QTLs each accounted for 10·8–12·0 % of the total variance.
Two significant QTLs for HS (qHS-C2 and qHS-D1b) were consistently detected on linkage groups C2 and D1b in the 2004 and 2005 data. QTL mapping using the RIL population therefore confirmed two of the three QTLs detected by ANOVA in the early generations of the same cross from which the RIL population derived (Sakamoto et al., 2004). Sakamoto et al. (2004) found another minor QTL on linkage group G, which was not confirmed in the RIL population tested in this study. Of the two QTLs detected, qHS-D1b had the more prominent effect, accounting for >40 % of the total variance, whereas qHS-C2 accounted for 13·7–18·5 of the total variance.
Interaction between QTLs
A pair-wise two-way ANOVA demonstrated that there was a consistently significant interaction (P < 0·01) between two QTLs for HS (qHS-C2 and qHS-D1b) in the 2004 and 2005 data. A combination of the soja alleles at both qHS-C2 and qHS-D1b affected HS more potently than the sum of their additive effects. No significant epistasis was detected in other pairs of QTLs.
Commonality of the QTLs for pod dehiscence, hard seededness and seed weight in different crosses
Table 3 presents a summary of the QTLs for PD, HS and SW detected in this and previous studies. In order to make a comparison with the QTLs detected in previous studies, non-significant QTLs with a LOD score of >2·5 are also included in Table 3. All of the crosses tested for PD identified a common QTL on linkage group J. It had the most marked effect in each cross, and accounted for approx. 20 to >50 % of the total variance. There were four additional QTLs in linkage groups D1b, E, L and N in this and previous studies (Saxe et al., 1996; Bailey et al., 1997). The effects of these QTLs varied with the crosses. A minor QTL, qPD-E, detected in this study was located near the QTL on linkage group E detected by Saxe et al. (1996).
Table 3.
Commonality of QTLs for pod dehiscence, hard seededness and seed weight in different crosses of soybean
| Parent* | Analysis method | Linkage group | Nearest marker | Position† (cM) | PVE or r2 | LOD | Reference | |
|---|---|---|---|---|---|---|---|---|
| Female | Male | |||||||
| Pod dehiscence | ||||||||
| A81-3560222 | PI468916 | IM | J | Sct_065 | 32 | 34·7 | Approx. 3·7 | Saxe et al. (1996) |
| (M) | (S) | J | A724 | 85 | 21·6 | Approx. 2·7 | ||
| D1b | B194-2 | 88 | 23·7 | Approx. 3·1 | ||||
| Young | PI1416937 | ANOVA | D1b | A725 | 9 | 5·3–7·2 | – | Bailey et al. (1997) |
| (M) | (M) | E | cr274-1 | 48–53 | 6·0–7·1 | – | ||
| J | B122-1 | 57 | 39·1–44·4 | – | ||||
| L | A489-1 | 95 | 5·0–5·6 | – | ||||
| N | A808n | 69 | 4·1–5·7 | – | ||||
| Toyomusume | Hayahikari | CIM | J | Sat_093 | 46 | >50·0 | 13·8–15·6 | Funatsuki et al. (2006) |
| (M) | (M) | |||||||
| Tokei 780 | Hidaka 4 | CIM | E | Sat_124 | 16 (32–40) | 9·6 | 2·7 | This study |
| (M) | (S) | J | Satt215 | 44 | 16·3–21·8 | 3·3–4·9 | ||
| Hard seededness | ||||||||
| A81-3560222 | PI468916 | ANOVA | A2 | I | 49 | 32·0 | – | Keim et al. (1990) |
| (M) | (S) | D1b | K411 | 119 | 13·0 | – | ||
| L | G173-1 | 87 | 15·0 | – | ||||
| N | K418 | 30 | 12·0 | – | ||||
| Misuzudaizu | Moshidou Gong 503 | CIM | C2 | Satt100 | 114 | 26·0 | 14·0 | Watanabe et al. (2004) |
| (M) | (M) | D1b | B142 | 119–132 | 10·6 | 6·3 | ||
| I | GM222b | 28–31 | 6·1 | 3·0 | ||||
| Tokei 780 | Hidaka 4 | CIM | C2 | AGG/CGC380 | 99–121 | 13·7–18·5 | 4·8–5·9 | This study |
| (M) | (S) | D1b | Satt459 | 119 | 42·5–47·8 | 12·5–13·7 | ||
| Seed weight‡ | ||||||||
| V71-370 | PI407·162 | ANOVA | A2 | T153 | 50 | 4·9–7·1 | – | Maughan et al. (1996) |
| (M) | (S) | B1 | A118 | 59 | 21·1–14·2 | – | ||
| (24·1 g) | (1·5 g) | G | A816 | 68 | 7·8–9·8 | – | ||
| J | K384 | 28 | 5·7–10·5 | – | ||||
| L | A023 | 37 | 4·3–8·7 | – | ||||
| L | K385 | 101 | 10·5–0·9 | – | ||||
| Kefeng 1 | Nannong 1138-2 | CIM | A2 | Satt525 | 97 | 6·7 | 3·4 | Zhang et al. (2004) |
| (M) | (M) | B1 | Satt509 | 33 | 10·8 | 3·8 | ||
| (8·1 g) | (19·3 g) | D2 | A611D | 10–23 | 9·2 | 4·5 | ||
| D2 | B146H | 23–25 | 11·4 | 4·8 | ||||
| Misuzudaizu | Moshidou Gong 503 | CIM | B2 | B124b | 84 | 6·1 | 3·1 | Watanabe et al. (2004) |
| (M) | (M) | C2 | Satt286 | 102 | 7·7 | 3·8 | ||
| (approx. 21 g) | (approx. 5 g) | H | A858a | 124 | 7·4 | 2·9 | ||
| K | Satt518 | 47 | 5·8 | 3·2 | ||||
| Tokei 780 | Hidaka 4 | CIM | D2 | Satt154 | 57 | 18·5–24·2 | 3·9–5·2 | This study |
| (M) | (S) | H | Satt442 | 47 | 12·0 | 2·7 | ||
| (26·4 g) | (3·2 g) | M | AGA/CAG540/560 | 67–95 | 10·8–11·5 | 2·7–2·9 | ||
| O | Sat 274 | 108·0 | 10·8–11·3 | 2·8 | ||||
* M and S indicate max and soja. A dehiscent or hard-seeded parent is underlined.
† Position in the consensus map (Cregan et al., 1999; Song et al., 2004). In the case of AFLP markers, the positions of flanking SSR markers are presented.
‡ Seed size of parents is given in parentheses.
Of the two significant QTLs for HS (qHS-C2 and qHS-D1b) detected in this study, qHS-D1b was positioned in almost the same region as the QTLs previously identified in crosses between max and soja (Keim et al., 1990b) and between the max and semi-wild max lines (Watanabe et al., 2004). Another significant QTL, qHS-C2, was also positioned in the region corresponding to the QTL detected in a cross between the max and semi-wild max lines (Watanabe et al., 2004). On the other hand, four additional QTLs for HS were detected on linkage groups A2, I, L and N in previous studies (Keim et al., 1990b; Watanabe et al., 2004). Of these, the QTL on linkage group A2, which was considered as the I locus inhibiting seed coat coloration itself, had the most prominent effect, with a PVE of 32 % in a cross between max and soja (Keim et al., 1990b). However, it was not observed in either the cross between max and soja tested in this study or the cross between the max and semi-wild max lines (Watanabe et al., 2004).
There are many reports on the QTLs for SW in soybean. Table 3 presents only the results obtained in three crosses in which the parents exhibited large differences in SW. The previous studies reported an involvement of 4–6 QTLs in the genetic control of SW in each population. As a whole, the QTLs for SW were scattered across 12 linkage groups (A2, B1, B2, C2, D2, G, H, J, K, L, M and O), and each accounted for around 10 % or less of the observed variation. The QTL corresponding to qSW-D2, the most marked QTL detected in this study, was observed only in a cross between the max lines (Zhang et al., 2004). No commonality was detected in the other QTLs across the crosses tested.
DISCUSSION
Segregation distortion
Segregation distortion of molecular markers has often been reported in the progeny of crossings of cultivated crops × wild progenitors in maize (Doebley et al., 1990; Doebley and Stec, 1993), rice (Cai and Morishima, 2002), pearl millet (Poncet et al., 2000, 2002) and soybean (Keim et al., 1990a; Yamanaka et al., 2001; Watanabe et al., 2004). Segregation distortion occurred in six linkage groups, i.e. C1, C2, E, G, L and M, in the RIL population tested in this study. Similarly, Yamanaka et al. (2001) found a highly significant (P < 0·01) segregation distortion in six linkage groups, i.e. A1, D1b, D2, E, J and L, in an F2 population of a cross between the max and semi-wild max lines, Misuzudaizu × Moshidou Gong 503. However, most of the distortions detected by Yamanaka et al. (2001) were not confirmed in the F8 RIL population generated from the same cross by SSD, in which the distortion occurred in four linkage groups, i.e. A2, I, L and M (Watanabe et al., 2004). Furthermore, there were only a few regions that consistently produced the segregation distortion across the two crosses tested by this study and in that of Watanabe et al. (2004). The observed segregation distortions may have occurred partly by chance because of the limited sample sizes in the segregating populations.
Xu et al. (1997) summarized the segregation distortion of molecular markers that has been reported in the literature, and highlighted possible factors, including the abortion of male or female gametophytes or the selective fertilization of gametes. Aberrant segregation ratios also result from non-deliberate selection against wild characters under cultivated conditions (Cai and Morishima, 2002). Cytogenetic studies in soybeans have revealed that paracentric inversions and reciprocal translocations existed between the cultivated soybean and certain wild soybean accessions (Palmer et al., 1987; Singh and Hymowitz, 1988; Palmer et al., 2000). Crossing over within inversions or interchange segments in plants heterozygous for paracentric inversions and/or reciprocal translocations leads to both pollen and ovule sterilities and restriction of recombination, because of formation of gametes with incomplete genetic complements. Pollen fertility and seed abortion were not observed in the F1 hybrid of the cross used in this study. However, the tight linkage unexpected from the recombination distance in the consensus map (Cregan et al., 1999; Song et al., 2004) was not observed for neighbouring SSR markers. Also, intentional selection against wild characters in experimental conditions can be excluded as a causal factor because the loss of wild traits, such as hard seededness and seed shattering, could be easily remedied by scarification of the seed coat and harvesting of the mature pods before shattering during development of the RIL population. A further study is needed to verify the possibility that the segregation distortion is caused by gametophyte genes, as frequently found in subspecific crosses of rice (Oka, 1988; Xu et al., 1997).
Genetic basis of pod dehiscence, hard seededness and twinning habit
CIM revealed that one or two significant QTLs were involved in the genetic control of each DRT. Of these QTLs, qPD-J for PD and qHS-D1b for HS were common across the different crosses tested so far (Table 2). In particular, qPD-J is a major QTL that contributes to a wide variation from the wild soybean to modern indehiscent soybean cultivars. Another commonly found QTL, qHS-D1b, differed in relative effects on hard seededness among the crosses. It had the most marked effect in the RIL population tested in this study, whereas the QTLs on linkage groups A2 and C2 (corresponding to qHS-C2 in this study) had larger effects in the other crosses (Keim et al., 1990b; Watanabe et al., 2004). In particular, the QTL on linkage group A2 detected by Keim et al. (1990b) was not found in the cross used in this study or in that used by Watanabe et al. (2004). Therefore, different sets of QTLs may contribute to the phenotypic difference in HS between the cultivated and wild soybeans to different degrees, as suggested in our earlier study (Sakamoto et al., 2004).
Two QTLs, qTH-G and qTH-D1b, are the first reported QTLs for TH in soybean. Each of the two QTLs was detected in both the 2004 and 2005 tests. MIL, a measure of the degree of internode elongation in the upper nodes, also characterized the difference in plant architecture between the parents used in the cross. qTH-G influenced the expression of MIL as well, whereas qTH-D1b did not. Rather, qMIL-C2 controlled MIL in the 2004 test. The detection of QTLs for plant architecture may therefore have depended on the growing stages in which morphological evaluation was performed. The DH and early-flowering habit derived from the max parent, on the other hand, had no developmental constraint on the expression of TH, because there was no significant phenotypic correlation between DH or FT and TH in the RIL population tested (data not presented). The DH was controlled exclusively by a major QTL (qDT-L) with a marked effect, most probably a major gene dt1. The expression of dt1 was therefore stable even in the genetic background segregating for the wild characters such as TH.
Seed size, a gigantism in soybean
Seed size evaluated as 100 seed weight was different from PD and HS in terms of the underlying genetic bases. SW in the RIL population tested produced a normal but shifted distribution toward smaller seed size classes. A lack of large-seeded phenotypes in the hybrid progeny was also observed in the F2 and F3 generations of a cross between max and soja (Maughan et al., 1996) and an F8 RIL population derived from a cross between the max and semi-wild max lines (Watanabe et al., 2004). In the present study and the study of Watanabe et al. (2004), there was little distorted segregation for markers proximal to the QTLs detected for SW. Thus, the frequency distribution observed in SW was not caused by unequal segregations of chromosomal regions harbouring the QTLs for SW. Maughan et al. (1996) suggested that partial dominance of the small-seeded allele at the QTLs produced the frequency distribution skewed toward the small-seeded phenotypes. However, this is not the case for the RIL population used in our study and the study of Watanabe et al. (2004), in which the number of heterozygotes was very small. The lack of large-seeded phenotypes might rather be attributed to epistasis between QTLs influencing seed size directly and indirectly.
A comparison of the QTLs detected so far among the different crosses may further differentiate a genetic basis for seed size from PD and HS. In contrast to PD and HS, common QTLs for seed size across the different crosses could not be identified (Table 2). Compiling the seed size QTLs reported so far, Hyten et al. (2004) found that a total of 26 QTLs for SW were scattered in 18 of the 20 soybean linkage groups, and there were no invariable QTLs across the cross combinations tested. Soybean varieties and/or landraces most probably have, by different combinations, the alleles of different effects at many QTLs. This may result in the transgressive segregation of seed size frequently observed in intervarietal crosses of soybean (Fasoula et al., 2004; Hyten et al., 2004). The lack of common QTLs for SW is in further sharp contrast to that of the Graminae species, such as rice, maize, sorghum and tetraploid wheat, where most of the QTLs for seed size existed commonly in homeologous chromosomes even across the species (Paterson et al., 1995; Peng et al., 2003). The absence of common major QTLs and the involvement of many QTLs with minor effects strongly suggest that evolutionary changes toward larger seed in soybean have resulted from an accumulation of minor changes at many QTLs influencing seed size directly and indirectly. Usage of a variety of landraces with different seed sizes in Asia might have contributed to the conservation of diversity for genetic control of seed size in soybean.
Implication of the genetic basis of domestication-related traits and the genetic diversity involved in soybean
The results obtained in this study suggest that most of the DRTs examined are controlled by one or two major QTLs and a number of genotype-dependent minor QTLs. This is in good agreement with the genetic basis for DRTs reported in many crop species as reviewed by Ross-Ibarra (2005). Another commonly found trend of the genetic basis of DRTs is a clustering of domestication-related QTLs. However, this did not hold true for soybean. In the RIL population tested, only four genomic regions harboured QTLs for different traits, i.e. QTLs for FT and HS in linkage group C2, QTLs for HS and TH in linkage group D1b, QTLs for FT and PD in linkage group J, and QTLs for MIL and DH in linkage group L. Minimal clustering of QTLs for DRTs observed in this study may be partly attributable to the fact that most of the traits examined, except for the four morphological traits associated with plant architecture, had no developmental constraint toward each other. Introgressions from the wild to cultivated soybeans can be relatively easily carried out without any large obstacle because the major QTLs for DRTs were present on only six of the 20 linkage groups, and there were few clusterings of those QTLs, although the role of minor QTLs undetected by CIM should not be underestimated.
The results obtained in this study also have an important implication concerning the genetic diversity embedded in the max germplasm. Compiling both their own data and the data presented in the literature, Wang et al. (2004) suggested that it would be difficult to unlock positive allelic diversity from the wild soybean. This is based on the findings that the useful QTLs from the wild soybean, such as the QTLs for high protein content (Sebolt et al., 2000), SCN resistance (Wang et al., 2001) and yield (Concibido et al., 2003), were already present in the max germplasm and/or were mapped in cultivated soybean populations as well (Wang et al., 2004). However, this is not contradictory to the findings that the introgression from the wild to cultivated soybeans repeatedly had occurred via hybridizations or independent domestications in various regions of East Asia, as has been indicated from a overlapping of geographical distributions of chloroplast and mitochondorial genome types between the cultivated and wild soybeans (Shimamoto et al., 1998, 2000; Abe et al., 1999; Xu et al., 2003). The findings obtained from this and previous studies on the genetic basis of DRTs may therefore indicate that some of the useful genes from the wild soybean, as exemplified by Wang et al. (2004), have been repeatedly introduced into the cultivated germplasm pools without marked obstacles and have been retained in a variety of soybean landraces in Asia. However, the wild soybean is still rich in novel and/or useful variants for soybean breeding in various traits, such as high tolerances to salt (Luo et al., 2005) and dehydration (Chen et al. 2006), seed storage protein electrophoretic variants (Fukuda et al., 2005), and high lutein content in seed (Kanamaru et al., 2006). The present results for QTL mapping of DRTs would be useful to introduce such unutilized genetic characteristics in the wild soybean into the cultivated genetic background by marker-assisted selection.
ACKNOWLEDGEMENTS
We thank Dr P. Srinives of Kasetsart University, Thailand, for his kind help in developing the recombinant inbred lines used in this study. This study was partly supported by a National Bioresource Project [Lotus and Glycine] of the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Abe J, Ohara M, Shimamoto Y. New electrophoretic mobility variants observed in wild soybean (Glycine soja) distributed in Japan and Korea. Soybean Genetics Newsletter. 1992;19:63–72. [Google Scholar]
- Abe J, Hasegawa A, Fukushi H, Mikami T, Ohara M, Shimamoto Y. Introgression between wild and cultivated soybeans of Japan revealed by RFLP analysis of chloroplast DNAs. Economic Botany. 1999;53:285–291. [Google Scholar]
- Bailey MA, Mian MAR, Carter TE, Jr, Ashley DA, Boerma HR. Pod dehiscence of soybean: identification of quantitative trait loci. Journal of Heredity. 1997;88:152–154. [Google Scholar]
- Box GEP, Cox DR. An analysis of transformations. Journal of the Royal Statistics Society, Series B. 1964;26:211–243. [Google Scholar]
- Broich S, Palmer RG. A cluster analysis of wild and domesticated soybean phenotypes. Euphytica. 1980;29:23–32. [Google Scholar]
- Broich S, Palmer RG. Evolutionary studies of the soybean: the frequency and distribution of alleles among collections of Glycine max and soja of various origin. Euphytica. 1981;30:55–64. [Google Scholar]
- Burke JM, Tang S, Knapp SJ, Rieseberg LH. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai HW, Morishima H. QTL clusters reflect character associations in wild and cultivated rice. Theoretical and Applied Genetics. 2002;104:1217–1228. doi: 10.1007/s00122-001-0819-7. [DOI] [PubMed] [Google Scholar]
- Chachalis D, Smith ML. Imbibition behavior of soybean (Glycine max (L.) Merrill) accessions with different testa characteristics. Seed Science and Technology. 2000;28:321–331. [Google Scholar]
- Chen YY, Chen PY, de los Reyes BG. Differential responses of the cultivated and wild species of soybean to dehydration stress. Crop Science. 2006;46:2041–2046. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concibido VC, Vallee BL, Mclaird P, Pineda N, Meyer J, Hummel L, et al. Introgression of a quantitative trait locus for yield from glycine soja into commercial soybean cultivars. Theoretical and Applied Genetics. 2003;106:575–582. doi: 10.1007/s00122-002-1071-5. [DOI] [PubMed] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, et al. An integrated genetic linkage map of the soybean genome. Crop Science. 1999;39:1464–1490. [Google Scholar]
- Doebley J, Stec A. Inheritace of the morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Wendel J, Edwards M. Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proceedings of the National Academy of Sciences, USA. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics. 2002;161:1713–1726. doi: 10.1093/genetics/161.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Fasoula VA, Harris DK, Boerma HR. Validation and designation of quantitative trait loci for seed protein, seed oil, and seed weight from two soybean populations. Crop Science. 2004;44 [Google Scholar]
- Fehr WR, Caviness CE, Burmood DT, Pennington JS. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Science. 1971;11:929–931. [Google Scholar]
- Fukuda T, Maruyama N, Kanazawa A, Abe J, Shimamoto Y, Hiemori M, et al. Molecular analysis and physicochemical properties of electrophoretic variants of wild soybean Glycine soja storage proteins. Journal of Agricultural and Food Chemistry. 2005;53:3658–3665. doi: 10.1021/jf0479620. [DOI] [PubMed] [Google Scholar]
- Funatsuki H, Ishimoto M, Tsuji H, Kawaguchi K, Hajika M, Fujino K. Simple sequence repeat markers linked to a major QTL controlling pod shattering in soybean. Plant Breeding. 2006;125:195–197. [Google Scholar]
- Gottlieb LD. Genetics and morphological evolution in plants. American Naturalist. 1984;123:681–709. [Google Scholar]
- Grandillo S, Tanksley SD. QTL analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theoretical and Applied Genetics. 1996;92:935–951. doi: 10.1007/BF00224033. [DOI] [PubMed] [Google Scholar]
- Harlan JR. Crops and man. 2nd edn. Madison, WI: American Society of Agronomy, Inc. and Crop Science Society of America, Inc; 1992. [Google Scholar]
- Hymowitz T. Speciation and cytogenetics. In: Boerma HR, JE Specht, editors. Soybeans: improvement, production, and uses. 3rd edn. Madison, WI: American Society of Agronomy, Inc. Crop Science Society of America, Inc. and Soil Science Society of America, Inc; 2004. pp. 97–136. [Google Scholar]
- Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, Stefaniak TR, Schmidt ME. Seed quality QTL in a prominent soybean population. Theoretical and Applied Genetics. 2004;109:552–561. doi: 10.1007/s00122-004-1661-5. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Wang SD, Abe J, Yamada T, Kitamura K. Identification and characterization of wild soybean (Glycine soja Sieb. et Zucc.) strains with high lutein content. Breeding Science. 2006;56:231–234. [Google Scholar]
- Keim P, Diers BW, Olson TC, Shoemaker RC. RFLP mapping in soybean: association between maker loci and variation in quantitative traits. Genetics. (a) 1990;126:735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Diers BW, Shoemaker RC. Genetic analysis of soybean hard seededness with molecular markers. Theoretical and Applied Genetics. (b) 1990;79:465–469. doi: 10.1007/BF00226154. [DOI] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common bean. Crop Science. 1996;36:1037–1045. [Google Scholar]
- Lande R. The response to selection on major and minor mutations affecting a metrical trait. Heredity. 1983;50:47–65. [Google Scholar]
- Lauter N, Doebley J. Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics. 2002;160:333–342. doi: 10.1093/genetics/160.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QY, Yu GJ, Liu YL. Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. Journal of Plant Physiology. 2005;162:1003–1012. doi: 10.1016/j.jplph.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Maughan PJ, Saghai Maroof MA, Buss GR. Molecular-marker analysis of seed-weight: genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theoretical and Applied Genetics. 1996;93:574–579. doi: 10.1007/BF00417950. [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Rai S, Charette M, Cober ER. Simple sequence repeat (SSR) markers linked to E1, E3, E4 and E7 maturity genes in soybean. Genome. 2003;46:1024–1036. doi: 10.1139/g03-079. [DOI] [PubMed] [Google Scholar]
- Mullin WJ, Xu W. Study of soybean seed coat components and their relationship to water absorption. Journal of Agricultural Food Chemistry. 2001;49:5331–5335. doi: 10.1021/jf010303s. [DOI] [PubMed] [Google Scholar]
- Ohashi H. Glycine max (L.) Merr. subsp. soja (Sieb and Zucc.) Ohashi. comb. nov. (In Japanese.) Journal of Japanese Botany. 1982;57:30. [Google Scholar]
- Oka HI. Origins of cultivated rice. Tokyo and Amsterdam: Japan Scientific Societies Press and Elsevier Science Publishers; 1988. [Google Scholar]
- Palmer RG, Sun H, Zhao LM. Genetics and cytology of chromosome inversions in soybean germplasm. Crop Science. 2000;40:683–687. [Google Scholar]
- Palmer RG, Newhouse KE, Graybosch RA, Delannay X. Chromosome structure of the wild soybean. Journal of Heredity. 1987;78:243–247. [Google Scholar]
- Paterson A, Lin Y, Li Z, Schertz K, Doebley J, Pinson S, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, Roder MS, Li Y, Nevo E, Korol A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proceedings of the National Academy of Sciences, USA. 2003;100:2489–2494. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet V, Lamy F, Devos KM, Gale MD, Sarr A, Robert T. Genetic control of domestication traits in pearl millet (Pennisetum glaucum L., Poaceae) Theoretical and Applied Genetics. 2000;100:147–159. doi: 10.1007/s00122-002-0889-1. [DOI] [PubMed] [Google Scholar]
- Poncet V, Martel E, Allouis S, Devos KM, Lamy F, Sarr A, Robert T. Comparative analysis of QTLs affecting domestication traits between two domesticated × wild pearl millet (Pennisetum glaucum L., Poaceae) crosses. Theoretical and Applied Genetics. 2002;104:965–975. doi: 10.1007/s00122-002-0889-1. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J. Quantitative trait loci and the study of plant domestication. Genetica. 2005;123:197–204. doi: 10.1007/s10709-004-2744-6. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Abe J, Kanazawa A, Shimamoto Y. Marker-assisted analysis for soybean hard seededness with isozyme and simple sequence repeat loci. Breeding Science. 2004;54:133–139. [Google Scholar]
- Saxe LA, Clark C, Lin SF, Lumpkin TA. Mapping the pod-shattering trait in soybean. Soybean Genetics Newsletter. 1996;23:250–253. [Google Scholar]
- Sebolt AM, Shoemaker RC, Diers BW. Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Science. 2000;40:1438–1444. [Google Scholar]
- Shimamoto Y, Fukushi H, Abe J, Kanazawa A, Gai JY, Gao Z, Xu DH. RFLPs of chloroplast and mitochondrial DNA in wild soybean, Glycine soja, growing in China. Genetic Resources and Crop Evolution. 1998;45:433–439. [Google Scholar]
- Shimamoto Y, Abe J, Gao Z, Gai JY, Thseng FS. Characterizing the cytoplasmic diversity and phyletic relationship of Chinese landraces of soybean, Glycine max, based on RFLPs of chloroplast and mitochondrial DNA. Genetic Resources and Crop Evolution. 2000;47:611–617. [Google Scholar]
- Singh RJ, Hymowitz T. The genomic relationship between Glycine max (L.) Merr. and G soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theoretical and Applied Genetics. 1988;76:705–711. doi: 10.1007/BF00303516. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2nd edn. San Francisco: WH Freeman and Company; 1981. [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theoretical and Applied Genetics. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Thseng FS, Hosokawa S. Significance of growth habit in soybean breeding. I. Varietal differences in characteristics of growth habit. Japanese Journal of Breeding. 1972;22:261–268. [Google Scholar]
- Van Ooijen JW. MapQTL 5, Software for the mapping of quantitative trait loci in experimental populations. Kyazma, BV: Wageningen; 2004. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Arelli PR, Shoemaker RC, Diers BW. Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theoretical and Applied Genetics. 2001;103:561–566. [Google Scholar]
- Wang D, Graef GL, Procopiuk AM, Diers BW. Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theoretical and Applied Genetics. 2004;108:458–467. doi: 10.1007/s00122-003-1449-z. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Tajuddin T, Yamanaka N, Hayashi M, Harada K. Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breeding Science. 2004;54:399–407. [Google Scholar]
- Xiong LZ, Liu KD, Dai XK, Xu CG, Zhang QSP. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theoretical and Applied Genetics. 1999;98:243–251. [Google Scholar]
- Xu DH, Abe J, Gai JY, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theoretical and Applied Genetics. 2003;105:645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu L, Xiao J, Huang N, McCouch SR. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.) Molecular and General Genetics. 1997;253:535–545. doi: 10.1007/s004380050355. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Ninomiya S, Hoshi M, Tsubokura Y, Yano M, Nagamura Y, Sasaki T, Harada K. An informative linkage map of soybean reveals QTLs for flowering time, leaf morphology and region of segregation distortion. DNA Research. 2001;8:61–72. doi: 10.1093/dnares/8.2.61. [DOI] [PubMed] [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theoretical and Applied Genetics. 2004;108:1131–1139. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]