Abstract
The plant pathogen Agrobacterium tumefaciens transforms plant cells by delivering its T-DNA into the plant cell nucleus where it integrates into the plant genome and causes tumor formation. A key role of VirE2-interacting protein 1 (VIP1) in the nuclear import of T-DNA during Agrobacterium-mediated plant transformation has been unravelled and VIP1 was shown to undergo nuclear localization upon phosphorylation by the mitogen-activated protein kinase MPK3. Here, we provide evidence that VIP1 encodes a functional bZIP transcription factor that stimulates stress-dependent gene expression by binding to VIP1 response elements (VREs), a DNA hexamer motif. VREs are overrepresented in promoters responding to activation of the MPK3 pathway such as Trxh8 and MYB44. Accordingly, plants overexpressing VIP1 accumulate high levels of Trxh8 and MYB44 transcripts, whereas stress-induced expression of these genes is impaired in mpk3 mutants. Trxh8 and MYB44 promoters are activated by VIP1 in a VRE-dependent manner. VIP1 strongly enhances expression from a synthetic promoter harboring multiple VRE copies and directly interacts with VREs in vitro and in vivo. Chromatin immunoprecipitation assays of the MYB44 promoter confirm that VIP1 binding to VREs is enhanced under conditions of MPK3 pathway stimulation. These results provide molecular insight into the cellular mechanism of target gene regulation by the MPK3 pathway.
Keywords: signal propagation, transcription factor, promoter motif, bZIP, Arabidopsis
The plant pathogen Agrobacterium tumefaciens transforms plant cells by delivering its transfer-DNA (T-DNA) into the plant cell nucleus, where it integrates into the plant genome. The subsequent growth of crown galls at the infection sites substantially interferes with plant development. There is ample experimental evidence demonstrating a key role of VIP1 (VirE2-interacting protein 1) in the nuclear import of T-DNA during Agrobacterium-mediated plant transformation (1, 2). VIP1 exhibits a stress-dependent subcellular localization; and phosphorylation of VIP1 at serine 79 by the stress-activated MAPK MPK3 (3) triggers its translocation from cytoplasm to the nucleus (2). Analysis of the vip1-1 mutant line, which produces only a truncated VIP1 protein, has shown that the N-terminal portion of VIP1 is sufficient to bind to and target VirE2 to the nucleus to facilitate transient genetic transformation by Agrobacterium (4). However, this is insufficient for stable genetic transformation and Agrobacterium-induced tumorigenesis (4). Apart from agrobacterial transformation, no in planta function has yet been ascribed to VIP1 in Arabidopsis. Considering that the only purpose of a plant protein would not merely lie in assisting pathogen invasion, we aimed to investigate the in planta function of VIP1 and the functional relevance of its stress-dependent nuclear translocation.
In this work, we focus on the molecular events immediately downstream of stress-activated VIP1. We report on the role of VIP1 as a transcriptional regulator of genes that are targets of the stress-activated MPK3 cascade. VIP1 mediates transcriptional induction of target genes by binding to a DNA motif, termed VRE for VIP1 response element. Multiple copies of VRE motifs are found in the promoters of various stress-responsive genes, and evidence is provided that VIP1 directly targets VRE motifs in vitro and in vivo. This work gives insight into the molecular mechanism how stress-induced MPK3 activation is linked to target gene induction.
Results and Discussion
PR1 Is an Indirect Target of VIP1.
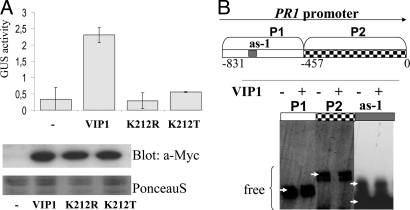
In transiently transformed Arabidopsis protoplasts, VIP1 shows a predominantly nuclear localization, and we found that nuclear VIP1 can activate the transcription of the pathogenesis-regulated protein 1 gene (PR1) (2), a major stress marker gene. A bZIP domain located in the C-terminal region classifies VIP1 as a putative transcription factor (Fig. S1). First, we addressed the question whether PR1 was directly regulated by VIP1. VIP1 is one of the 75 members belonging to the bZIP family of Arabidopsis thaliana. A phylogenetic study places VIP1 into the so far uncharacterized subgroup I (5). The 12 members of this family carry a characteristic lysine (K) instead of the otherwise highly conserved arginine residue (K212 in VIP1) within the bZIP domain. The nature of this particular residue is a determinant for the binding preference of bZIP family members, with arginine-type members preferentially binding to palindromic, and lysine-type members preferentially binding to nonpalindromic DNA elements (6, 7). If VIP1 encodes a functional bZIP transcription factor, mutation at K212 might affect its transactivating activity. To test this idea, K212 of VIP1 was replaced by the related residue arginine (R), or the nonsimilar threonine (T) residue, to potentially alter the DNA-binding preference or DNA-binding capacity of the protein, respectively. In protoplast transient expression experiments, only wild-type VIP1, but not its mutant derivatives K212R or K212T, efficiently activated the PR1 promoter::GUS (β-glucuronidase) reporter construct (Fig. 1A).
Fig. 1.
Indirect induction of PR1 by VIP1. (A) bZIP residue Lys-212 is essential for VIP1-induced PR1 expression. Protoplasts were transfected with the PR1::GUS reporter construct alone or in combination with constructs for overexpression of VIP1-Myc or VIP1 K212-Myc mutant variants. Given are mean values and standard deviations of GUS activity (nmol 4-MU) min−1 (mg protein)−1; n = 6. Transgene expression was visualized by immunoblotting with anti-Myc antibody. Equal loading was visualized by staining of the membrane with PonceauS. (B) VIP1 does not bind to the PR1 promoter (EMSA). The indicated PR1 promoter fragments or the as-1 element were biotin-labeled and incubated without or with recombinant VIP1 protein. Nonbound DNA fragments are indicated by arrows. No band shift was observed.
PR1 gene expression is regulated through binding of proteins belonging to the TGA subfamily of bZIP transcription factors to specific promoter motifs, including the as-1 element (consisting of subelements LS5 and LS7) and LS4 (8). Mutations at LS4, LS7, and/or LS5 did not affect VIP1 activation of PR1 promoter variants in protoplast cotransfection studies (data not shown). Moreover, VIP1 was unable to induce an as-1 element-containing synthetic promoter or the cauliflower mosaic virus CaMV 35S promoter, which harbors as-1 elements (data not shown). Electrophoretic mobility shift assays (EMSAs) also showed no detectable binding of recombinant VIP1 to fragments of the PR1 promoter or to the as-1 element alone (Fig. 1B).
Together, these findings suggest that VIP1 is a functional bZIP transcription factor, but that, unlike TGA proteins, VIP1 most probably acts on PR1 expression in an indirect manner.
PR1 is known as a late stress-responsive gene. Considering the rapid activation of MPK3 upon stress that is followed by VIP1 phosphorylation and nuclear translocation within minutes (2), good candidates for VIP1 direct target genes might therefore rather be found among early stress-responsive genes.
Identification of VRE as a DNA Motif Binding to VIP1.
In contrast to the subgroup of TGAs and several other family members in Arabidopsis (5, 9) little is known about the subfamily I of bZIPs. Unlike most other bZIP proteins, members of subfamily I are predicted to form homo- rather than to heterodimers (10). Co-immunoprecipitation experiments (Fig. S2) and bimolecular fluorescence complementation (BiFC) analysis (4), confirm homodimerization activity of VIP1. Consequently, additional proteins may not necessarily be required for VIP1 to bind to DNA. The target DNA motifs of bZIP transcription factors related to VIP1 (subfamily I) have been identified in other species, such as tomato VSF-1 (GCTCCGTTG) (11) and tobacco RSG (TCCAGCTTGA, TCCAACTTGGA) (6). However, the dissimilarity in VSF-1 and RSG target DNA motifs suggests that, despite high homology in the bZIP domains, the DNA binding preferences of VSF-1 and RSG are not conserved. Moreover, no target DNA motifs are yet known for any Arabidopsis bZIP subfamily I member.
In an attempt to identify DNA elements targeted by VIP1, we carried out random DNA binding selection assays (RDSAs). To this end, chitin-immobilized recombinant VIP1 protein was incubated with random 17- or 18-bp-long DNA fragments flanked by defined primer annealing sites (SI Text). VIP1-bound DNA fragments were PCR-amplified and used as input for another RDSA cycle with a second aliquot of VIP1 protein. By raising the concentration of nonspecific competitor, double-stranded DNA poly(dIdC), the stringency was increased in each subsequent purification cycle. A total of five cycles were performed, and the amplified DNA fragments were subsequently cloned and sequenced.
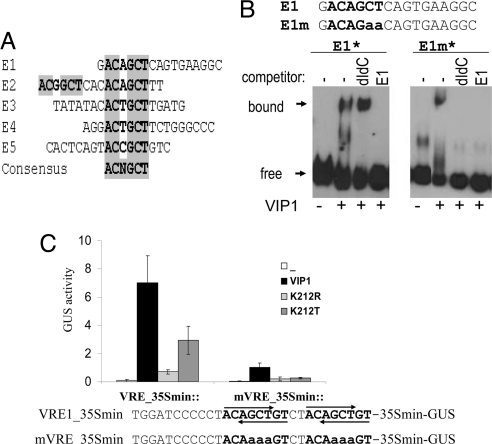
Of the seven independent candidate elements isolated, five (E1–E5; Fig. 2A) contained the common motif ACNGCT. Moreover, E2 carries two copies of ACNGCT, and two candidate fragments were isolated in duplicates, indicating the selectivity of the RDSA procedure.
Fig. 2.
Identification and binding of VIP1 to VREs. (A) Alignment of potential VIP1-target DNA elements isolated by RDSA. The consensus motif shared by five candidate fragments is highlighted. (B) VIP1 binds to VRE sequences (EMSA). Binding of VIP1 to candidate motif E1 isolated by RDSA (Left) or element E1m, carrying a mutation at the ACNGCT motif (Right). Biotin-labeled (*) E1 or E1m were incubated with or without recombinant VIP1 in the absence or presence of an excess of either nonspecific (dIdC) or specific inhibitor (nonlabeled E1). (C) VIP1-induced expression from a synthetic multiple-VRE-containing promoter. Protoplasts were transfected with synthetic promoter-GUS reporter gene constructs containing four copies of VRE (or mutated VRE-mVRE), alone or in combination with 35S::VIP1-Myc or 35S::VIP1 K212-Myc mutant variants and analyzed as described in Fig. 1A. Lower: Sequence of the promoter region. VRE sequences are shown in bold, and their orientation is indicated by arrows.
To test whether VIP1 can directly interact with the consensus motif, EMSA was performed using recombinant VIP1 protein and biotin-labeled DNA fragments. A clear band shift indicated the DNA-binding capability of VIP1 to the E1 fragment (Fig. 2B). The binding specificity of VIP1 for the ACNGCT core sequence was tested in EMSA with the E1m DNA fragment, which is identical to E1 except for a mutation in the ACNGCT motif (ACNGaa). EMSAs showed that although VIP1 can also bind to biotin-labeled E1m, this binding is abolished in the presence of the nonspecific competitor poly(dIdC) as well as by a 20-fold excess of nonlabeled E1 (Fig. 2B). In contrast, binding of VIP1 to labeled E1 is only substantially reduced in the presence of the specific competitor (nonlabeled E1) but not by the nonspecific competitor poly(dIdC). These results strongly indicate that VIP1 directly targets the ACNGCT DNA motif, which we hitherto call VRE. The VRE sequence does not match any characterized regulatory DNA motif, nor does it contain the TGAC core sequence commonly targeted by various bZIP proteins (12). Consistent with the predicted preference for nonpalindromic DNA elements (7), VRE is devoid of a palindromic core.
VIP1 Induces Transcription from a Synthetic Promoter Carrying VRE Motifs.
To test whether VIP1 can directly regulate VRE-containing promoters, we generated two synthetic promoter-reporter gene constructs. Four tandem copies of VRE, two in either orientation, were fused to a minimal promoter fragment derived from the CaMV35S promoter to give VRE::GUS. A mutated derivative containing two mismatches in each of the four VRE copies was generated to give mVRE::GUS (Fig. 2C). Protoplasts transfected with the reporter gene constructs alone showed no detectable GUS activity (Fig. 2C). However, constitutive coexpression of VIP1 strongly induced GUS expression from the VRE promoter, but only poorly from the mVRE promoter (Fig. 2C). VIP1 mutant variants in which the characteristic lysine residue (K212) that confers DNA-binding specificity had been replaced by an arginine (K212R) or a threonine (K212T) did not activate expression from either promoter. These findings substantiate the results from RDSA and EMSA experiments and suggest that VIP1-binding to VRE boxes is directly responsible for inducing gene expression. In line with the notion that PR1 expression is regulated by VIP1 only indirectly, no VRE sequences are found in the PR1 promoter.
VRE Defines a DNA-Binding Motif in Promoters of Stress Genes.
We then wondered whether the VRE motif could be a bona fide regulatory motif. MPK3 is activated by multiple biotic and abiotic stresses (reviewed in references 3 and 13). Based on the stress-activated protein kinase MPK3-triggered rapid nuclear translocation of VIP1 in planta (2), we hypothesized that VIP1 might bind to promoters of genes involved in the early stress response. Apart from a small number of uncharacterized candidates isolated from a protein chip-based screen for targets of MPK3 in vitro (14, 15), little is known about the steps of early stress signaling immediately downstream of MPK3. Thus, VIP1 is so far the only transcription factor with in vivo evidence for a direct regulation by MPK3. If VIP1 was a major mediator of MPK3-mediated gene expression and if VREs were the major motifs targeted by VIP1, one might expect to find an accumulation of VRE motifs in the promoters of genes responsive to the signals activating the MPK3 pathway. To test this theory, we performed a statistical analysis of A. thaliana promoters, exploiting publicly available microarray data resources.
Although in eukaryotic genes regulatory elements can be distributed anywhere within approximately 1,500 bp upstream of the transcription start sites, they are commonly found within the proximal 500 bps (16). We therefore compared the abundance of VRE sequences (in both forward and reverse orientations) in the 500-bp promoter regions of A. thaliana genes with reported stress-regulated transcript abundance in published microarrays to the overall abundance in all Arabidopsis genes. Because bZIP proteins form dimers, with both monomers binding to DNA, we expected VIP1 target promoters to carry at least two copies of VREs. Indeed, there were no significant differences in the abundance of stress- or nonstress-regulated promoters carrying a single VRE copy (data not shown), whereas the proportion of promoters carrying two or more VRE copies was significantly higher in stress-regulated genes retrieved from several independent transcriptome studies (Table 1). There was no apparent bias for the orientation of the motif (ACNGCT or AGCNGT).
Table 1.
VRE1 overrepresentation in promoters of stress-responsive genes
| Experimental condition | Total number of differentially expressed genes | Proportion of promoters with two or more VRE | P value | Reference |
|---|---|---|---|---|
| Total genome | 33,282 | 1,946 (5.8%) | 1.0 | |
| Stress datasets (under conditions when MPK3 is activated) | ||||
| Transient response to flg22 | 42 | (6) 14.3% | 0.0198* | 17 |
| Early systemic wound response | 14 | (4) 28.6% | 0.0003*** | 18 |
| Early response to cold, mannitol and NaCl | 66 | (11) 16.7% | 0.0002*** | 19 |
| Common response to cold, osmotic stress, wounding and biotic stress | 197 | (18) 9.1% | 0.049** | 20 |
| Common response to 9 or more abiotic or biotic stress treatments | 182 | (16) 8.8% | 0.0905* | 21 |
| Altered expression in mkp1 | 21 | (3) 16.7% | 0.0002*** | 22 |
| Control datasets (transcriptome studies of conditions not affecting MPK3 activity) | ||||
| Pollen-enriched | 456 | 27 (5.9%) | 0.9463 | 23 |
| Response to methyl jasmonate | 975 | 50 (5.1%) | 0.3388 | 24 |
| Response to ACC (ethylene) | 314 | 18 (5.7%) | 0.9311 | 24 |
The proportion of genes containing at least two VRE copies in their 500 bp promoter regions was compared between all Arabidopsis genes and datasets from published microarrays. The statistical significance, as calculated by Chi2 test, is indicated as * (significant),
** (highly significant), and
*** (extremely significant).
A gene expression study of the early and late wound response in local and systemic leaves identified 14 genes that are induced systemically early after wounding (18). Four of these genes (28.6%) carry multiple VRE copies in their promoters, a significantly higher proportion than would be expected from the overall abundance of such promoters in the Arabidopsis genome (5.8%). Since this is only a small dataset, one should be careful to generalize from these findings. However, we also found proportionally more multiple VRE-containing promoters among the dataset of genes with reported early transient induction by the bacterial elicitor flagellin-derived peptide flg22 (17). Of the 42 genes whose expression is induced after 30 min, but declined after 60 min of treatment, six (14.3%) harbor multiple VRE sequences in their promoters. It is interesting to note the time correlation between MPK3 activity and the expression of VRE-containing genes. MPK3 activity increases rapidly upon flg22 treatment and sharply declines after 30 min (25). Transcription factors that are directly activated by MPK3 are therefore likely to contribute to transient flg22-triggered changes in gene expression.
Three independent genome-wide studies have established clusters of genes that commonly respond to multiple stress treatments (19–21). Among these clusters, genes with multiple VREs in their 500-bp promoter region are clearly overrepresented (Table 1). VREs are also more abundant in promoters of genes differentially expressed in mkp1 plants, which are impaired in the MPK3-inactivation via dual-specific phosphatase MKP1 (22). To assess the significance of the overrepresentation of multiple VRE-containing promoters in the statistical analysis of these stress-related transcriptome datasets, we also included datasets obtained under conditions when MPK3 is not activated or found to play a prominent role (23, 24) (Table 1). Unlike their overrepresentation in datasets of stress-responsive genes (8.8–28.6%), multiple VRE-containing promoters in these control datasets (5.1–5.7%) show an abundance that is close to that expected for the entire Arabidopsis genome (5.8%).
Investigation of VRE Regulation in Two Potential VIP1 Target Genes.
Based on the above observations, we hypothesized that VREs may serve as transcriptional regulatory motifs in at least a subset of stress-responsive genes. For the selection of putative target genes of the MPK3-VIP1 pathway, we applied two criteria: such genes should be (i) responsive to conditions that correlate with MPK3 activation and (ii) carry at least two VRE copies in their promoter regions. Clearly, this approach excludes promoters which are bound by possible heterodimers of VIP1 and other bZIP-like proteins recognizing distinct DNA motifs.
For a more detailed analysis of the contribution and mechanism how VRE motifs function in the context of MPK3-mediated signaling by VIP1, we selected two genes, thioredoxin Trxh8 (At1g69880) and the transcription factor MYB44 (At5g67300). Trxh8 and/or MYB44 gene expression correlates with activation of MPK3 in the response to a diversity of stresses, e.g., treatment with the plant pathogen Pseudomonas syringae, various pathogen-associated molecular patterns, wounding, and oxidative stress (18, 25–27).
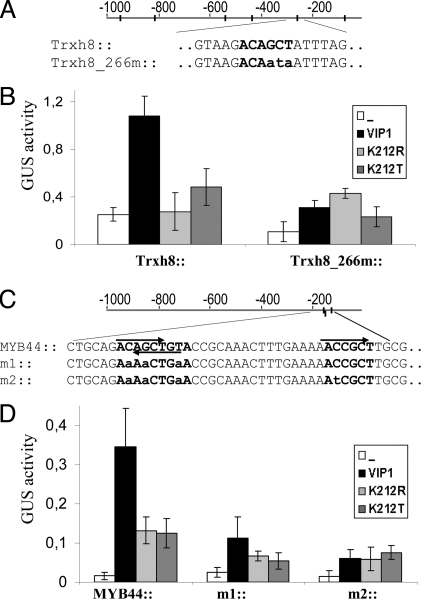
The 1,000-bp promoter region of Trxh8 harbors six VRE copies (Fig. 3A). Protoplasts transfected with a 1,100-bp Trxh8 promoter-driven GUS construct displayed higher reporter gene activity when cotransfected with a construct for constitutive overexpression of VIP1 (35S::VIP1) (Fig. 3B). We then tested whether VIP1 activated the Trxh8 promoter in a VRE-dependent manner. As statistical analyses indicated the presence of regulatory DNA elements to be positioned most often at regions 200–300 bp upstream of the translation start site (28), we mutated the VRE motif that is located 266 bp upstream of the Trxh8 transcription start site to give Trxh8_266m::GUS. Compared with protoplasts transfected with a wild-type Trxh8::GUS construct, Trxh8_266m::GUS activity was significantly compromised upon cotransfection with 35S::VIP1 (Fig. 3B). Consistent with our concept that the lysine residue K212 is essential for VIP1 DNA binding, overexpression of VIP1 K212 mutant variants (K212R or K212T) did not activate the Trxh8 promoter (Fig. 3B).
Fig. 3.
VIP1 target genes. (A and C) Schematic presentation of the Trxh8 and MYB44 promoter regions. Promoter regions matching the VRE1 consensus (ACNGCT or its reverse complement, AGCNGT) are highlighted, respective mutations of the promoter constructs are shown in lowercase. Note that in the MYB44 promoter two oppositely-oriented copies of VRE1 are overlapping. (B and D) Protoplast cotransfection. Protoplasts were transfected with Trxh8 (B) or MYB44 (D) native and mutated promoter::GUS constructs in the absence or presence of VIP1 or Lys-212 mutant variants K212R, K212T, and analyses as described in Fig. 1A.
The second potential VIP1 target gene, MYB44, is one of the 197 core stress responsive genes reported by Ma and Bohnert (20). The MYB44 promoter region harbors three VRE copies, two of which are overlapping in opposite orientation (Fig. 3C). Protoplast cotransfection studies revealed that a 137-bp MYB44 promoter fragment containing the three VRE copies was sufficient to drive VIP1-induced GUS reporter gene expression. VRE mutation (MYB44m1, MYB44m2) rendered the promoter nonresponsive to activation by VIP1 (Fig. 3D). Similar to Trxh8 promoter activation, induction of MYB44 expression by VIP1 depends on VIP1 residue Lys-212. Taken together, these results are consistent with the assumption that Trxh8 and MYB44 are direct VIP1 targets and that direct binding through the bZIP domain of VIP1 to its cognate VRE sequences is necessary for promoter activation.
Defining the VIP1 Signaling Pathway.
According to our hypothesis, MYB44 and Trxh8 should be stress-regulated target genes of VIP1, whose nuclear localization depends on prior phosphorylation by stress-activated MPK3. Consequently, mutant plants lacking MPK3 or VIP1 would be expected to be impaired in the stress-induced expression of these genes. We analyzed MYB44 and Trxh8 gene expression at 5, 10, and 20 min of treatment with flg22. In wild-type plants, MYB44 and Trxh8 transcripts rapidly accumulated upon flg22 treatment (Fig. 4A). Similar stress gene induction was observed in the previously characterized vip1 mutant line, vip1-1 (SALK_001014) (Fig. S3). [The only other available putative vip1 T-DNA insertion line (SALK_148460) turned out to express wild-type levels of the complete VIP1 transcript.] vip1-1 forms a C-terminally truncated protein in which the MPK3 phosphorylation site, as well as the entire bZIP domain, are retained and is apparently not affected in its transcriptional activation capacity. Thus, vip1-1 mutants were found unsuitable for testing our hypothesis. We therefore focused on the analysis of mpk3 null mutants.
Fig. 4.
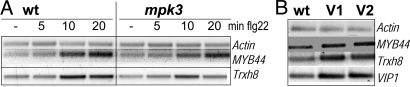
MPK3-dependent stress responsiveness of VIP1 target genes Trxh8 and MYB44. (A) Flg22-induced expression of VIP1 target genes in wild-type and mpk3 mutant plants. Fourteen-day-old seedlings grown in Petri dishes were adjusted for 24 h in liquid medium before application of water or 2.5 μM flg22. Samples were frozen after 0, 5, 10, and 20 min and assessed by semiquantitative RT-PCR. Actin and MYB44 transcripts were amplified in a multiplex PCR. (B) VIP1 target gene expression in VIP1 overexpressing plants. Semiquantitative RT-PCR analysis of expression levels of the indicated genes in 14-day-old seedlings of wild-type and two stably transformed VIP1-HAo/e lines (V1, V2).
Compared with wild-type plants, we consistently observed lower flg22-induced MYB44 and Trxh8 transcript accumulation in mpk3 mutant plants (Fig. 4A). Stress gene induction was not completely blocked in mpk3, most probably because MPK3 can be partially compensated for by the highly homologous MAPK MPK6. Previous in vitro kinase assays had revealed that VIP1 can, although very poorly, also be phosphorylated by MPK6 (data not shown). In addition, MPK3-VIP1-mediated stress gene induction might be partially masked by the contribution of additional regulatory elements targeted by other stress-related transcription factors.
Complimentary to studying VIP1 target gene expression in vip1 or mpk3 mutant plants, we sought further confidence of VIP1 being an activator of MYB44 and Trxh8 expression by a gain-of-function approach. To this end, transgenic plants constitutively overexpressing hemagglutining-tagged VIP1 (VIP1-HAo/e) were generated. In all six lines tested (shown for two lines in Fig. 4B), overexpression of the VIP1-HA transgene correlated with pronounced accumulation of MYB44 and Trxh8 transcripts.
Taken together, the observed impairment of stress-triggered induction of two candidate VIP1 target genes, MYB44 and Trxh8, in mpk3 null mutants as well as the elevated transcript levels of these genes in VIP1-HA overexpressing plants suggest a direct regulation via the following pathway [stress→MPK3→VIP1→VRE promoter activation→stress gene induction]. That MYB44 and Trxh8 transcript levels are enhanced in nontreated VIP1-HA overexpressing plants is likely to be because of a basal nuclear level of VIP1. This, in turn, may be explained by the strong activity of the constitutive promoter driving VIP1 expression or a basal level of MPK3 activity. Upon stress treatment, additional VIP1 molecules are recruited from the cytoplasm to the nucleus. This assumption is consistent with our previously reported microscopy analysis, where a fraction of the transgenic VIP1 protein was found in the nucleus already in nontreated VIP1-YFP-expressing plants (2).
Chromatin Immunoprecipitation Reveals Stress-Dependent in Vivo Binding of VIP1 to the MYB44 Promoter.
To investigate whether VIP1 directly interacts with target promoters in planta, VIP1-HA overexpressing plants were subjected to chromatin immunoprecipitation (ChIP) analysis and compared with wild-type plants (Fig. 5).
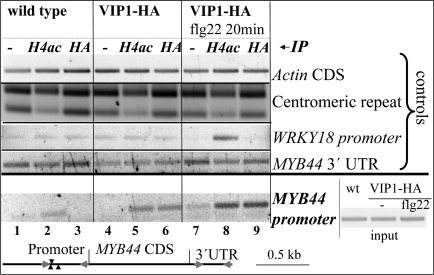
Fig. 5.
ChIP of the MYB44 promoter region comprising the VRE motifs. Chromatin of wild-type (wt) or VIP1-HA overexpressing plants (−/+ application of a 20-min treatment with flg22) was immunoprecipitated without antibody (mock, −) or with antibodies directed against histone H4ac or HA. PCR with primers binding to the actin coding region, a 180-bp repetitive centromeric region, or a WRKY18 promoter region served as controls for equal DNA content, for successful ChIP, and effective flg22 response, respectively. Lower: relative position of the primers used for MYB44 promoter amplification. Right: “Input,” PCR amplification products of “input” DNA samples [chromatin isolated from wt, VIP1o/e (−/+ flg22) before immunoprecipitation] with MYB44 promoter-specific primers.
ChIP performed on wild-type plants without (mock control) or with anti-HA antibody yielded no detectable enrichment of a MYB44 promoter region (spanning the VRE1-containing region) (Fig. 5, lanes 1 and 3, respectively). In contrast, we observed MYB44 promoter amplification products when ChIP was performed with anti-histone H4ac antibody. This antibody recognizes acetylated histone H4 and enriches for expressed genes [as evidenced by the shift of amplification products of a repetitive centromeric region (29) in the anti H4ac samples] (Fig. 5, lane 2).
In contrast, ChIP with either anti-HA or anti-histone H4ac antibody yielded detectable MYB44 promoter amplification products in VIP1-HAo/e plants (Fig. 5, lanes 5 and 6, respectively), consistent with the elevated MYB44 transcript levels in these plants (Fig. 4B).
Because MPK3-dependent phosphorylation of VIP1 results in enhanced activation of transcription by VIP1, we also tested whether the VIP1 interaction with the MYB44 promoter will increase under MPK3-activating conditions. VIP1-HAo/e plants were stimulated with flg22 for 20 min before chromatin isolation. ChIP analysis with anti-HA antibody resulted in stronger accumulation of MYB44 promoter-specific amplification products in flg22-treated compared with nontreated samples of VIP1-HAo/e plants (Fig. 5, lanes 9 and 6, respectively), indicating more efficient binding of VIP1 to the MYB44 promoter. Interestingly, when ChIP was performed with anti-histone H4ac from flg22-treated VIP1-HAo/e plants and compared with nontreated conditions, a much stronger amplification was equally obtained (Fig. 5, lanes 8 and 5, respectively). These results indicate that activation of the MYB44 promoter by flg22 is correlated with enhanced association of acetylated histone H4 with the MYB44 promoter. The enhanced association of histone H4ac or VIP1-HA with the MYB44 promoter upon flg22 treatment was not because of unequal reaction conditions as evidenced by ChIP with the same antibodies, but PCR performed with primers for the actin coding region or a 180-bp centromeric region (Fig. 5). PCR amplification of a WRKY18 promoter region served as an additional control. The activity of the early stress-responsive WRKY18 promoter is regulated via W-boxes recognized by WRKY proteins (30). Enrichment of a WRKY18 promoter region was observed in samples of flg22-treated VIP1-HAo/e plants immunoprecipitated with anti-histone H4ac-antibody, but not with anti-HA antibody. This indicates that VIP1o/e plants responded to flg22 treatment with activation of the WRKY18 promoter, but that this activation was not mediated through binding of VIP1.
In summary, our ChIP experiments provide evidence that VIP1 can directly bind to the MYB44 promoter in vivo. Furthermore, binding is enhanced under conditions of flg22-induced MPK3 activation. These data are consistent with the observed enhanced MYB44 transcript levels in VIP1 overexpressing plants and the inducibility of MYB44 expression by flg22.
MAPK-targeted transcription factors are good candidates for regulating gene expression of components involved in the primary/immediate stress response (e.g., redox-regulatory enzymes such as thioredoxins) and of additional transcription factors, which in turn activate respective subsets of target genes encoding components involved in stress adaptation.
By acting directly downstream of MPK3, VIP1 is a potent transducer of the stress signaling MAPK cascade. Through the transcriptional induction of genes encoding transcription factors themselves, such as MYB44, VIP1 may also be involved in mediating later than the immediate early responses characterized by the transient activation pattern of the MAP kinase. The presence of multiple VREs in the promotors of several stress-responsive genes suggests that VREs might also serve to coordinately activate a larger set of genes by VIP1. It is likely that the signaling pathways for responses to individual stresses bifurcate downstream of VIP1, a possible scenario could be through heterodimerization with additional transcriptional activators and/or repressors that are regulated by separate signaling pathways.
Since even in nontreated plants, a minor pool of VIP1 is found in the nucleus (2), VIP1 may be contributing to the background levels of activation of promoters harboring VREs.
VIP1 joins the group of Arabidopsis bZIP proteins with known DNA target motifs. VRE is a DNA element. VIP1 is the so far only characterized member of the bZIP subfamily I. For few Arabidopsis bZIP proteins, target motifs and putative target genes have been identified (8, 9). Unlike VIP1, these proteins display a constitutive nuclear localization and are not subject to any known posttranslational control.
A recent study reports on a B3-DNA binding protein, NtWIF, whose transactivation activity is regulated through phosphorylation by WIPK, the putative MPK3 ortholog in tobacco (31). A number of stress-related putative target genes, including WIPK, have been identified by microarray analysis. MPK3 is also subject to transcriptional control (13). However, a self-amplifying circuit of MPK3 signaling via VIP1—similar to NtWIF-induced WIPK gene expression (31)—appears unlikely, since no VRE sequences are found in the MPK3 promoter.
A previous study (32) has shown that heterologous overexpression of VIP1 in tobacco results in developmental abnormalities, including growth retardation and impaired differentiation. These effects may be related to aberrant activation/repression of VIP1-targeted promoters, since VIP1 in tobacco locates exclusively to the nucleus. As no transcriptional analysis has been performed on these plants, the identity of VIP1-targeted tobacco promoters remains elusive. If VIP1 overexpression activated stress gene expression also in tobacco, this might account for the developmental abnormalities displayed by these plants, since constitutive activation of stress responses is often accompanied by developmental defects (30). Alternatively, the phenotype found in VIP1 overexpressing tobacco plants might be the consequence of an imbalance of endogenous bZIP proteins (subfamily I-type) that compete with VIP1 for their true dimerizing partners
Moreover, Avivi et al. (32) observed that VIP1 gene expression is enhanced in de-differentiating Arabidopsis cells (undergoing protoplasting); and we found ectopically expressed VIP1-YFP predominantly in the nucleus (2). VIP1 may thus be involved in the maintenance of enhanced basal expression of stress-responsive genes in protoplasts and thereby confer a certain preparedness against challenging conditions, as de-differentiating cells are (because of the lack of a cell wall) particularly susceptible to stress.
In summary, VIP1 is a protein incorporating two distinct functions. It mediates the nuclear import of Agrobacterium T-DNA, thereby assisting plant transformation. The second function involves transcriptional regulation of stress-responsive genes. Both VIP1 functions rely on stress-triggered nuclear translocation of VIP1 and are mediated through phosphorylation by MPK3. In our study, we have also identified VRE as a DNA motif that is bound by VIP1 in vivo and whose abundance in stress-responsive promoters correlates with MPK3 activation. These results provide evidence that one response to stress-triggered MPK3 activation is mediated by VRE-dependent stress gene expression through phosphorylation-dependent control of VIP1. The potential impact of VIP1-mediated stress gene activation in stress tolerance will be addressed in future studies.
The question arises how Agrobacterium can take advantage of the stress-triggered nuclear translocation of VIP1 if nuclear VIP1 activates the defense response. In fact, numerous stress-response genes are induced in the early response to Agrobacterium infection (33). Subsequent expression of these genes is suppressed later during infection with strains that can transfer Vir proteins and T-DNA (33). Once it has fulfilled its function as T-DNA transporter, VIP1 undergoes proteasomal degradation triggered by Agrobacterium VirF (34).
Thus, the initial stress gene activation upon Agrobacterium contact might—at least partially—be attributed to the action of nuclear VIP1, whereas because of its proteasomal degradation later in the infection process, VIP1 cannot activate defense genes any more.
Methods
Plant Material and Treatment.
A. thaliana Col-O and CaMV35S::VIP1-HA overexpressing plants were grown on plates containing ½ Murashige and Skoog (MS) medium (Duchefa), 1% sucrose and 0.8% agar at 24 °C and a 16 h photoperiod. For flg22 treatment, 14-day-old wild-type seedlings were adapted overnight in liquid ½ MS medium containing 1% sucrose. Flg22 was added to a final concentration of 2.5 μM. Seedlings were frozen at 0, 5, 10, and 20 min after treatment.
Protoplast Transfection and GUS Quantification.
Arabidopsis protoplasts preparation, transfection, protein extraction, GUS activity quantification, and immunoblotting were performed as described in ref. 2.
Labeling of RDSA Candidates for EMSA.
Inserts of sequenced clones were PCR-amplified with primers RDSA_1fo/re or RDSA_2fo/re (Table S1), respectively, and labeled using the biotin 3′end labeling kit (Pierce). Efficiency of labeling was checked by dot blot analysis according to the manufacturer's instructions. The following labeled oligonucleotides were annealed E1fo/E1re; E1mfo/E1mre; as-1fo/as-1re.
Electrophoretic Mobility Shift Assay.
Recombinant VIP1 was incubated with biotin-labeled DNA fragments in RDSA binding buffer for 40 min at 4 °C. The reactions were loaded on a 6% TBE gel that had been prerun for 1 h at 4 °C. Blotting and detection were performed using the Light Shift Chemiluminescence kit (Pierce). For competition analysis, 500 ng poly(dIdC) or a 20-fold excess of nonlabeled RDSA candidate fragment E1 were added to the binding reactions.
Chromatin Immunoprecipitation.
Four-week-old wild-type and VIP1-HAo/e plants were subjected to ChIP analysis with rabbit polyclonal antibody directed against acetylated Histone H4 (Upstate) or monoclonal mouse anti-hemagglutinin antibody following a protocol described in http://www.epigenome-noe.net/researchtools/protocol.php?protid = 13.
Additional Experimental Procedures.
A detailed description of plasmids and cloning, RT-PCR analysis, expression and purification of VIP1 recombinant protein, the Random DNA Selection assay, and VRE motif abundance calculation can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Christiane Gatz and Sebastian Pape for providing PR1 promoter mutant constructs and for critically reading the manuscript. This work was supported by grants from the Austrian Science Fund and the Forschungsschwerpunkt of the University of Vienna.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905599106/DCSupplemental.
References
- 1.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 3.Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends Plants Sci. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakoby M, et al. bZIP transcription factors in Arabidopsis. Trends Plants Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 6.Fukazawa J, et al. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aukerman MJ, Schmidt RJ, Burr B, Burr FA. An arginine to lysine substitution in the bZIP domain of an opaque-2 mutant in maize abolishes specific DNA binding. Genes Dev. 1991;5:310–320. doi: 10.1101/gad.5.2.310. [DOI] [PubMed] [Google Scholar]
- 8.Johnson C, Boden E, Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15:1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weltmeier F, et al. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006;25:3133–3143. doi: 10.1038/sj.emboj.7601206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deppmann CD, Alvania RS, Taparowsky EJ. Cross-species annotation of basic leucine zipper factor interactions: Insight into the evolution of closed interaction networks. Mol Biol Evol. 2006;23:1480–1492. doi: 10.1093/molbev/msl022. [DOI] [PubMed] [Google Scholar]
- 11.Ringli C, Keller B. Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol Biol. 1998;37:977–988. doi: 10.1023/a:1006030007333. [DOI] [PubMed] [Google Scholar]
- 12.Foster R, Izawa T, Chua N. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- 13.Pitzschke A, Hirt H. Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol. 2009;149:606–615. doi: 10.1104/pp.108.131557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feilner T, et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics. 2005;4:1558–1568. doi: 10.1074/mcp.M500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Popescu SC, et al. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23:80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caselle M, Di Cunto F, Provero P. Correlating overrepresented upstream motifs to gene expression: A computational approach to regulatory element discovery in eukaryotes. BMC Bioinformatics. 2002;3:7. doi: 10.1186/1471-2105-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro L, et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R. Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol. 2004;55:165–181. doi: 10.1007/s11103-004-0112-7. [DOI] [PubMed] [Google Scholar]
- 19.Kreps JA, et al. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 2007;8:R49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kant P, et al. Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ. 2008;31:697–714. doi: 10.1111/j.1365-3040.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- 22.Ulm R, et al. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21:6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goda H, et al. The AtGenExpress hormone and chemical treatment dataset: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 25.Livaja M, Zeidler D, von Rad U, Durner J. Transcriptional responses of Arabidopsis thaliana to the bacteria-derived PAMPs harpin and lipopolysaccharide. Immunobiology. 2008;213:161–171. doi: 10.1016/j.imbio.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Gust AA, et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem. 2007;282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- 27.Gadjev I, et al. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki M, Ketterling MG, McCarty DR. Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol. 2005;139:437–447. doi: 10.1104/pp.104.058412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson LM, Cao X, Jacobsen SE. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129:706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung KM, Sano H. Transactivation of wound-responsive genes containing the core sequence of the auxin-responsive element by a wound-induced protein kinase-activated transcription factor in tobacco plants. Plant Mol Biol. 2007;65:763–773. doi: 10.1007/s11103-007-9240-1. [DOI] [PubMed] [Google Scholar]
- 32.Avivi Y, et al. Reorganization of specific chromosomal domains and activation of silent genes in plant cells acquiring pluripotentiality. Dev Dyn. 2004;230:12–22. doi: 10.1002/dvdy.20006. [DOI] [PubMed] [Google Scholar]
- 33.Veena, Jiang H, Doerge RW, Gelvin SB. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003;35:219–236. doi: 10.1046/j.1365-313x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- 34.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.