Abstract
Background and Aims
The changes that occur during the domestication of crops such as maize and common bean appear to be controlled by relatively few genes. This study investigates the genetic basis of domestication in pea (Pisum sativum) and compares the genes involved with those determined to be important in common bean domestication.
Methods
Quantitative trait loci and classical genetic analysis are used to investigate and identify the genes modified at three stages of the domestication process. Five recombinant inbred populations involving crosses between different lines representing different stages are examined.
Key Results
A minimum of 15 known genes, in addition to a relatively few major quantitative trait loci, are identified as being critical to the domestication process. These genes control traits such as pod dehiscence, seed dormancy, seed size and other seed quality characters, stem height, root mass, and harvest index. Several of the genes have pleiotropic effects that in species possessing a more rudimentary genetic characterization might have been interpreted as clusters of genes. Very little evidence for gene clustering was found in pea. When compared with common bean, pea has used a different set of genes to produce the same or similar phenotypic changes.
Conclusions
Similar to results for common bean, relatively few genes appear to have been modified during the domestication of pea. However, the genes involved are different, and there does not appear to be a common genetic basis to ‘domestication syndrome’ in the Fabaceae.
Key words: Domestication syndrome, genetics, Pisum sativum, Phaseolus vulgaris, seed dispersal, seed dormancy, roots
INTRODUCTION
The domestication of many seed crops has involved similar modifications in a set of traits including seed dispersal, seed dormancy, gigantism and increased harvest index. Hammer (1984) first used the term ‘domestication syndrome’ to describe this suite of changes in seed crops, and the term has been popularized by Harlan (1992) and others (e.g. Koinange et al., 1996). In the last 20 years the genes modified during the domestication of a diverse set of crops such as maize (Zea mays), rice (Oryza sativa) and common bean (Phaseolus vulgaris) have begun to be identified. The best studied case of these crops is maize, in which a relatively few genes are responsible for the significant morphological and physiological differences between wild and cultivated forms (Doebley et al., 1990; Doebley and Stec, 1991, 1993; Doebley, 1995; Bomblies and Doebley, 2006). In several additional studies, usually involving a quantitative trait locus (QTL) analysis of many traits associated with the domestication process, the QTLs identified have tended to cluster on the genome, suggesting that either relatively few genes with pleiotropic effects are involved or that the alleles selected during domestication have tended to represent closely linked loci in three or four regions of the genome (Xiong et al., 1999; Poncet et al., 2000; Peng et al., 2003; Li et al., 2006). These findings led several researchers to postulate that because domestication occurred relatively rapidly on an evolutionary scale, mutations with large effects would be selected, and these would often be at linked loci owing to the greater ease in maintaining linked mutations in a selection programme (Koinange et al., 1996).
The Fabaceae possesses the greatest number of domesticated crops of any plant family (Harlan, 1992) with 41 domesticated species. Some legume species, such as alfalfa and clover, have been domesticated for their use as fodder. However, most are cultivated for their highly nutritious seeds, which formed an important component of the diet of many early civilizations. Most legumes grown for their seeds, including pea, lentil (Lens culinaris), chickpea (Cicer arietinum), common bean, cowpea (Vigna unguiculata, soybean (Glycine max), pigeon pea (Cajanus cajan) and lupin (Lupinus angustifolius and others), can be expected to exhibit the type of modifications typical of the domestication syndrome. Thus, the Fabaceae may be an excellent system to examine to what extent parallel changes in morphology are determined by parallel mutations in genotype, i.e. are the same genes mutated or do plants have many alternative genetic pathways for obtaining the same phenotype?
An investigation of this question in the Poaceae (Paterson et al., 1995) suggested that the convergent evolution observed during domestication in sorghum, rice and maize was produced by mutations in many of the same genes. This finding led the authors to predict that identifying the genetic basis of domestication-related changes in minor crops in the Poaceae would be relatively easily achieved by extrapolation from findings in rice or maize. If the same is true for the Fabaceae, the most efficient approach to understanding the genetic changes that occurred during the domestication of lupin may be to study intensively a better characterized crop such as common bean. The answer to this question thus has considerable significance for legume breeders.
The initial problem was to choose which legume crops to compare. The most popular legume model for the study of domestication has been the common bean (Gepts, 1990; Sonnante et al., 1994; Blair et al., 2006), which has been shown to have two centres of domestication, one in Central America and one in the Andes (Gepts and Debouck, 1991). Koinange et al. (1996) performed a detailed QTL analysis of a recombinant inbred population derived from a wild × cultivated cross. They found that the QTLs for the various traits measured tended to cluster into several regions on the bean linkage map. Several of the genes identified by Koinange et al. (1996) as central to the domestication process have now been cloned and sequenced (Anthony et al., 1990; Kwak et al., 2006). Thus, in the present study, common bean was selected as the base species against which other species would be compared.
Pea was chosen as the second species for two reasons. First, this crop has been the object of many genetic studies (Blixt, 1972; Weeden and Muehlbauer, 2004). Although an exhaustive analysis of the genetic changes it has undergone during domestication has not been performed, many of the traits typically modified as part of the domestication syndrome have been at least partially analysed (Table 1) and at least 11 loci have been identified (the locus A affects both seed dormancy and seed quality and thus appears twice in Table 1). The Dpo1 locus was identified relatively early as a primary factor controlling pod dehiscence (Blixt, 1972). Flowering time is controlled by at least six loci (Murfet and Reid, 1985), although not all of them are important in domestication. Numerous genes or QTLs have been identified that influence plant habit (Blixt, 1972), seed size (Timmerman-Vaughan et al., 1996) and seed quality (Blixt, 1972). More recent studies have added to this list of genes controlling domestication-syndrome traits (Weeden et al., 2002; Timmerman-Vaughan et al., 2005), and molecular studies have now identified the coding sequences of many of these genes.
Table 1.
Previous results on genetic studies on traits forming the domestication syndrome of pea*
| Wild-type phenotype | Cultivated phenotype | Number of loci identified | Locus symbol(s) |
|---|---|---|---|
| Dehiscent pods | Indehiscent pods | 1 | Dpo |
| Dormancy present | Dormancy lacking | 1 | A |
| Tall | Dwarf | 1 | Le |
| Many basal branches | Few basal branches | 1 (minimum) | Rms series |
| Small seeds | Large seeds | several | QTL |
| Poor seed quality† | Good seed quality | 4 (minimum) | R, A, Pl, Gty |
| Long-day flowering | Day neutral flowering | 4 | Sn, Hr, Lf, E |
* See text for references.
† Seed quality involves taste, colour and texture genes.
The second reason for choosing pea is that several apparently intermediate stages for the domestication of pea are available in germplasm collections. The species is native to the Middle East, particularly the area between Turkey and Iraq, and domestication probably was initiated as soon as humans began to till the soil in that area (Zohary and Hopf, 1973). Although it may be debated as to whether any accessions of Pisum sativum ssp. elatius, the presumed wild ancestor of the cultivated pea, are truly wild in the sense that they have never been grown by humans, this taxon is sufficiently close to the wild ancestor to provide a reasonable starting point for the domestication process. The germplasm identified by the taxonomic label P. sativum ssp. abyssinicum appears to be a primitive landrace that displays several traits (indehiscent pods, smooth pods, thin testa) that are usually associated with initial steps in the domestication process. This subspecies is known to have a very narrow genetic diversity (Vershinin et al., 2003) and probably was transported to what are now the highlands of Ethiopia some 4000–5000 years ago when trade up the Nile extended all the way to this region. Presumably, this landrace became isolated when the trade route disappeared with the fall of the Kingdom of Egypt perhaps 3000 years ago and may provide a reasonable snapshot of the progress that had been made in the domestication of pea after some 5000 years of cultivation. A number of more advanced landraces of pea exist in various countries, but the most divergent and distinctive after P. s. ssp. abyssinicum is that found in the foothills and higher slopes of Afghanistan, Nepal, Iran and Pakistan. This landrace has been described as the ‘Afghanistan’ type (Weeden and Wolko, 1988) because most of the accessions representing this type in the USDA Pisum germplasm collection come from Afghanistan. The final stage in the domestication of pea is represented by the modern cultivars of the fresh market pea of commerce.
The approach used in the present study was simply to make crosses between lines representing the different stages of domestication for pea, determine which genes or QTLs were segregating in each population and compare these genes with those identified in common bean. The goals were to determine (1) how many genes were involved in the domestication of pea, (2) if these showed the same clustering that Koinange et al. (1996) observed in bean and (3) if mutants at the same loci in both species had been selected.
MATERIALS AND METHODS
Plant material
A set of five recombinant inbred populations (Table 2) were developed and analysed for segregating morphological and physiological traits. The populations represent different combinations of crosses between the four stages of domestication available in P. sativum. The 87-18&19 population represents the widest cross (P. sativum ssp. elatius × P. s. ssp. sativum) possible. This population was used to generate the base data for the consensus linkage map for the species (Weeden et al., 1998). Over 1000 segregating markers have been mapped in this population, representing an average distance between markers of <1 cM. The M × J population is similar to 87-18&19 in the categories being crossed, but it was made in the reciprocal direction (cultivated × wild) to test for cytoplasmic factors that might be involved in the domestication of pea. The JI1794 line was used in both the above crosses because it was a representative of the northern ‘humile’ type that produces fertile hybrids with, and is the likely wild ancestor of, domesticated pea (Palmer et al., 1985). Both of the crosses gave highly fertile F1 and F2 generations. Observations on plant height, dehiscent pod trait and flowering time have been made on these populations for each generation since the F4.
Table 2.
Recombinant inbred populations investigated
| Cross designation | Parental lines | Domestication stages involved | No. of lines | Generation analysed |
|---|---|---|---|---|
| 87-18&19 | JI1794 × Slow | Wild × cultivated | 53 | F12 |
| M × J | MN313 × JI1794 | Cultivated × wild | 50 | F6 |
| C × Ab | Cultivated × WL808 | Cultivated × primitive | 143 | BC1F4 |
| B06-100 | WL808 × JI261 | Primitive × wild | 30 | F4 |
| C × Af | CMG × PI220174 | Cultivated × landrace | 120 | F5 |
The crosses with the primitive landrace, P. sativum ssp. abyssinicum, produced a semi-sterile hybrid, partially as a result of a chromosomal rearrangement known to be present in this subspecies. To minimize sterility issues and loss of inbred lines as much as possible in subsequent generations, the hybrid from the cultivated × P. s. ssp. abyssinicum cross was backcrossed to the cultivated type, and the BC1 generation was then selfed to form the recombinant inbred (RI) population. The hybrid produced from a P. sativum ssp. abyssinicum × P. s. ssp. elatius cross was also semi-sterile, and the resulting F2-derived RI population contained several weak or semi-sterile lines. The cultivated × advanced landrace (C × Af) population was vigorous and fully fertile.
Each of these populations was examined for segregating morphological features known to differ between the wild and cultivated types (Table 1). Pod dehiscence was measured either by direct observation of the pods on the plant or by drying freshly harvested pods in an incubator (Weeden et al., 2002). Seed dormancy was determined by placing five seeds from a line between moistened filter papers in a Petri dish and noting the number of seeds that imbibed after 48 h. Seeds that were still hard after 48 h had their testa nicked and were allowed to sit for additional 24 h to confirm that they would imbibe after testa scarification. For seed weight determination, a sample of at least ten healthy seeds from a line was weighed to the nearest 0·1 g. In most cases, the determination was made on at least two successive generations of each line and an average estimate of weight per 100 seeds calculated. Seeds from each line were scored for testa smoothness, presence of anthocyanins, black hilum and round or wrinkled phenotype. For the C × Ab population, each line was grown under long day (≥16 h light) as well as short day (≤12 h light) conditions, and both days to flowering (DTF) and nodes to flowering (NTF) were determined under both conditions. In the 87-18&19, M × J and C × Ab populations, roots were measured, weighed and compared with stem weights as described in Weeden and Moffet (2002).
Genetic analyses
Linkage maps were developed for each population using morphological, isozyme and DNA markers. Morphological markers were scored using known phenotypic ranges for each genotype (Blixt, 1972). Allozyme variation was determined according to Weeden and Marx (1987). Random amplified polymorphic DNA (RAPD) analysis was performed as per Torres et al. (1993) and sequence tagged site (STS) analysis as per Brauner et al. (2002) and Moffet (2006). In those populations with less saturated maps, the markers were selected to provide thorough coverage of the chromosomes. Because each population consisted of relatively few inbred lines, an extensive QTL analysis was not performed. Instead, major QTLs were tested for by performing ANOVA for each quantitatively measured trait against a selection of segregating markers distributed every 10 cM along the linkage map. When a clear correlation was identified, such as in the case with plant height and the region containing Le, the inbred population was divided into two groups based on the allele at the nearest anchor locus, and each subgroup was subjected to a similar ANOVA analysis over the remainder of the linkage map.
RESULTS
Wild × cultivated crosses
The traits segregating along with the number of genes identified controlling these traits in the wide (wild by cultivated and reciprocal) crosses 87-18&19 and M × J are presented in Table 3. Analysis of pod dehiscence revealed four QTLs in the 87-18&19 RI population and only two in the M × J population. In both populations one QTL was placed at a position indistinguishable from that for the locus Dpo1 on Linkage Group (LG) III. Another QTL in both populations co-segregated with Np, also on LG III. In the glasshouse, Np plants form a large number of pustules or even a greatly thickened pod wall (Fig. 1) that appears to interfere with the natural dehiscence of the pod. Hence, it is probable that the second QTL on LG III is the same as Np. A third QTL in the 87-18&19 population co-segregated with the yellow pod gene, gp. The gp allele reduces pod wall sclenchyma and probably is the source of this QTL. The Gp locus was polymorphic only in 87-18&19. The final QTL, observable only in 87-18&19, was found on LG VII, and was designated Dpo2.
Table 3.
Genetic differences identified in two populations derived from crosses between wild pea (JI1794) and cultivated types
| Trait | Cross, number of loci and locus symbol* |
|
|---|---|---|
| 87-18&19 | M × J | |
| Pod dehiscence | 4 (Dpo1, Dpo2, Np, Gp) | 2 (Dpo1, Np) |
| Seed dormancy | 2 (R, S) | 2 (A, R) |
| Plant height | 2 (Le, GA2βOH) | 2 (Le, GA2βOH) |
| Basal branching | 1 (Rms1) | 1 (Rms1) |
| Seed weight | 4 (R, Np, QTL-I, -IV) | 2 (R, Np) |
| Seed quality | 3 (Gty, Pl, R) | 4 (Gty, Pl, R, A) |
| Flowering response | 4 (E, Sn, Le, QTL-V) | 3 (E, Sn, Le) |
| Pod neoplasm | 1 (Np) | 1 (Np) |
| Root mass | 1 (Le) | 1 (Le) |
| Root/shoot ratio | 2 (QTL-III, QTL-VII) | 1 (QTL-VII) |
* Quantitative trait loci discussed in the text are identified in the table as ‘QTL’ followed by the linkage group on which they mapped.
Fig. 1.

Photograph of pea pods with considerable growth of undifferentiated tissue (neoplasms) caused by expression of the Np allele. Such expression usually occurs only when plants are grown in the glasshouse. In the field, Np expression is suppressed except under specific conditions such as bruchid infestation.
Seed dormancy gave two QTLs in the two populations, but only one was shared. This QTL co-segregated with Mendel's round/wrinkled locus (R) and appeared to be identical to it because wrinkled seeds tend to develop cracks in the testa more readily than round seeds. The second QTL in the 87-18&19 population was identified as the gene s that causes seeds to stick together in the pod (Blixt, 1972). Separating the seeds usually causes small fractures in the testa. The second QTL in the M × J population mapped to the A locus. As the A locus was monomorphic in the 87-18&19 population and the S locus was not segregating in the M × J population, the QTLs identified for seed dormancy could all be assigned to known genes and provided consistent results.
Plant habit traits segregating included height, the presence of basal branches and leaflet shape (round vs. lanceolate). In both RI populations, the region on LG III just distal to Tpip contained a major QTL for internode length (plant height), reflecting segregation at Le (Fig 2). A second QTL for overall plant height centred very near the STS for gibberellin 2 β-hydroxylase (GA2βOH) on LG IV in both populations. JI1794 contained the recessive allele for this second QTL, causing this line to have a shorter stature than the hybrid and domesticated germplasm possessing the Le allele. A QTL for leaf shape (as measured by width/length ratio) also mapped to this region. Basal branching appeared to be monogenic in both populations, mapping near to the vicilin5 locus (Fig. 2) on the end of LG III opposite Le. The position of this gene suggested that Rms1 is responsible for the polymorphism.
Fig. 2.
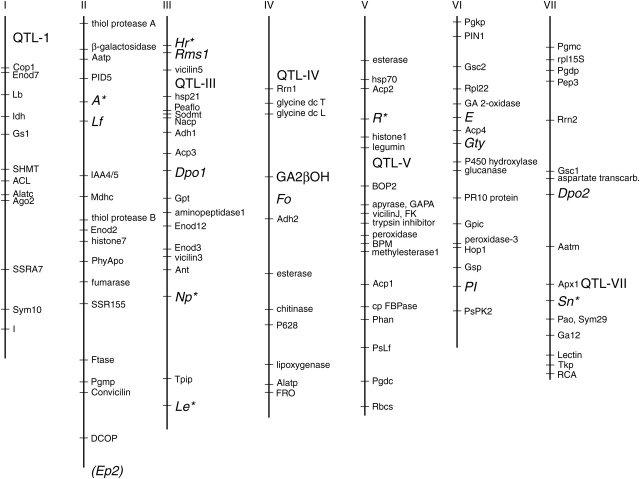
The linkage groups of Pisum sativum (n = 7). Symbols and relative positions of loci and QTLs influencing morphological or physiological changes occurring during the domestication of the species are given in large print to the right of the respective linkage group. Anchor markers are shown in small print along the linkage groups. An asterisk following a locus symbol indicates that the locus had pleiotropic effects on several traits modified during domestication of pea.
Seed size in both populations gave QTLs near R on LG V and Np on LG III. In the 87-18&19 population two additional QTLs were observed, one near Rrn1 on LG IV and one near Cop1 on LG I (Fig. 2). The effect of the R and Np regions was particularly clear in the M × J population (Table 4), where the largest and smallest seed size classes are the recombinant genotypes.
Table 4.
Effect of variation at the loci Np and R on the average seed weight in the M × J population
| Genotype of RIL | Average seed weight (± s.d.)* |
|---|---|
| np/np, round seed | 16·2 ± 2·0 g/100 seeds |
| np/np, wrinkled seed | 14·9 ± 2·4 g/100 seeds |
| Np/Np, round seed | 12·8 ± 2·3 g/100 seeds |
| Np/Np, wrinkled seed | 11·9 ± 2·2 g/100 seeds |
* Differences between np and Np categories are significant in all comparisons. Differences between r and R categories are not significant at P = 0·05.
All seed quality characters investigated were monogenic and could be identified with genes previously identified in pea. The rough seed coat in JI1794 is produced by a dominant allele at locus Gty on LG VI (Blixt, 1974). Black hilum is conditioned by a dominant allele at the locus Pl, also on LG VI. The round/wrinkled polymorphism and the spotted vs. clear testa are both controlled by loci (R and A, respectively) described by Mendel (1865). Each of these segregated as expected in the M × J population, and only the first three loci segregated in the 87-18&19 population, both parents of this cross possessing the dominant gene at the A locus.
QTL analysis of flowering time identified four regions affecting the trait in the 87-18&19 population and three regions in the M × J population (Table 3, Fig. 2). The three regions in the latter population each correlated with a different locus known to influence time of flowering (Hr, E and Le). The fourth QTL in the 87-18&19 population mapped to a position on LG V that lacks a known flowering gene.
Similar results from the two populations were also obtained for the root analysis. A major QTL for root mass co-segregated with Le in both populations. As in the parental types, the larger root mass was associated with dwarf plants. No other significant QTLs were identified for root mass. The 87-18&19 population gave two QTLs for root/shoot ratio, one near Sn on LG VII and one near Vicilin5 on LG III. Only the QTL near Sn was observed in the M × J population. Curiously, the root mass QTL at Le did not show up as a root/shoot QTL in either population, indicating that dwarf plants have greater shoot mass despite their shorter stature.
The direction of the cross did not appear to make a difference in the segregation of domestication-associated characters, suggesting that neither the mitochondrial nor the plastid genome has been significantly modified during domestication. In the 87-18&19 F2 and F3 generations, several seedlings lacking chlorophyll were observed, indicating some cytoplasmic incompatibilities in this cross. However, white or pale yellow seedlings have not been produced in any of the inbred lines since the F3.
Primitive × wild (population B06-100)
The semi-sterility associated with many lines in this population interfered with the genetic analysis of several traits in this population. However, important conclusions can still be made despite distorted segregation patterns at many marker loci. Those traits identified as polymorphic in the crosses between wild and cultivated lines were examined for segregation in this RI population (Table 5). JI261 has dehiscent pods typical of wild pea, whereas P. s. ssp. abyssincum possesses indehiscent pods. This trait segregated in the RI population, and the segregation pattern correlated with the region on LG III where the major gene (Dpo1) controlling pod dehiscence in the wild × cultivated crosses is located. This region is on one of the chromosomes involved in the reciprocal translocation that characterizes P. s. ssp. abyssinicum and exhibited a distorted segregation ratio. Other QTLs affecting the dehiscent pod trait were not resolved.
Table 5.
Behaviour of domestication-syndrome characters in the recombinant inbred populations derived from crosses other than between wild and cultivated lines
| Primitive × wild (B06-100) |
| Monomorphic traits: internode length (Le), basal branching (Rms1), black hilum (Pl), round seed (R), coloured flowers (A), root mass, root/shoot ratio |
| Polymorphic traits: pod dehiscence (Dpo1), seed dormancy (Ep2), plant height (GA2βOH), seed weight (QTL not resolved), testa texture (Gty), pod neoplasm (Np), flowering time (QTL not resolved) |
| Cultivated × primitive (C × Ab) |
| Monomorphic traits: pod dehiscence (dpo1), seed dormancy (Ep2), plant height (GA2βOH), seed weight (no significant QTL), testa texture (gty), pod neoplasm (np) |
| Polymorphic traits: internode length (Le), basal branching (Rms1), black hilum (Pl), round seed (R), coloured flowers (A), flowering time (Le, Lf, Sn, E), root mass |
| Cultivated × Afghanistan (C × Af) |
| Monomorphic traits: pod dehiscence (dpo1), seed dormancy, plant height (GA2βOH), seed weight (no significant QTL), testa texture (gty), pod neoplasm (np) |
| Polymorphic traits: internode length (Le), basal branching (Rms1), black hilum (Pl), round seed (R), coloured flowers (A), flowering time (Hr) |
Seed dormancy also appeared to display segregation in this population. JI261 has a thick testa and will not germinate unless the intactness of the testa is disrupted by scarification. Seeds from P. s. ssp abyssincum usually do not require such treatment before germination. Freshly harvested seed from all lines in this population would not germinate without scarification. However, with time (3–12 months) some of the lines lost this dormancy character. The loss of dormancy showed correlation with markers on LG II but could not be mapped unambiguously. This QTL was tentatively labelled Ep2, a gene controlling testa thickness identified by Kaznowski (1926), and its association with LG II is indicated in Fig. 2 by placing it at the base of the linkage group.
Two aspects of plant habit segregated in this population. All plants possessed long internodes (Le), but the lines showed the same segregation for overall plant height that was observed in the wild × cultivated RI populations. This QTL mapped on LG IV near GA2βOH (Table 5). The second morphological polymorphism segregating was the strongly serrate leaflets produce by the Ser allele in P. s. ssp. abyssincum (Weeden and Ambrose, 2004). As expected, this polymorphism displays monogenic segregation and mapped very near the breakpoint for the translocation involving LG III. However, the segregation at Ser obscured a possible segregation in leaflet shape observed in the wild × cultivated crosses. Both parents and all RI lines displayed basal branching (Table 5).
The seeds of P. s. ssp. abyssincum are significantly larger than P. s. ssp. elatius line JI261. However, the low vigour of some of the lines made analysis of seed weight problematic. No obvious correlations between seed weight and regions of the linkage map were detected. The only other aspect of seed quality that clearly changed between ssp. elatius and ssp. abyssinicum was a transition from a rough to a smooth testa. Genetic analysis mapped this difference to LG VI, confirming that the difference was the Gty/gty polymorphism seen in the wild × cultivated crosses. The Np locus also segregated in this population.
Node to flowering was significantly earlier in ssp. abyssinicum than in ssp. elatius (node 13 ± 1 vs. node 17 ± 2, respectively). The genetic basis of this change has yet to be determined in this population.
Cultivated × primitive (backcross RI population C × Ab)
The backcross RI population displayed semi-sterility (low productivity and incomplete filling of pods) in some lines as well as distortion in some regions of the genome from the expected 1 : 1 allelic ratio. However, genetic analysis was possible for most traits. Both parents possessed indehiscent pods and the character was monomorphic in the RI population, confirming that the same loci (Dpo1 and Dpo2) had mutations in both parental lines (Table 5). The data on seed dormancy provided ambiguous results, and no clear QTLs were identified for this character in this cross.
Two of the plant habit loci detected in the wild × cultivated populations (Le and Rms1) segregated in the backcross RI population, as did the Ser locus. Segregation in the region near GA2βOH did not appear to influence plant height, nor were QTLs observed for seed dormancy or seed weight. The seed quality loci Pl, R and A each segregated as expected from the known phenotype of the parents, whereas no evidence for Np expression was observed.
The genes influencing flowering time were examined in this population under both short-day and long-day photoperiod. The primitive landrace (P. s. ssp. abyssinicum) displayed considerable delay in flowering under short-day conditions (>50 DTF) compared with long photoperiod (approximately 32 DTF) and was suspected of having the genotype Sn, Hr. However, no evidence could be found for segregation at Hr in the RI population. Instead, the major QTLs for flowering time were found at Sn and Le, with minor QTLs at positions corresponding to Lf and E. The allele at Lf appears to be unique to the subspecies, bestowing an early-flowering phenotype under long-day photoperiods.
As anticipated from the findings for the crosses involving the wild subspecies, where no difference in root mass was observed, segregation of root mass in the C × Ab population gave a single large QTL that co-segregated with Le. Shoot weights were not obtained for most of the lines, precluding an analysis of root/shoot ratios.
Cultivated × Afghanistan (C × Af)
This population showed no segregation for pod dehiscence or for seed dormancy. A clear monogenic ratio was observed in segregation for plant height, with Le being the polymorphic locus. Basal branching segregated, and Rms1 was postulated to be the locus responsible based on the placement of the branching gene on the upper end of LG III. Seed size varied slightly among the lines, but significant QTLs could not be identified. The only two seed quality traits segregating were those controlled by the A and R loci. The flowering gene, Hr, also segregated. Root mass and root/shoot data have not been taken on this population.
Distribution of domestication genes on the pea linkage map
The distribution of the 15 genes and QTLs influencing the traits modified during domestication of pea is shown in Fig. 2. There is at least one gene on each linkage group, with the longest linkage group (III) possessing the greatest number of genes. There does not appear to be a clustering of genes involved in the control of domestication-syndrome traits. The only possible tight cluster of loci affecting domestication traits would include Hr, Rms1 and the QTL controlling root/shoot ratio in that region if it is not a pleiotropic effect of Hr.
Comparison of genes with common bean
Several genes in bean are associated with the domestication syndrome. These are Fin (controlling determinancy), Ppd (photoperiod sensitivity), P (anthocyanin in testa), St (pod string) and Phs (phaseolin/seed weight). In pea, potential orthologues or homologues of four of these genes include det, sn, dne, sin and those coding several seed proteins. The most interesting of these is det, almost certainly the orthologue of the fin gene in common bean (Moffet, 2006). Although fin is a critical component of the domestication process in common bean, det in not important in the domestication of pea. Similarly, pea genes p and v reduce pod wall sclerenchyma, a part of the mechanism by which pods became indehiscent in bean. However, neither p nor v was involved in the early steps in the domestication of pea in which dpo1 and dpo2 were selected to avoid pod shattering. The loss of the ‘string’ also contributed to pod indehiscence in common bean. A ‘stringless’ gene (sin) is also present in pea, but it also appears not to have been involved in the original solution to pod shattering during domestication.
The loss of seed dormancy in pea is far more significant than what has been determined for common bean. The wild-type seed in pea will not germinate for a year or more without scarification (my pers. observ.), while Koinange et al. (1996) report only a 30 % drop in germination rate in common bean. The mechanism (testa impermeability) may be the same, but a comparable gene to the thick testa locus (Ep2) in pea has not been found in bean. The loss of anthocyanin pigmentation in the seed coat is found in both pea and common bean. However, this parallel appears to involve different genes. Although the DNA sequence of neither the a mutation (pea) or the p mutation (bean) has been determined, the a gene is almost certainly upstream of p, given that it blocks the synthesis of anthocyanin and many flavonoids throughout the plant, whereas p simply eliminates anthocyanins from the testa.
Although the phaseolin protein gene is an important component of seed weight in bean, none of the QTLs for seed weight in pea has mapped near seed protein genes such as legumin or vicilin. One of the seed weight QTLs in pea maps near the rDNA array (Timmerman-Vaughan et al., 1996), as does a similar QTL in lentil (Abbo et al., 1992), suggesting that expression and interaction of ribosomal RNAs may influence seed weight in these genera. No such correlation has been identified in Phaseolus.
Another aspect of the domestication syndrome in common bean is a significant reduction of the number of nodes on the main stem accompanied by a small increase in internode length. The dwarfing mechanism in pea involves virtually the opposite approach, with the internode length, not the number of internodes, being affected by the substitution of le for Le. Loss of photoperiod sensitivity was part of the domestication process in both crops, but this trait is difficult to compare because pea requires long daylength to flower, while bean requires short daylength. An early-flowering gene has been identified and used in pea (lf) that is different from either of the genes in pea that modify photosensitivity (hr and sn). This gene is not orthologous to any of the flowering genes important in the domestication of common bean. The actual coding sequences of pea flowering genes Ppd, sn and dne have yet to be identified, precluding the examination and evaluation of homologous relationships between the genes bestowing day neutrality in bean and pea.
DISCUSSION
Approximately 20 genes or QTLs are responsible for the modifications of plant form and function that accompanied the domestication of pea. This number is comparable with estimates for the number of genes involved in the domestication of common bean, maize and rice (Li et al., 2006) but much fewer than those estimated for sunflower (Burke et al., 2002). Most of the genes identified in pea are well characterized, making it possible in many cases to identify pleiotropic actions of these genes. Thus, we know that the substitution of a for A in pea improved seed quality and reduced seed dormancy. Loss of Np increased seed size (at least under certain conditions) but also reduced tolerance to bruchid attack. The recessive r allele improves seed quality (sweetness) but appears to reduce seed size. Homozygosity for the dwarfing gene may increase root mass, and two of the photoperiod response genes, Sn and Hr, are either closely linked to genes that influence root/shoot ratio or are directly involved themselves.
Thus, in at least three instances (A, R and Np) it is not necessary to postulate a closely linked set of genes that form a co-adapted complex in order to explain this clustering of effects on the linkage map. Only the region that includes the photoperiod-sensitivity gene Hr, the branching gene Rms1 and possibly the root/shoot ratio QTL is a candidate for a co-adapted complex of genes being involved in the domestication process. Whether the root/shoot QTL is actually a pleiotropic effect of Hr will have to await the identification of the gene and additional physiological studies on the partitioning of photosynthate between roots and shoots. In summary, the distribution of domestication genes on the pea genome appears to be random, in contrast to findings in common bean (Koinange et al., 1996; Li et al., 2006).
The availability of pea germplasm representing several stages during the domestication process permits us to develop a time line for the process as well as infer the sequence of steps followed in this process. Of the eight categories of traits outlined by Koinange et al. (1996) for the domestication of common bean, four had been at least partially changed in pea by about 5000 years ago. The indehiscent pod trait appears to have been fixed in domesticated germplasm by that time, and the longer-term seed dormancy also appears to have been eliminated. Gigantism, in the form of seed weight, had increased about two-fold, although there are Pisum sativum lines with significantly smaller seeds, and it may be that the JI1794 accession already was a product of some selection for larger seed. Arguing against this possibility is the fact that the wild species Pisum fulvum has seeds approximately the same size as JI1794. Earliness may also have been selected before the split of P. s. ssp. abyssinicum from the main track of pea domestication because it flowers relatively early. However, the allele responsible for earliness in this subspecies appears not to be present in the remaining domesticated germplasm, and an alternative explanation would be that the allele was selected after the divergence.
The remaining four categories of traits were not modified in the initial stage of domestication. The dwarf pea habit probably only appeared in the last 500–1000 years, along with those types that were photoperiod-insensitive and white flowered. Increases in seed weight, number of seeds per pod (not measured in this study) and harvest index also occurred in the last 1000 years and at least some of the variation in seed weight is controlled by pleiotropic effects of other genes selected during domestication. Although no data on harvest index are presented here, it is probable that genes controlling seed weight, plant height, branching habit and flowering time all contribute to variation and improvement in this character.
The result that root mass and root/shoot ratio have increased during domestication is somewhat surprising considering that much of the progress made in increasing yields during the last 50 years has been by reducing plant vegetative mass (usually stem tissue) relative to seed or fruit (increasing harvest index). It appears that in pea, a major limiting factor for yield in wild-type and primitive domesticated forms may be either nutrients or water supplied through the roots. Modification of stem height or photoperiod response may have allowed more photosynthate to be diverted to the roots, allowing this organ to explore more soil volume. In support of this hypothesis, Kelly and Spanswick (1997) found that variation at the Sn locus had an effect on rate and duration of seed growth and suggested that this effect was due to an action of Sn on the partitioning of carbon resources between vegetative and reproductive portions of the plant. If Sn affects this partitioning, it may also have a role in determining partitioning of photosynthate between roots and shoots. Alternatively, the larger roots may be a result of greater overall vigour of the genotype, with breeders selecting for a smaller stem in order to increase harvest index but not focusing on the root volume.
In several cases the analysis of a domestication-syndrome trait clearly indicated that other genes, unrelated to domestication, were segregating in the population and influencing the expression of the trait. Examples include the effect of the yellow pod (gp) and neoplasm (Np) alleles on pod dehiscence and the r and s genes on seed dormancy in the 87-18&19 population. For the pod dehiscence character, two other QTLs could be identified, and one of these corresponded to the locus, Dpo1, previously shown to differ between wild and cultivated types. However, the effects of r and s completely obscured the effects of genes from JI1794 that produced the thick testa phenotype. Hence, the use of several different RI populations provided the ability to analyse for the presence of several QTLs in different genetic backgrounds and expose the action of some that were hidden in the wild by cultivated crosses.
Differences in the loci identified in the analyses of the two wild × cultivated crosses were present. Presumably, if the trait being examined was altered during the domestication process, both crosses should give the same results unless the cytoplasm has an influence on the expression of the trait. No cytoplasmic effect could be confirmed here. It appeared that the inconsistencies were due to the differences in genotype of the cultivated lines used in the crosses, the saturation of the genetic maps available for the two populations (only slightly over 300 segregating markers were available for the M × J population) and the number of replicate analyses performed on each population for traits such as pod dehiscence and flowering time (for the 87-18&19 population eight generations of data are available, whereas for the M × J population only two generations have been analysed thoroughly). Because the 87-18&19 population was far more thoroughly investigated, the results from this population were assumed to be more reliable.
The most surprising finding of this study is that despite many parallels in the modifications during domestication between pea and common bean, no genes that were involved in the domestication of both crops were identified. Problems with seed dispersal, growth habit, earliness, seed quality and seed pigmentation all appear to involve different suites of genes in pea compared with bean. The case for seed dormancy, gigantism and particularly the loss of photoperiod sensitivity is less clear and may involve homologous or orthologous sequences. Resolution of these issues probably will require the identification of the coding sequence of the gene affected in one crop followed by the mapping of that sequence in the other. It is unfortunate that common bean has not proven to be a very useful model for understanding the genetics of domestication in pea, or vice versa. However, it is encouraging from a breeder's perspective to find that there are at least several ways to modify unwanted characters such a pod dehiscence and plant habit and possibly avoid some of the detrimental effects accompanying the substitution of certain alleles for others.
ACKNOWLEDGEMENTS
I would like to thank the late Dr G. A. Marx for initiating my interest in pea domestication and two anonymous reviewers for a careful reading and helpful comments on the initial submission. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Abbo S, Ladizinsky G, Weeden NF. Genetic analysis and linkage study of seed weight in lentil. Euphytica. 1992;58:259–266. [Google Scholar]
- Anthony JL, Vonder Haar RA, Hall TC. Nucleotide sequence of an alpha-phaseolin gene from Phaseolus vulgaris. Nucleic Acids Research. 1990;18:3396. doi: 10.1093/nar/18.11.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair MW, Iriarte G, Beebe S. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean × wild common bean (Phaseolus vulgaris L.) cross. Theoretical and Applied Genetics. 2006;112:1149–1163. doi: 10.1007/s00122-006-0217-2. [DOI] [PubMed] [Google Scholar]
- Blixt S. Mutation genetics in Pisum. Agri Hort. Genetica. 1972;30:1–293. [Google Scholar]
- Blixt S. The pea. In: King RC, editor. Handbook of genetics. Vol. 2. New York: Plenum Press; 1974. pp. 181–221. [Google Scholar]
- Bomblies K, Doebley JF. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics. 2006;172:519–531. doi: 10.1534/genetics.105.048595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner S, Murphy RL, Walling JG, Przyborowski J, Weeden NF. STS markers for comparative mapping in legumes. Journal of the American Society of Horticulture Science. 2002;127:616–622. [Google Scholar]
- Burke JM, Tang S, Knapp SJ, Rieseberg LH. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. Genetic dissection of the morphological evolution of maize. Aliso. 1995;14:297–304. (1996) [Google Scholar]
- Doebley J, Stec A. Genetic analysis of the morphological differences between maize and teosinte. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A. Inheritance of the morphological differences between aize teosinte: comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Wendel J, Edwards M. Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proceedings of the National Academy of Sciences USA. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P. Biochemical evidence bearing on the domestication of Phaseolus beans. Economic Botany. 1990;44(3S):28–38. [Google Scholar]
- Gepts P, Debouck DG. Origin, domestication, and evolution of the common bean, Phaseolus vulgaris. In: Voysest O, Van Schoonhoven A, editors. Common beans: research for crop improvement. Wallingford, UK: CAB; 1991. pp. 7–53. [Google Scholar]
- Harlan JR. Crops and man. Madison, WI: American Society of Agronomy; 1992. [Google Scholar]
- Hammer K. The domestication syndrome. Kulturpflanze. 1984;32:11–34. (in German) [Google Scholar]
- Kaznowski L. Recherches sur le pois. (Pisum) Mémoires de l'Institut national polonais d'économie rurale á Pulawy. 1926;7:1–91. [Google Scholar]
- Kelly MO, Spanswick RM. Maternal, single-gene regulation of assimilate partitioning in pea. Plant Physiology. 1997;114:1055–1059. doi: 10.1104/pp.114.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common bean. Crop Science. 1996;36:1037–1045. [Google Scholar]
- Kwak M, Kami JA, Gepts P. Plant & Animal Genome XIV. 2006. Identification of the determinacy gene (Fin) and its evolution during domestication in common bean (Phaseolus vulgaris L.) poster 447. The abstract is available at: http://www.intl-pag.org/14/abstracts/PAG14_P447.html . [Google Scholar]
- Li C, Zhou A, Sang T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytologist. 2006;170:185–194. doi: 10.1111/j.1469-8137.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- Mendel G. Versuche ueber Pflanzen-Hybriden. Verhandlungen des Naturforschenden den Vereines in Bruenn. 1865;4:3–47. [Google Scholar]
- Moffet M. Bozeman, MT: Montana State University; 2006. Identifying regions of conserved synteny between pea (Pisum spp.), lentil (Lens spp.), and bean (Phaseolus spp.). MS thesis. [Google Scholar]
- Murfet IC, Reid JB. The control of flowering and internode length in Pisum. In: Hebblethwaite PD, Heath MC, Dawkins TCK, editors. The pea crop. London: Butterworths; 1985. pp. 67–80. [Google Scholar]
- Palmer JD, Jorgensen RA, Thompson WF. Chloroplast DNA variation and evolution in Pisum patterns of change and phylogenetic analysis. Genetics. 1985;109:195–214. doi: 10.1093/genetics/109.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Lin Y-R, Li Z, Schertz KF, Doebley J, Pinson SRM, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, Roeder MS, Li Y, Nevo E, Korol AK. Domestication quantitative trait loci in Triticum diococcoides, the progenitor of wheat. Proceedings of the National Academy of Sciences USA. 2003;100:2489–2494. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet V, Lamy F, Devos KM, Gale MD, Sarr A, Robert T. Genetic control of domestication traits in pearl millet (Pennisetum glaucum L., Poaceae) Theoretical and Applied Genetics. 2000;100:147–159. doi: 10.1007/s00122-002-0889-1. [DOI] [PubMed] [Google Scholar]
- Sonnante G, Stockton T, Nodari RO, Becerra Velasquez VL, Gepts P. Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.) Theoretical and Applied Genetics. 1994;89:629–635. doi: 10.1007/BF00222458. [DOI] [PubMed] [Google Scholar]
- Timmerman-Vaughan GM, McCallum JA, Frew TJ, Weeden NF, Russell AC. Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum L.) Theoretical and Applied Genetics. 1996;93:431–439. doi: 10.1007/BF00223187. [DOI] [PubMed] [Google Scholar]
- Timmerman-Vaughan GM, Mills A, Whitfield C, Frew T, Butler R, Murray S, et al. Linkage mapping of QTL for seed yield, yield components and developmental traits in pea. Crop Science. 2005;45:1336–1344. [Google Scholar]
- Torres AM, Weeden NF, Martin A. Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theoretical and Applied Genetics. 1993;85:937–945. doi: 10.1007/BF00215032. [DOI] [PubMed] [Google Scholar]
- Vershinin AV, Allnutt TR, Knox MR, Ambrose MJ, Ellis THN. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Molecular Biological Evolution. 2003;20:2067–2075. doi: 10.1093/molbev/msg220. [DOI] [PubMed] [Google Scholar]
- Weeden NF, Ambrose MJ. Ser appears to be the serrate leaflet locus mapped on linkage group III. Pisum Genetics. 2004;36:25–26. [Google Scholar]
- Weeden NF, Marx GA. Further genetic analysis and linkage relationships of isozyme loci in pea. Journal of Heredity. 1987;78:153–159. [Google Scholar]
- Weeden NF, Moffet M. Identification of genes affecting root mass and root/shoot ratio in a JI1794 × ‘Slow’ RIL population. Pisum Genetics. 2002;34:28–31. [Google Scholar]
- Weeden NF, Muehlbauer FJ. Genomics and genetic improvement in the cool season pulse crops pea, lentils and chickpea. In: Wilson RF, Stalker HT, Brummer EC, editors. Legume crop genomics. Champaign, IL: AOCS Press; 2004. pp. 83–96. [Google Scholar]
- Weeden NF, Wolko B. Measurement of genetic diversity in pea accessions collected near the center of origin of domesticated pea. Rome: IPBGR; 1988. [Google Scholar]
- Weeden NF, Ellis THN, Timmerman-Vaughan GM, Swiecicki WK, Rozov SM, Berdnikov VA. A consensus linkage map for Pisum sativum. Pisum Genetics. 1998;30:1–4. [Google Scholar]
- Weeden NF, Brauner S, Przyborowski JA. Genetic analysis of pod dehiscence in pea (Pisum sativum L.) Cellular and Molecular Biology Letters. 2002;7:657–663. [PubMed] [Google Scholar]
- Xiong LZ, Liu KD, Dai XK, Xu CG, Zhang Q. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theoretical and Applied Genetics. 1999;989:243–251. [Google Scholar]
- Zohary D, Hopf M. Domestication of pulses in the Old World. Science. 1973;182:887–894. doi: 10.1126/science.182.4115.887. [DOI] [PubMed] [Google Scholar]