Abstract
Backgroud and Aims
The Caesalpinia hintonii group comprises six species of endemic shrubs or trees, C. epifanioi, C. hintonii, C. laxa, C. macvaughii, C. melanadenia and C. oyamae, found in scattered patches of seasonally dry forest in the Río Balsas depression and the neighbouring Tehuacán–Cuicatlán valley, which are part of the Mexican morphotectonic province of Sierra Madre del Sur. An evaluation is made of phylogeographic patterns and genetic diversity with a phylogenetic analysis of the C. hintonii complex in order to study the dynamics of speciation in this endemic group of legumes.
Methods
A phylogeographic study based on four highly variable non-coding plastid regions (trnL intron, trnL-F intergenic spacer, trnH-psbA intergenic spacer, and accD-psaI intergenic spacer) was carried out for the Caesalpinia hintonii complex. Five of the six taxa of the C. hintonii complex were included.
Key Results and Conclusions
The plastid analyses involving multiple accessions of each taxon from throughout their ranges resolved C. epifanioi and C. hintonii as well-supported clusters, but C. oyamae has two unexpectedly divergent lineages. Two well-supported geographic clades: eastern (C. epifanioi, C. melanadenia and C. oyamae) and western (C. hintonii and C. macvaughii) were established. The analyses performed provide evidence of recent morphostatic radiation in C. oyamae resulting from isolation and local adaptation. This pattern of genetic differentiation without morphological divergence may be a model that fits many groups of tropical woody taxa inhabiting similarly dry forests subjected to shifting selection.
Key words: Caesalpinia hintonii complex, legumes, Mesoamerica, Mexico, plant phylogeography, population differentiation, seasonally dry forest
INTRODUCTION
All currently observed distribution patterns and evolutionary processes are the product of the interplay of historical and ecological factors. A substantial body of evidence indicates that climatic changes have led to major reshuffling of species ranges, dispersal and colonization (Hewitt, 1996, 2001; Dynesius and Jansson, 2000). These have left a distinct imprint on the genetic structure and variation of many species (e.g. Taberlet et al., 1998; Austin et al., 2002). For many taxa, climatic changes during glacial periods have caused range adjustments and extinctions (Hewitt, 2000; Kropf et al., 2002), population subdivision and divergence (Wakeley, 2000), and in some cases radiation (Sáez et al., 2003).
Phylogeographic studies for plant species in Europe (e.g. Dumolin-Lapègue et al., 1997; Petit et al., 1997; Comes and Kadereit, 1998; King and Ferris, 1998; Taberlet et al., 1998; Abbott et al., 2000; Kropf et al., 2002), North America (e.g. Sewell et al., 1996; Tremblay and Schoen, 1999) and the tropics (e.g. Dutech et al., 2000; Caron et al., 2000; Ogden and Thorpe, 2002; Knapp and Mallet, 2003), have mainly focused on identifying refugia and tracing routes by which glaciated territories were recolonized. However, much less is known about deserts (Riddle et al., 2002) and even less about rates of plant speciation and radiation in seasonally dry forests during these periods. The main reason for this is probably the difficulty of unravelling the spatial genetic history of species in these refuges as compared with northern regions where climatic changes have been better characterized.
Within areas of seasonally dry forest, a complex of vegetation types occurs, depending on local climate, soil and topography (Pennington et al., 2000). Thus, the complexity of patterns expected in seasonally dry forest is due to contraction–expansion cycles that took place with limited geographical displacement as compared with northern areas. Distributions of taxa in the same place should be historically related and display common patterns resulting from orogenic and widespread ecological events. Therefore, because seasonally dry forest species have similar ecological requirements, major geographical barriers may have led to similarities in their refuge areas and migration routes, even if their migration was asynchronous.
There is evidence from the distribution of species that disjunct areas of seasonally dry forests have an historical link. Pennington et al. (2000, 2004) suggested that the contemporary, fragmented distribution of seasonally dry forests in the Neotropics must be considered in relation to the climatic fluctuations of the Quaternary. However, for South American seasonally dry forests, population migration and long-distance dispersal are other explanations for their current biogeographic pattern (Mayle, 2004). In Mexico, it is well known that a wide area of disjunct seasonally dry forest occupying the Río Balsas Depression (RBD) contains a diverse endemic biota (Sousa and Delgado-Salinas, 1993; Becerra and Venable, 1999) in which some of these species represent old and others recent elements of the Mexican flora (Sousa and Soto, 1987). The nature of this endemism and role of climatic changes in evolution and the speciation rates of this diverse biota have so far not been explored.
The Caesalpinia hintonii complex (Contreras, 1991; Lewis, 1998) comprises six allopatric taxa, C. epifanioi, C. hintonii, C. laxa, C. macvaughii, C. melanadenia, and the recently described C. oyamae (Sotuyo and Lewis, 2007), distributed in patches along the RBD and Tehuacán–Cuicatlán Valley (TCV; Fig. 1). Previous genetic analyses of this group of taxa have shown a clear population structure and apparently recent speciation (Sotuyo et al., 2004) based on morphological similarity among species of the complex. However, given that the taxa of the C. hintonii complex are restricted to dry areas, that their flowers are bee- (Centris and Xylocopa) or hummingbird-pollinated, and that they appear to be dispersal limited (with dehiscent seed pods), their observed local endemicity suggests ancient fragmentation of a previously more-widespread ancestor. Moreover, morphological and genetic differences among species in the C. hintonii complex may provide evidence of timing and patterns of distribution of several other groups of taxa in the RBD and TCV.
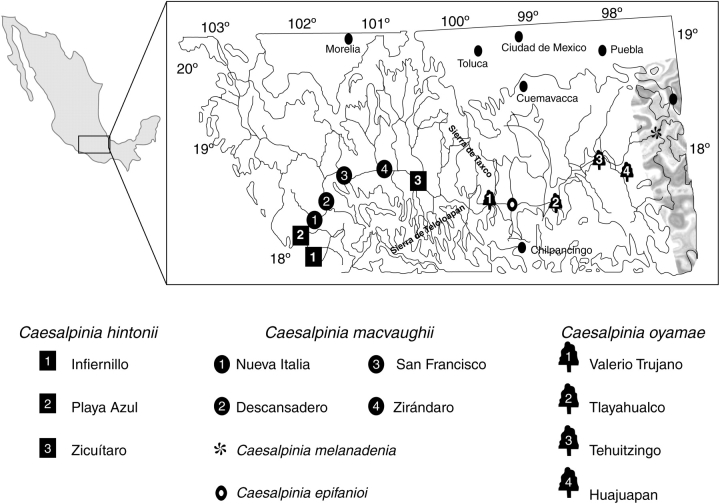
Fig. 1.
Geographical distribution of the Caesalpinia hintonii complex. The shaded area is part of the Tehuacán–Cuicatlán valley.
Earlier results based on isozyme data (Sotuyo et al., 2004) demonstrated that Caesalpinia hintonii diverged genetically as a by-product of local adaptations to different habitats and that the genetic structure of these populations was in close accordance with their spatial distribution. Nevertheless, no conclusive explanation of the observed genetic structure of the six species currently recognized in the complex in the RBD and TCV exists due to the lack of a temporal framework in which to interpret results.
The Caesalpinia hintonii complex does not have a fossil record (Ramírez and Cevallos-Ferriz, 2000), and dispersal time and distance rates are unknown. If populations expanded and experienced an increase in inter-population gene flow during glaciations, as predicted by refugial theory (Haffer, 1969), the following would be expected: no significant genetic structure among populations; no isolation by distance effect; and no clear phylogenetic structure among populations. In contrast, if populations remained fragmented throughout these periods, clear genetic structure and isolation by distance, as well as a highly structured phylogenetic tree would be expected, reflecting long-standing genetic divergence among populations.
Under the latter scenario, two hypotheses should be considered: vicariance versus dispersal. Under the vicariance approach, existence of several allopatric lineages in the RBD and TCV might reflect fragmentation of the ancestral population, and, thus, species distributed in the two regions would be sister taxa. On the other hand, under the dispersal hypothesis allopatric taxa should not necessarily be linked temporally. Dispersal and mixing of lineages during glaciations should have homogenized their haplotypes, and these taxa should exhibit lower genetic diversity and divergence than those taxa not exposed to climatic displacements and homogenization. Support for either of the two scenarios is evaluated in this study by molecular phylogenetic reconstruction, inferring the phylogeographic patterns and genetic diversity of the Caesalpinia hintonii complex. To achieve this, plastid DNA sequence variation was examined in one intron and three intergenic spacers, all of which are known to be variable at the species level in several legume genera (e.g. Bruneau et al., 2001), and microstructural changes, primarily those involving insertion/deletion events (indels) that have been reported as especially frequent in these regions. Plastids are generally maternally inherited in angiosperms and, therefore, dispersed by seeds only. Because colonization of new habitats occurs through seeds, plastid DNA markers provide information on past changes in species distribution that is unaffected by subsequent pollen movement.
MATERIALS AND METHODS
Study sites and species sampling
The Río Balsas Depression (RBD) and Tehuacán–Cuicatlán Valley (TCV) subprovinces are located in the Mexican morphotectonic province of Sierra Madre del Sur and include parts of the following states: Michoacán, Guerrero, Morelos, Puebla and adjacent Oaxaca (Fig. 1). The RBD is an east–west oriented basin that forms the lowlands of the Balsas River drainage basin surrounded by mountains, some of which exceed 2000 m and have an horizon that extends towards the east of the TCV in Puebla and Oaxaca states. These subprovinces together form one of the most geologically complicated regions of Mexico (Ferrusquía-Villafranca, 1993). The isolated TCV is located on the east side of the RBD and is considered to be an ecological island containing assemblages of endemic species.
A total of 60 individuals from 13 populations from the C. hintonii complex were analysed (Table 1). Caesalpinia laxa was not included in the study because it was not available for sampling. Eight species (19 individuals) were specified as outgroups: C. caladenia, C. coccinea, C. eriostachys, C. exostemma subsp. exostemma, C. hughesii, C. nelsonii, C. mexicana, and their sister taxon C. pannosa. These taxa were selected because a previous phylogenetic analysis resolved the latter and C. exostemma as sister to C. hintonii (Simpson et al., 2003). All taxa were sequenced for four plastid regions. Voucher specimens were deposited at either MEXU or K (Table 1).
Table 1.
Localities of the Caesalpinia hintonii complex and outgroup specimens included in this study
| Species/population/abbreviation | State | n* | Co-ordinates (N, W) | Elevation (m) | Voucher specimens | Genbank number |
|---|---|---|---|---|---|---|
| C. oyamae (Coya) | 25 | |||||
| Huajuapan (HU) | Oaxaca | 4 | 17°52′56, 98°05′ | 1325 | Sotuyo & González 17; Sotuyo et al. 72, 74, 75 | DQ208808-DQ208806, DQ208736-DQ298733, DQ208880-DQ208877 |
| Tehuitzingo (TE) | Puebla | 5 | 18°20′, 98°117′ | 1100 | Sotuyo et al. 65, 66, 67, 68 | DQ208805-DQ208801, DQ208732-DQ208728, DQ208876-DQ208772 |
| Tlayahualco (TL) | Guerrero | 9 | 17°50′, 99°06′ | 660 | Sotuyo et al. 55, 57 | DQ208800-DQ208792, DQ208727-DQ208719, DQ208870-DQ208863 |
| Valerio Trujano (VT) | Guerrero | 7 | 17°50′, 99°36′ | 500 | Sotuyo & Sotuyo 18; Sotuyo et al. 58, 59, 60 | DQ208790-DQ208785, DQ208717-DQ208712, DQ208862-DQ208856 |
| C. hintonii (Chin) | 15 | |||||
| Zicuítaro (ZIC) | Guerrero | 8 | 18°26′, 100°51′ | 300 | Sotuyo et al. 50, 52 | DQ208757-DQ208751, DQ208897-DQ208890, DQ208824-DQ208817 |
| Infiernillo (IN) | Michoacán | 6 | 18°24′4, 101°55′ | 240 | Sotuyo et al. 41, 46, 47, 48 | DQ208749-DQ208743, DQ208815-DQ208809, DQ208887-DQ208881 |
| Playa azul (PA) | Michoacán | 1 | 18°24′4, 101°55′ | 200 | Sotuyo et al. s.n. | DQ208816, DQ208750, DQ208889 |
| C. macvaughii (Cmac) | 9 | |||||
| Descansadero (DE) | Michoacán | 3 | 18°38′, 101°57′ | 450 | Contreras et al. s.n. | DQ208840-DQ208836, DQ208762-DQ208760, DQ208912-DQ208910 |
| Nueva Italia (NIT) | Michoacán | 1 | 19°00′, 102°04′ | 400 | Steinmann 3175 | DQ208843, DQ208916, |
| San Francisco (SFC) | Michoacán | 1 | 18°38′, 101°59′ | 330 | Steinmann 3176 | DQ208766, DQ208917 |
| Zirándaro (ZIR) | Guerrero | 4 | 18°32′, 101°13′ | 270 | Sotuyo et al. 8, 8bis | DQ208842-DQ208840, DQ208767, DQ208765-DQ208763, DQ208909, DQ208765-DQ2088913 |
| C. epifanioi (Cepi) | 6 | |||||
| Mezcala | Guerrero | 6 | 18°17′, 98°05′ | 500 | Sotuyo & Sotuyo 20, Sotuyo et al. 63 | DQ208830-DQ208825, DQ208742-DQ208737, DQ208903-DQ208898 |
| C. melanadenia (Cmel) | 5 | |||||
| Zapotitlán (ZAP) | Puebla | 5 | 18°19′, 97°27′ | 1500 | Sotuyo & González s.n. | DQ208835-DQ208831, DQ208772-DQ208768, DQ208908-DQ208904 |
| C. pannosa | Baja California Sur | 12 | 24º09’, 110º56’ | 10-700 | Contreras 2050, 2696, 2703, 2715, 2721, 2726, 2727; Hawkins 87, Lewis et al. 2047, 2051; Carter 2124; Gentry 4365; Wiggins 15688 | DQ208855-DQ208844, DQ208784-DQ208773, DQ208930-DQ208919 |
| C. caladenia | Michoacan | 1 | Not available | 220-410 | Contreras 2818 | EF177383, EF177390, EF177397 |
| C. coccinea | Oaxaca | 1 | 15º55’, 95º48’ | 100 | Lewis 1803 | EF177386, EF177393, EF177400 |
| C. eriostachys | Guerrero | 1 | 18º13’, 101º47’ | 450 | MacQueen 449 | EF177389, EF177396, EF177403 |
| C. exostemma subsp. exostemma | Guatemala | 1 | 14º59’, 89º40’ | 180-190 | Lewis et al. 1712 (Chase 15196) | EF177388, EF177395, EF177402 |
| C. hughesii | Oaxaca | 1 | Not available | 15-130 | Lewis 1795 | EF177384, EF177391, EF177398 |
| C. mexicana | Nuevo León | 1 | 24º51’, 99º24’ | 350 | Delgado 01-2114 | EF177387, EF177394, EF177401 |
| C. nelsonii | Oaxaca | 1 | 18°45’, 098°10’ | 100-500 | Sotuyo s.n. (Chase 18851) | EF177385, EF177392, EF177399 |
* n = sampled number.
Molecular methods
Total DNA was isolated from frozen or silica gel-dried leaf tissue (Chase and Hills, 1991) or herbarium samples using a modified 2× CTAB miniprep protocol (Doyle and Doyle, 1987). Polymerase chain reaction (PCR) was used to amplify the trnL intron, trnL-F spacer (primer information for both regions in Taberlet et al., 1991), the trnH-psbA intergenic spacer (Hamilton, 1999), and the accD-psaI spacer (Mendenhall, 1994). For trnL and trnL-F, the following programme was used: initial denaturing at 94 °C for 2 min, 30 cycles of denaturing at 94 °C for 20 s, annealing at 58 °C for 20 s, extension at 72 °C for 20 s, and a final extension at 72 °C for 5 min. For trnH-psbA and accD-psaI, an initial denaturing at 94 °C for 2 min, 30 cycles of denaturing at 94 °C for 45 s, annealing at 49 °C for 1 min, extension at 72 °C for 1·15 min, and a final extension at 72 °C for 7 min was used. PCR amplifications were purified with QIAquick silica columns following the manufacturer's protocol (Qiagen, Inc.). Purified PCR products were sequenced on an Applied Biosystems, Inc. (ABI), 3100 automated sequencer using the ABI 3·0 cycle-sequencing protocol. Individual sequences were edited and assembled using Sequencher 4·1 (Gene Codes, Corp.). Complete sequences were aligned by eye following the guidelines of Kelchner (2000). Sequences reported here have been deposited in the NCBI GenBank database.
Phylogenetic analyses
All phylogenetic studies were conducted using PAUP version 4·0b8 (Swofford, 2001). Phylogenetic relationships were established using the maximum parsimony criterion. Because the plastid regions used are inherited without recombination and share a single history, data from all four regions were not analysed separately and were directly combined. The heuristic search employed 100 random additions of sequences with tree bisection reconnection branch-swapping, which was restricted to 1000 retained trees. Bootstrap resampling (Felsenstein, 1985) was applied to assess support for individual nodes using tree bisection reconnection swapping and 1000 bootstrap replications. DELTRAN character optimization was used to illustrate branch lengths throughout, due to reported errors with ACCTRAN optimization in PAUP version 4·0b. In some individuals, two different inverted repeats in the trnH-psbA spacer were found; these were eliminated for the analyses. Indels were coded separately as binary characters using the simple coding of Simmons and Ochoterena (2000).
RESULTS
The lengths of the aligned sequence matrices were as follows: trnL, 612 bp; trnL-F, 518 bp; trnH-psbA, 654 bp; accD-psaI 803 bp. The base frequencies were: AccD-psaI, A = 0·310, C = 0·163, G = 0·153, T = 0·372; trnH-psbA, A = 0·330, C = 0·135, G = 0·137, T = 0·395; trnL, trnLF, A = 0·340, C = 0·169, G = 0·172, T = 0·317. Levels of sequence divergence for the accD-psaI showed values that ranged from 0·003 to 0·021 in the C. hintonii complex and from 0·004 to 0·019 between the other Caesalpinia species. The trnH-psbA region ranged from 0·005 to 0·033 among the C. hintonii complex species and from 0·019 to 0·065 in the other species. The trnL, trnL-F regions showed less divergence with a range of 0·002–0·019 in the C. hintonii complex and 0·016–0·33 in the rest of the species. Caesalpinia caladenia was the least divergent (accD-psaI 0·005–0·017; trnL, trnL-F 0·016–0·021; trnH-psbA 0·009–0·033), while C. nelsonii was the most divergent (accD-psaI 0·009–0·022; trnL, trnL-F 0·016–0·035; trnH-psbA 0·036–0·047). Caesalpinia oyamae showed similar values (accD-psaI 0·009–0·021; trnL, trnL-F 0·010–0·035; trnH- psbA 0·008–0·047).
Diversity in the C. hintonii complex was high, and numerous indels (109) were present throughout the four plastid regions. Ninety-four of these indels (86 %) are phylogenetically informative. Four indels were identified for the C. hintonii complex relative to C. pannosa, three in trnH-psbA (3, 25 and 28 bp) and one in trnL (6 bp). One synapomorphic indel region was found in C. hintonii and C. macvaughii (73 bp). Seven were shared by C. oyamae, C. melanadenia, C. epifanioi and C. pannosa; these are synapomorphic for C. hintonii and C. macvaughii, which is why that branch is so long (Fig 2A). Six were identified in trnH-psbA (4, 39, 34, 4, 6 and 6 bp), and one in trnL-F (23 bp). Caesalpinia hintonii shared one accD-psaI indel (8 bp) with C. pannosa. One shared trnL-F indel was found in C. hintonii, C. macvaughii and C. pannosa (23 bp), whereas one accD-psaI indel was synapomorphic in all C. hintonii populations (12 bp).
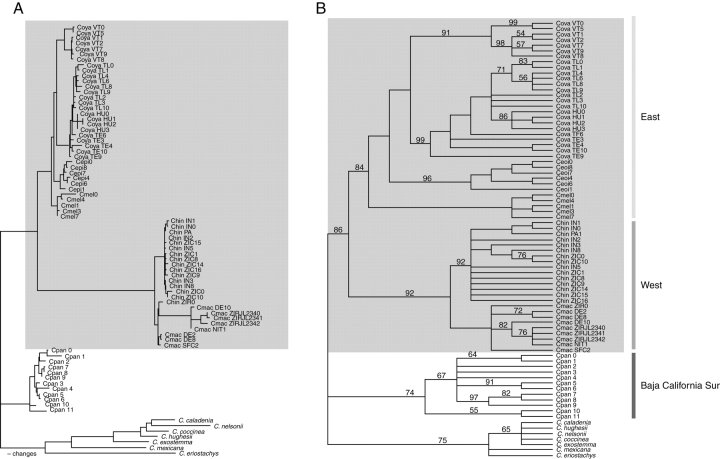
Fig. 2.
Trees produced by simultaneous analysis of sequences from all four plastid DNA regions. (A) One of the most-parsimonious trees showing the number of changes for the Caesalpinia hintonii complex based on accD-psaI, trnL, trnL-F and, trnH-psaI plastid regions including indels (DELTRAN optimization). (B) Strict consensus tree of all equally most-parsimonious trees found in the 1000 replicates of random taxon-addition; numbers above branches are bootstrap percentages >50. The C. hintonii complex is in a grey square. The abbreviations are given in Table 1.
Phylogenetic analyses
The strict consensus trees for each region analysed showed the same topology and were combined. The combined plastid matrix (2602 bp) yielded trees of 575 steps long, with a consistency index (CI) of 0·68 and retention index (RI) of 0·86. The indels matrix yielded trees of 1535 steps long, CI = 0·53 and RI = 0·91. Of the 828 sites examined, 61 were parsimony informative. Indels were less homoplasious than non-indel characters (HI = 0·0328 vs. HI = 0·44, respectively). The combined plastid matrix including gaps has 766 potentially parsimony informative sites and produced trees of 2141 steps long, with CI = 0·56 and RI = 0·91. The tree shows that the C. hintonii complex contains two geographically matching lineages, one eastern (84 % bootstrap support) and the other western (92 % bootstrap support) parts of the RBD (Fig. 2B).
The eastern RBD region cluster includes C. oyamae, C. epifanioi and C. melanadenia. Included in this eastern cluster, the Valerio Trujano population of C. oyamae is isolated from the other three eastern populations (91 %, 99 % bootstrap support). Within the eastern lineage, C. melanadenia, restricted to TCV, was poorly supported, although in all shortest trees the four accessions clustered together. The western region cluster containing Caesalpinia hintonii and C. macvaughii populations is well supported (bootstrap 92 %). The western lineage also contains a diversified and weakly supported C. macvaughii.
DISCUSSION
Phylogenetic analyses
At the molecular level, the most striking difference between species of Caesalpinia in the RBD–TCV was the clear evidence of isolation between most populations. This study shows that the Caesalpinia hintonii complex possesses high levels of genetic divergence over its entire range, which could indicate that the places where the constituent populations currently exist could be refugia isolated by changes in climate. Previous studies of other plant species have implicated vicariance with climate change in the speciation process (Riddle et al., 2002) and, thus, the origin of genetic structure among populations (Vijverberg et al., 2000). When vicariance has occurred, more distant haplotype relationships are expected. Conversely, recent immigrants, under a secondary contact scenario, would present haplotypes identical to the donor population(s). Therefore, the population structure shown by the C. hintonii complex could be the result of populations having been isolated in two or more glacial refugia, followed by population expansion and a subsequent long period without secondary contact as the Neotropics became wetter and cooler (Pennington et al., 2000).
Two major lineages were found in the RBD and TCV (Fig. 2) that fit the two physiographic units proposed by Miranda (1947). The separation of the eastern and western lineages of the Caesalpinia hintonii complex may well correspond to fragmentation that was triggered by the development of the Sierra of Taxco-Teloloapan, which divided the RBD (Cabral-Cano et al., 2000). This most prominent physical barrier appears to be a clear obstacle to the movement of some Caesalpinia taxa; populations of C. hintonii and C. macvaughii apparently have not exchanged plastids (Tsitrone et al., 2003) since their establishment on the western side of the RBD. Likewise C. oyamae, C. epifanioi and C. melanadenia are restricted to the eastern flank of the Depression. Furthermore, volcanism and geological perturbations in the Trans-Mexican Volcanic Belt (Ferrari et al., 1999) must have had a considerable influence on population divergence within C. oyamae in the RBD.
Within the western lineage, the molecular markers used in this study indicate that both populations (Infiernillo and Zicuítaro) of Caesalpinia hintonii are genetically a close-knit homogeneous group, even though they are morphologically distinct (Contreras, 1991; Lewis, 1998). The genetic similarity of these populations, also supported by allozyme analysis (Sotuyo et al., 2004), indicates that their morphological differences are the result of changes in only a few genes or phenotypic plasticity.
In contrast, populations of the eastern lineage species proved to be morphologically homogeneous (Sotuyo and Lewis, 2007) but genetically distinct. Caesalpinia epifanioi and C. melanadenia, the first restricted to eastern RBD, the latter endemic to the TCV, proved to be both morphologically and genetically different. In contrast, results of this study indicate that there are two distinct population assemblages in Caesalpinia oyamae (Fig. 2), which could each be considered cryptic species following the model of recent and rapid speciation demonstrated by Weinberg et al. (1992). Evidence for speciation in C. oyamae populations was revealed by the high level of well-supported phylogenetic structure. However, allozyme analysis indicated the same degree of genetic isolation as the plastid DNA analyses (Sotuyo et al., 2004) except in the Huajuapan population (C. oyamae) which was not sampled for that study. Studies on a diverse range of taxa suggest that selection due to shifts in ecology or invasion of new habitats can cause extremely rapid divergence and might play a prominent role in speciation (Orr and Smith, 1998). Proposed cryptic species in this eastern lineage may indicate the presence of optimal phenotypes that are subject to strong stabilizing selection and thus remain morphologically similar (e.g. Shaw, 2001; McDaniel and Shaw, 2003).
However, relationships of the eastern populations are suggestive of a recent origin of Caesalpinia oyamae, as would be expected for genotypes at the limits of a recently expanded range (Posada and Crandall, 2001). Plastid DNA variation also corresponds closely to geographical distribution, which is likely to reflect dispersals that involve interruption of gene flow among populations. Dispersals, together with genetic drift in the early stages of local establishment, may result in decreases in genetic variation within populations and in increases between populations (Okada et al., 1997).
The results presented in this study provide evidence of reproductive isolation of lineages in allopatry. Explanations for cryptic radiation have ranged from high mutation rate to developmental constraints and long-term environmental stability. It has been further suggested that stabilizing selection (Hansen, 1997) and lack of additive genetic variance for morphological traits (McDaniel and Shaw, 2003) may play an important role in maintaining morphological stasis if the environment has been stable over long periods of time. In contrast, Sheldon (1996) has argued that morphological stasis should occur in unstable, changing environments (e.g. changes in climate), whereas habitats which have been stable for a long time should promote gradual morphological evolution. His model predicts that initial major environmental perturbations select for generalist phenotypes that are well adapted to subsequent environmental fluctuations. If Sheldon's assumption is correct, this process might help to explain why morphostasis has occurred in taxa such as C. oyamae.
The phylogeographic complexity of the eastern clade suggests that species expansion of the Caesalpinia hintonii complex in the region considerably predated the contemporary expansion of the species across the RBD and/or that greater geological heterogeneity of the Trans-Mexican Volcanic Belt has caused a reduction in gene flow leading to increased population subdivision. There are no previous phylogeographic studies in plants within the RBD region. However, a phylogeographic study of pocket gophers demonstrated a clear phylogeographic structure in the Trans-Mexican Volcanic Belt and Mesa del Norte of the Mexican Plateau that was associated with a recent expansion and displacement of existing populations by volcanic activity (Desmastes et al., 2002).
Divergence into two geographically distinct clades remains evident in the C. hintonii complex. Taken together, the present analysis supports the hypothesis that isolated populations of C. oyamae in the RBD diverged in refugia during times of climatic change and that the geographical variation reflects patterns of recolonization. The C. hintonii complex fits the model of Pennington et al. (2004) better because all the species are dispersal limited.
Taxonomic implications and concluding remarks
The Caesalpinia hintonii complex currently comprises six essentially allopatric taxa: C. epifanioi, C. hintonii, C. laxa, C. macvaughii, C. melanadenia and C. oyamae. Caesalpinia oyamae is the most morphologicaly similar species to C. hintonii; in the past they were considered the same species. The present results support the hypothesis that both species are genetically distinct lineages in which gene flow between the two species by seeds is likely to have ceased long ago. In contrast, the two species are morphologically separable by differences in flower size and colour, hardly significant diagnostic characters (Sotuyo et al., 2004).
Furthermore plastid DNA analysis established two lineages within Caesalpinia oyamae that are morphologically identical but genetically divergent. Should these two geographically distinct lineages within C. oyamae also be formally recognized as species despite their morphological similarity? We consider it unwise to take this course, unless some morphological evidence can be found that facilitates recognition of taxonomic distinctiveness.
Diversification in the Caesalpinia hintonii complex is clearly linked to environmental conditions, orogeny, volcanism and glaciations, suggesting that a more detailed analysis will reveal more data that facilitate taxonomic decisions. The apparent transformation of a single, regionally endemic, molecularly variable ancestor into a complex of at least eight species is noteworthy. Each species now occupies different ecological environments in the Mexican morphotectonic province of Sierra Madre del Sur. It appears likely that other endemic species complexes such as the C. hintonii complex will be shown to be an amalgam of isolated lineages. Therefore, genetic differentiation without morphological differentiation may prove wider in plants than previously acknowledged and, with more studies like the present one, may become established as another differentiation pattern that has led to species diversification in the RBD. In addition, studies like this one offer an exciting way of analysing intricate taxonomic group complexes.
ACKNOWLEDGEMENTS
We thank the Molecular Systematics staff at the Jodrell Laboratory, Royal Botanic Gardens, Kew, and Laura Márquez (IBUNAM) for their technical assistance and Victor Steinmann (INECOL) for his help collecting C. macvaughii. The research was supported by Conacyt doctoral scholarship number 15291811144 to Solange Sotuyo, DGEP-UNAM and the Royal Botanic Gardens, Kew. We are grateful to the editor Professor Warren Hauk and two anonymous reviewers for their comments.
LITERATURE CITED
- Abbott RJ, Smith LC, Milne RI, et al. Molecular analysis of plant migration and refugia in the Arctic. Science. 2000;289:1343–1346. doi: 10.1126/science.289.5483.1343. [DOI] [PubMed] [Google Scholar]
- Austin JD, Lougheed SC, Neidrauer L, Chek AA, Boag PT. Cryptic lineages in a small frog: the post-glacial history of the spring peeper, Pseudacris crucifer (Anura: Hylidae) Molecular Phylogenetics and Evolution. 2002;25:316–329. doi: 10.1016/s1055-7903(02)00260-9. [DOI] [PubMed] [Google Scholar]
- Becerra J, Venable L. Nuclear ribosomal DNA and its implications for evolutionary trends in Mexican Bursera (Burseraceae) American Journal of Botany. 1999;86:1047–1057. [PubMed] [Google Scholar]
- Bruneau A, Forest F, Herendeen PS, Klitgaard BB, Lewis GP. Phylogenetic relationships in the Caesalpinioideae (Leguminosae) as inferred from chloroplast trnL intron sequences. Systematic Botany. 2001;26:487–514. [Google Scholar]
- Cabral-Cano E, Lang HR, Harrison CGA. Stratigraphic assessment of the Arcelia-Teloloapan area, southern Mexico: implications for southern Mexico's post-Neocomian tectonic evolution. American Earth Science. 2000;13:443–457. [Google Scholar]
- Caron H, Dumas S, Marque G, Messier C, Bandou E, Petit RJ, et al. Spatial and temporal distribution of chloroplast DNA polymorphism in a tropical tree species. Molecular Ecology. 2000;9:1089–1098. doi: 10.1046/j.1365-294x.2000.00970.x. [DOI] [PubMed] [Google Scholar]
- Chase MW, Hills HG. Silica gel: an ideal dessicant for preserving field-collected leaves for use in molecular studies. Taxon. 1991;40:215–220. [Google Scholar]
- Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Sciences. 1998;3:432–438. [Google Scholar]
- Contreras JL. Contribución al conocimiento del género Caesalpinia (Leguminosae: Caesalpinioideae) en el estado de Guerrero. México: Facultad de Ciencias, UNAM; 1991. BSc Thesis. [Google Scholar]
- Desmastes JW, Spradling TA, Hafner DJ, Reed DL. Systematics and phylogeography of pocket gophers in the genera Crarogeomys and Pappogeomys. Molecular Phylogenetics and Evolution. 2002;22:144–154. doi: 10.1006/mpev.2001.1044. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Dumolin-Lapègue S, Demesure B, Le Corre V, Petit RJ. Phylogeographic structure of white oaks throughout the European continent. Genetics. 1997;146:1475–1487. doi: 10.1093/genetics/146.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutech C, Maggia L, Joly HI. Chloroplast diversity in Vouacapoua americana (Caesalpiniaceae), a Neotropical forest tree. Molecular Ecology. 2000;9:1427–1432. doi: 10.1046/j.1365-294x.2000.01027.x. [DOI] [PubMed] [Google Scholar]
- Dynesius M, Jansson R. Evolutionary consequences of changes in species' geographical distribution driven by Milankovitch climate oscillations. Proceedings of the National Academy of Sciences of the USA. 2000;97:9115–9120. doi: 10.1073/pnas.97.16.9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferrari L, López-Martínez M, Aguirre-Díaz G, Carrasco-Núñez G. Space-time patterns of Cenozoic arc volcanism in central Mexico: from the Sierra Madre Occidental to the Mexican Volcanic Belt. Geology. 1999;27:303–306. [Google Scholar]
- Ferrusquía-Villafranca I. Geology of Mexico: a synopsis. In: Ramamoorthy AP, Bye R, Lot A, Fa J, editors. Biological diversity in Mexico: origins and distribution. New York, NY: Oxford University Press; 1993. pp. 3–108. [Google Scholar]
- Haffer J. Speciation in Amazonian birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Hamilton MB. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology. 1999;8:521–523. [PubMed] [Google Scholar]
- Hansen TF. Stabilizing selection and the comparative analysis of adaptation. Evolution. 1997;51:1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Speciation, hybrid zones and phylogeography or seeing genes in space and time. Molecular Ecology. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- Kelchner SA. The evolution of noncoding chloroplast DNA and its application in plant systematics. Annals of Missouri Botanical Garden. 2000;87:482–498. [Google Scholar]
- King RA, Ferris C. Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Molecular Ecology. 1998;7:1151–1161. [Google Scholar]
- Knapp S, Mallet J. Refuting refugia? Science. 2003;300:71–72. doi: 10.1126/science.1083007. [DOI] [PubMed] [Google Scholar]
- Kropf M, Kadereit JW, Comes HP. Late Quaternary distributional stasis in the submediterranean mountain plant Anthyllis montana L. (Fabaceae) inferred from ITS sequences and amplified fragment length polymorphism markers. Molecular Ecology. 2002;11:447–463. doi: 10.1046/j.1365-294x.2002.01446.x. [DOI] [PubMed] [Google Scholar]
- Lewis GP. Caesalpinia: a revision of the Poincianella-Erythrostemon Group. London: Royal Botanic Gardens, Kew; 1998. [Google Scholar]
- McDaniel SF, Shaw JA. Phylogeographic structure and cryptic speciation in the trans-Antarctic moss Pyrrhobryum minioides. Evolution. 2003;57:205–215. doi: 10.1111/j.0014-3820.2003.tb00256.x. [DOI] [PubMed] [Google Scholar]
- Mayle FE. Assesment of the Neotropical dry forest refugia hypothesis in the light of paleoecological data and vegetation model simulations. Journal of Quaternary Science. 2004;19:713–720. [Google Scholar]
- Mendenhall M. Austin, TX: University of Texas; 1994. Phylogeny of Baptisia and Thermopsis (Leguminosae) as inferred from chloroplast DNA and nuclear ribosomal DNA sequences, secondary chemistry, and morphology. PhD Thesis. [Google Scholar]
- Miranda F. Estudios sobre la vegetación de México – rasgos de la vegetación en la Cuenca del Río Balsas. Revista de la Sociedad Mexicana de Historia Natural. 1947;VIII:1–4. Tomo. [Google Scholar]
- Ogden R, Thorpe RS. Molecular evidence for ecological speciation in tropical habitats. Proceedings of the National Academy of Sciences of the USA. 2002;99:13612–13615. doi: 10.1073/pnas.212248499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Whitkins R, Lowrey TK. Genetics of adaptative radiation in Hawaiian and Cook Islands species of Tetramolopium (Asteraceae: Astereae). I. Nuclear RFLP marker diversity. American Journal of Botany. 1997;84:1236–1246. [PubMed] [Google Scholar]
- Orr M, Smith TB. Ecology and speciation. Trends in Ecology and Evolution. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- Pennington T, Prado DE, Pendry CA. Neotropical seasonally dry forests and the Quaternary vegetation changes. Journal of Biogeography. 2000;27:261–273. [Google Scholar]
- Pennington T, Lavin M, Prado DE, Pendry CA, Pell SK, Butterworth CA. Historical climate change and speciation: Neotropical seasonally dry forest plants show patterns of both Tertiary and Quaternary diversification. Philosophical Transactions of the Royal Society of London B. 2004;359:515–537. doi: 10.1098/rstb.2003.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Pineau ED, Demesure B, Bacilieri R, Ducousso A, Kremer A. Chloroplast DNA footprints of postglacial recolonization by oaks. Proceedings of the National Academy of Sciences of the USA. 1997;94:9996–10001. doi: 10.1073/pnas.94.18.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Intraspecific phylogenetics: trees grafting into networks. Trends in Ecology and Evolution. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Ramírez JL, Cevallos-Ferriz SR. Consideraciones sobre las angiospermas (plantas con flor) fósiles de México. GEOS. 2000:433–444. Diciembre. [Google Scholar]
- Riddle BR, Hafner DJ, Alexander LF, Jaeger JR. Cryptic vicariance in the historical assembly of Baja California Peninsular desert biota. Proceedings of the National Academy of Sciences of the USA. 2002;97:14438–14443. doi: 10.1073/pnas.250413397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez A, Probert I, Geisen M, Quinn P, Young J, Medlin L. Pseudo-cryptic speciation in coccolithophores. Proceedings of the National Academy of Sciences, USA. 2003;12:7163–7168. doi: 10.1073/pnas.1132069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell MM, Parks CR, Chase MW. Intraspecific chloroplast DNA variation and biogeography of North American Liriodendron L. (Magnoliaceae) Evolution. 1996;50:1147–1154. doi: 10.1111/j.1558-5646.1996.tb02355.x. [DOI] [PubMed] [Google Scholar]
- Shaw AJ. Molecular phylogeography and cryptic speciation in bryophytes. Journal of Biogeography. 2001;28:253–261. [Google Scholar]
- Sheldon P. Plus ça change – a model for stasis and evolution in different environments. Palaeogeography, Palaeoclimatology and Palaeoecology. 1996;127:209–227. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Simpson B, Latkin L, Weeks A. Progress towards resolving the relationships of the Caesalpinia group (Caesalpinieae: Caesalpinioideae: Leguminosae) In: Klitgaard B, Bruneau A, editors. Advances in legume systematics. Vol. 10. London: Royal Botanic Gardens, Kew; 2003. pp. 123–148. Higher Level Systematics. [Google Scholar]
- Sotuyo S, Lewis GP. A new species of Caesalpinia from the Rio Balsas Depression, Mexico and an updated taxonomic circumscription of the Caesalpinia hintonii complex (Leguminosae: Caesalpinioideae: Caesalpinieae: Poincianella Group) Brittonia. 2007;59:33–36. [Google Scholar]
- Sotuyo S, Contreras JL, Delgado-Salinas A, Oyama K. Genetic structure of the endemic Caesalpinia hintonii complex (Leguminosae: Caesalpinioideae) Plant Systematics and Evolution. 2004;247:131–143. [Google Scholar]
- Sousa MS, Delgado-Salinas A. Mexican Leguminosae: Phytogeography, endemism, and origins. In: Ramamoorthy AP, Bye R, Lot A, Fa J, editors. Biological diversity in Mexico: origins and distribution. New York, NY: Oxford University Press; 1993. pp. 459–511. [Google Scholar]
- Sousa MS, Soto JC. Nuevos taxa de Lonchocarpus (Leguminosae) de las cuencas baja y media del Río Balsas, México. Anales del Instituto de Biología, UNAM, serie Botánica. 1987;58:69–86. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 2001. Ver. 4·0b8. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of the chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Tremblay NO, Schoen DJ. Molecular phylogeography of Dryas integrifolia: glacial refugia and postglacial recolonization. Molecular Ecology. 1999;8:1187–1198. doi: 10.1046/j.1365-294x.1999.00680.x. [DOI] [PubMed] [Google Scholar]
- Tsitrone A, Kirkpatrick M, Levin DA. A model for chloroplast capture. Evolution. 2003;57:1776–1782. doi: 10.1111/j.0014-3820.2003.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Vijverberg K, Kuperus P, Breeuwer JA, Bachmann K. Incipient radiation of New Zealand and Australian Microseris (Asteraceae): an amplified fragment length polymorphism (AFLP) study. Journal of Evolutionary Biology. 2000;13:997–1007. [Google Scholar]
- Wakeley J. The effects of subdivision on the genetic divergence of populations and species. Evolution. 2000;54:1092–1101. doi: 10.1111/j.0014-3820.2000.tb00545.x. [DOI] [PubMed] [Google Scholar]
- Weinberg JR, Starczak VR, Jora P. Evidence for rapid speciation following a founder event in the laboratory. Evolution. 1992;46:1214–1220. doi: 10.1111/j.1558-5646.1992.tb00629.x. [DOI] [PubMed] [Google Scholar]