Abstract
Recent findings have highlighted roles played by innate cellular factors in restricting intracellular viral replication. In this review, we discuss in brief the activities of apolipoprotein B mRNA-editing enzyme 3G (APOBEC3G), bone marrow stromal cell antigen 2 (BST-2), cyclophilin A, tripartite motif protein 5 alpha (Trim5α), and cellular microRNAs as examples of host restriction factors that target HIV-1. We point to countermeasures encoded by HIV-1 for moderating the potency of these cellular restriction functions.
A brief overview of HIV-1/acquired immune deficiency syndrome (AIDS)
AIDS was first recognized nearly 30 years ago. The initial description was followed shortly by the discovery and characterization of its causative agent, the human immunodeficiency virus, HIV-1. Today, approximately 33 million people worldwide are infected with HIV. Each year, 2.5 million people become newly infected and 2 million others die from AIDS. While there are several effective drugs for treating HIV/AIDS, ongoing attempts to develop a useful HIV-1 vaccine and a protective antiviral microbicide face significant challenges and seem unlikely to be successful in the near future [1]. In this setting, a fuller understanding of the innate restriction mechanisms in human cells that modulate HIV-1 replication is worthwhile.
HIV-1 infects CD4+ T-cells. The virus encodes nine genes; three are regarded as 'structural' genes (Gag, Pol, Env), while the other six are considered 'accessory' genes (Tat, Rev, Nef, Vpr, Vpu, Vif). Steps in HIV-1 replication, including the interaction of the viral envelope protein (gp120) with the cellular CD4 receptor, reverse transcription to generate proviral DNA, integration, RNA transcription, viral protein synthesis, virion assembly and egress have been reviewed elsewhere [2-5]. Here, we discuss in brief the recent findings on apolipoprotein B mRNA-editing enzyme 3G (APOBEC3G), bone marrow stromal cell antigen 2 (BST-2), cyclophilin A, tripartite motif protein 5 alpha (Trim5α) and cellular microRNAs (miRNAs) as examples of host restriction factors [6-8] that target intracellular HIV-1 replication.
APOBEC and Vif
APOBEC3 (A3) genes are unique to mammals and encompass a family of cytidine deaminases that are now believed to play an important role in the intrinsic or innate host immune response to control retroviruses, retrotransposons, hepadnaviruses, foamy viruses and, perhaps, even some DNA viruses such as human papillomavirus (reviewed in [6,9]). A3 genes have arisen through gene duplication and their number varies from one gene in mice to seven genes in humans [10]. They contain either one or two zinc coordinating domains. In enzymes containing two zinc coordinating domains, generally only one (in most cases it is the C-terminal domain) is catalytically active.
All of the A3 genes are catalytically active. However, there is an ongoing discussion on the functional importance of A3 catalytic activity for antiviral effects. For instance, the inhibition of parvoviruses and retrotransposons by A3A was found to be deaminase-independent [11-13]. Deaminase-independent inhibition by A3G was also reported for other viruses such as HTLV-1 and hepatitis B virus [14-17]. Finally, A3G and A3F were shown to inhibit HIV replication in a deaminase-independent manner (reviewed in [6]). However, most of the data supporting deaminase-independent mechanisms result from a transient overexpression of A3 proteins or are based on in vitro assays. Indeed, there is strong evidence that HIV-1 restriction does require A3G deaminase activity when the protein is not transiently overexpressed [18-20]. A3G is a powerful inhibitor of HIV-1 and several studies showed that only a few molecules of packaged A3G are sufficient to inhibit virus replication [20,21]. On the other hand, the inhibition of HIV-1 replication appears to require a minimum A3G threshold level. This is suggested by the observation that HIV-1 carrying a partially defective Vif gene was found to replicate, albeit with delayed kinetics, in A3G expressing CEM cells, a human cell line originally isolated from an acute lymphoblastic leukemia [22]. Under those conditions, viral DNA showed clear evidence of hypermutation whereas viral RNA was largely unaffected, suggesting a mechanism of purifying selection [22].
A3 proteins are packaged into viral particles through an interaction with viral Gag protein and viral or cellular RNA [23]. Vif neutralizes the antiviral activity of A3G and A3F by inhibiting their packaging into viral particles. This involves a proteasome mediated degradation of A3 proteins as well as the degradation-independent mechanism(s) [24]. Endogenous A3G appears to be much less sensitive to degradation by Vif than transiently expressed protein [25]. While the relative contribution of degradation-dependent or independent mechanisms is still being debated, it is generally accepted that the inhibition of A3G and A3F by whatever mechanism involves a direct physical interaction with Vif (Figure 1). The regions in Vif, important for binding to A3G and A3F, appear to be overlapping but not identical. They map to the N-terminal region of Vif and appear to be discontinuous [26-31]. Interestingly, the regions in A3G and A3F important for interaction with Vif are also distinct. In A3G, residues of 126-132 in the N-terminal region were found to be sufficient for Vif binding, while Vif appears to interact with a C-terminal region of the A3F encompassing residues 283-300 [32-34]. In fact, that A3F region is sufficient to enable Vif binding and A3F degradation, whereas degradation of A3G by Vif requires a region larger than that necessary for Vif binding [32].
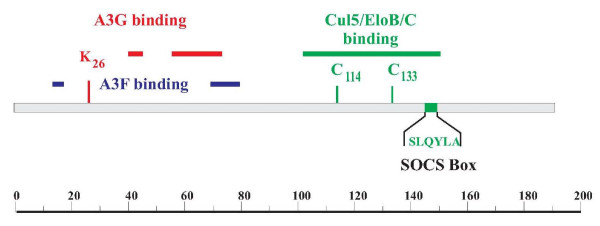
Figure 1.
Protein interaction domains in HIV-1 Vif. The regions responsible for interaction with A3G (red) and A3F (blue) are discontinuous. They overlap but are not identical. The region important for interaction with the Cul5 ubiquitin ligase complex is marked in green and includes a conserved histidine-cysteine-cysteine-histidine motif and a suppressor of cytokine signaling Box domain. The scale at the bottom indicates amino acid positions.
The structural determination of full-length A3G has so far been unsuccessful. This is mostly due to the insolubility of the purified protein. Structural data are currently available only for monomeric forms of the C-terminal catalytic domain [35-38]. However, biochemical analyses demonstrated that A3G forms oligomeric structures that are mediated or stabilized by RNA [39-41]. Huthoff et al. have identified oligomerization mutants of A3G that were packaging incompetently and had lost their antiviral activities [41]. Opi et al., on the other hand, identified an oligomerization mutant that remained packaging competent and retained antiviral activity [39]. Thus, even though A3G is likely to be recruited into viral particles as an oligomer [42], A3G oligomerization does not appear to be critical for antiviral activity, at least in transient expression assays.
One of the intriguing properties of A3G is its ability to assume different intracellular configurations. In activated cells and T cell lines, A3G exists as a mixture of low molecular mass (LMM) and high molecular mass (HMM) complexes, and it has been proposed that only the LMM form of A3G has antiviral activity [43]. A3G exerts its antiviral effect during the early phase of virus replication. In order to do so, the protein has to be packaged into virions from the virus-producing cell. A3G expressed in target cells does not generally inhibit virus replication. A notable exception is resting CD4+ T cells. In these cells A3G exists predominantly in the LMM configuration and was shown to impose a potent post-entry restriction on HIV infection [43]. This view was recently challenged by Kamata et al. who could not confirm the involvement of A3G in the post-entry restriction of HIV-1 [44]. Thus, the relative contribution of A3G to the resistance of quiescent T cells to HIV infection remains to be resolved.
Vpu and BST-2
It is generally accepted that Vpu regulates the detachment of otherwise complete and infectious virions from the cell surface [45,46]. Interestingly, the ability to enhance virus release is not unique to Vpu. In fact, some HIV-2 isolates have been known to encode a Vpu-like activity in their Env glycoproteins [47-50]. Intriguingly, a similar Vpu-like activity has now also been shown for the Nef protein of several simian immunodeficiency viruses, including SIVmac and SIVagm [51,52].
Vpu-dependent virus release is host cell-dependent [53]. Indeed, several other host factors have been identified whose overexpression was associated with reduced virus release. These include the Vpu binding protein UBP [54], TASK-1, a human potassium channel protein, [55] and the recently identified host factors BST-2 (also referred to as tetherin, CD317, or HM1.24 [56,57]) and CAML (calcium modulating cyclophilin ligand) [58]. Among those, BST-2 is of particular interest since its cell type-specific expression most closely matches that of Vpu-dependent cell types. Silencing the BST-2 expression in HeLa cells by siRNA or shRNA (short hairpin ribonucleic acid) rendered virus release from these cells Vpu-independent [56,57].
A functional Vpu-BST-2 interaction was first reported in a quantitative membrane proteomics study where Vpu expressed from an adenovirus vector was found to reduce cellular expression of BST-2 in HeLa cells [59]. Intriguingly, BST-2 expression is cell type dependent and is induced by interferon treatment [56,57,60,61] which is consistent with the previous observation that cell lines that did not normally require Vpu for efficient virus release became Vpu-dependent upon being treated with interferon [60].
How does BST-2 inhibit virus release? BST-2 is a 30-36 kDa type II transmembrane glycoprotein [62]. The protein has both an N-terminal transmembrane domain and a C-terminal glycosyl-phosphatidylinositol (GPI) anchor [63] (Figure 2). This has led to a model, in which BST-2, by means of its N-terminal transmembrane domain and its C-terminal GPI anchor, tethers otherwise fully detached virions to the producer cell [56]. Such a model is consistent with the observation that Vpu-defective particles can be released by protease treatment [46] or by physical force [45]. Particles released by the latter method were found to be fully infectious [45]. The tethering model is elegant and simple but it still awaits formal experimental validation. Somewhat unexpectedly, Vpu-deficient virions that were separated from the cell surface by physical force did not contain detectable amounts of BST-2 [61] even though the electron micrograph data imply that BST-2 not only tethers particles to the virus producing cell but crosslinks virions among each other [45,56].
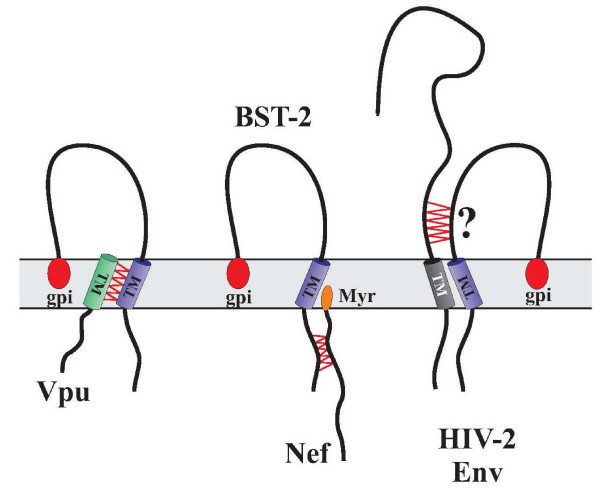
Figure 2.
Interaction of BST-2 with HIV-1 proteins. Vpu interacts with BST-2 through its transmembrane domain. Interaction of BST-2 with Nef involves the cytoplasmic domain. The interaction of HIV-2 Env with BST-2 has not been mapped, but may involve the ectodomain. Proposed interaction points are indicated by red zig-zag lines. Myr: Nef myristoylationl; gpi: glycosyl-phosphatidylinositol anchor.
How does Vpu counteract the BST-2 imposed restriction of virus release? Recent data suggest that the BST-2 transmembrane (TM) domain is critical for interference by Vpu [64-66] consistent with the previous observation of the importance of the Vpu TM domain for the regulation of virus release [67-69]. This suggests a direct physical interaction of Vpu and BST-2. Indeed, Vpu was found to interact with BST-2 in HeLa cells [70]. It is unclear how Vpu inhibits BST-2. One simple way of interference would be to down regulate the protein from the cell surface. Indeed, several studies have noted that Vpu can down regulate BST-2 [57,61,71] although Vpu does not seem to increase the rate of BST-2 endocytosis [72]. This suggests that Vpu may affect the resupply or the surface delivery of BST-2, a function that could be exerted from an intracellular location such as the trans-Golgi compartment [70,73]. Interestingly, Vpu was found to reduce total cellular levels of endogenous as well as exogenously expressed BST-2 [59,61]. How this is accomplished is not yet completely clear. However, it has been suggested that antagonism of BST-2 by Vpu involves proteasomal degradation by Vpu [74,75]. It was also reported to encompass a β-TrCP (beta transducing repeat containing protein)-dependent endo-lysosomal pathway [70,72]. The involvement of β-TrCP in the virus release activity of Vpu necessitates the conservation of Vpu's TrCP binding motif. However, mutation of this motif was previously found to only partially affect the virus release activity of Vpu [57,76,77] and in more recent studies the mutation of the TrCP-binding site in Vpu (Vpu26) did not affect replication kinetics in a variety of cell types, including the peripheral blood mononuclear cells [61]. Expression of the TrCP binding-deficient Vpu26 had no effect on BST-2 stability. To the contrary, it appeared to stabilize or even increase the BST-2 surface expression [61]. Thus, more work needs to be done to sort out which of the effects of Vpu on BST-2 (surface expression, degradation or intracellular sequestration) are critical for enabling enhanced virus release.
The effects of BST-2 are not restricted to lentiviruses. They have also been shown to affect Ebola virus release [78,79]. Furthermore, the inhibition of HIV-1 virus release is not limited to human BST-2 but is also observed in mouse, rat and monkey BST-2 [65,74]. However, only the human BST-2 is sensitive to Vpu. Finally, the ability of HIV-2 Env and SIV (simian immunodeficiency virus) Nef to antagonize BST-2 appears to involve cell surface down modulation of BST-2. However, unlike Vpu, neither Nef nor HIV-2 seems to affect the stability of BST-2 [51,70]. Although Vpu, targets the BST-2 TM domain, Nef targets the cytoplasmic domain (Figure 2) [51,52]. The site(s) in BST-2 critical for its sensitivity to HIV-2 Env have not yet been mapped. Recently, however, it was noted that the Vpu-like activity of HIV-2 Env was regulated by a single amino acid difference in the ectodomain of its TM subunit [80]. It will, therefore, be interesting to see if HIV-2 Env targets the BST-2 ectodomain.
A final point concerns the question of whether or not BST-2 deserves the label of restriction factor. It is true that BST-2 potently inhibits the release of viral particles and that there does seem to be a selective pressure to maintain Vpu expression, at least in the SHIV (simian-human immunodeficiency virus) model [81]. However, Vpu is not absolutely essential for viral replication and there are primary HIV-1 isolates that lack a functional Vpu gene due to the mutation of the initiation codon [82]. The inhibition of Vpu will reduce virus release but that does not prevent viral spread. Instead, virus replication will simply shift from a cell-free mode to a cell-to-cell mode. This finding likely explains why Vpu-deficient viruses replicate in tissue culture with the same kinetics as wild type virus [45,83-85]. Thus, BST-2 is a restriction factor in the sense that it limits the release of viral particles from infected cells. However, it may not be a restriction factor in the sense that it blocks the spread of virus in an infected individual.
Cyclophilin
In the early 1990s cyclophilin A (Cypa) was reported to be a cellular factor that binds HIV-1 capsid protein (CA), but not the CA of other lentiviruses such as SIVmac239 [86-88]. Recently, it was shown that it also interacts with the CA of some HIV-2 isolates, with SIVagmtan and with FIV (feline immunodeficiency virus) [89-91]. Cypa is a small globular protein that binds to proline-containing peptides via its hydrophobic pocket and is believed to have subtle, but global, effects on protein folding within cells. Among the specific cellular functions of Cypa, it regulates the nuclear export of Zpr1 (zinc finger protein 1) in yeast [92] and the kinase activity of Itch (inducible T-cell kinase) in CD4+ T cells [93]. As a complex with the immunosuppressive drug cyclosporine, it is a potent inhibitor of the CA-dependent phosphatase calcineurin in T cells [94]. Interaction with HIV-1 CA does, indeed, involve a critical proline residue [95] and binding of CypA catalyzes a conformational change in CA [96].
CypA acts within the target cell soon after HIV-1 infection [95,97,98], in some cases to promote infection and in others to inhibit infection [97,99,100]. Sometimes the effect is at the level of reverse transcription [95]. At other times the effect of CypA is evident later, perhaps at the level of nuclear entry [100,101]. It is generally thought that CA indirectly regulates reverse transcription via its effects on an ill-defined process called uncoating (Figure 3). However, CA remains associated with nascent viral cDNA, may dock to the nuclear pore and does seem to be a critical determinant in HIV-1's ability to cross the nuclear pore or even to integrate into host chromosomal DNA [100,102-104]. The simplest explanation for the diverse effects of CypA is that it regulates CA interaction with host factors, either uncoating factors, nuclear pore components or restriction factors that act in a similar fashion to TRIM5 [105-107] (Figure 3).
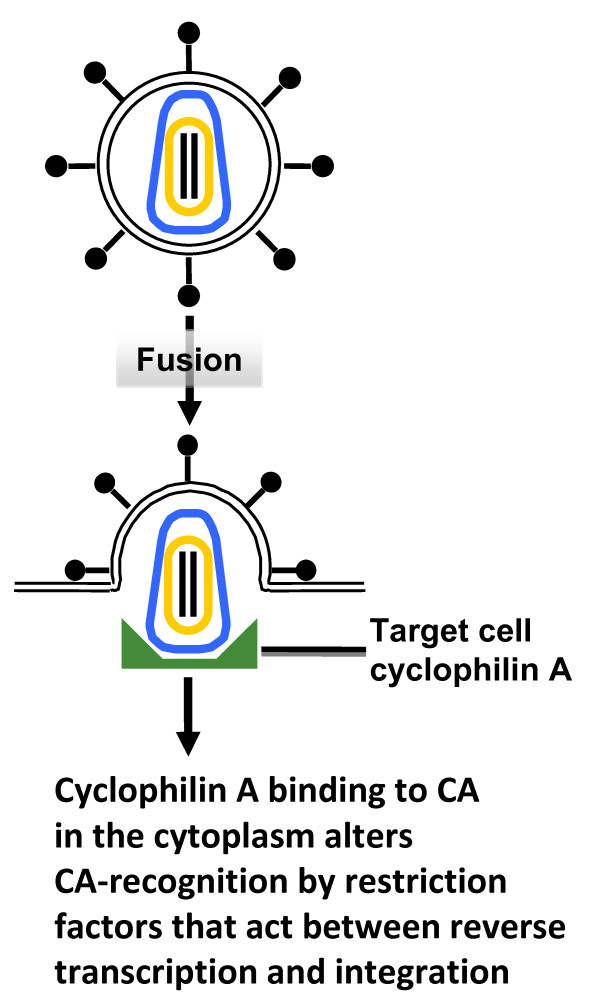
Figure 3.
The effect of host cell cyclophilin A on HIV-1 transduction. Host cell cyclophilin A (shown in green) binds to the HIV-1 CA and regulates CA-recognition by host cell restriction factors. In some cases, cyclophilin A promotes HIV-1 CA recognition by these factors (for example, by rhesus macaque TRIM5alpha). In other cases, human cells for example, cyclophilin A seems to prevent recognition by these factors. The effect of cyclophilin A is, therefore, determined by the effect of the restriction factor, which may act at the time of reverse transcription, nuclear import, or integration.
As discussed below, characterization of the varied effects of CypA steered investigators to the discovery of TRIM5 as an innate, species-specific anti-HIV-1 factor in the cells of non-human primates [106,107]. In the future, the proviral and antiviral effects of CypA in different human cell types may guide the discovery of similar factors acting in human cells [107-110]. A more detailed structural analysis of the HIV-1 CA lattice [111] may reveal how CypA alters the CA conformation and, thus, the accessibility of antiviral factors such as TRIM5. A modest inhibition of viral load by cyclophilin A-binding drugs (non-immunosuppressive analogues of cyclosporine) was seen in some individuals with HIV-1 infection [112], leaving open the possibility that such drugs might play a role in the clinic in the future.
TRIM5
Cells from South American owl monkeys potently inhibit HIV-1 transduction [113]. This HIV-1 restriction activity was examined in great detail because of its apparent dependence upon CypA [105-107]. These studies led to the cloning of an essential factor, the TRIM5-CypA fusion gene [106]. A similar anti-HIV-1 restriction activity in rhesus macaque cells was shown to be due to TRIM5alpha [114]. TRIM5alpha orthologues with anti-retroviral activity have been observed in most primate species, as well as in cows [115,116] and rabbits [117]. The gene is deleted in dogs and truncated in cats [118]. Rodents have multiple copies of the gene, though antiviral activity has not been demonstrated [119]. The owl monkey gene was created by retrotransposition of a CypA cDNA into the TRIM5 locus [106,120,121]. Amazingly, a TRIM5-CypA fusion allele was created by an independent retrotransposition event in Asian macaques [122-125].
TRIM5 is one of the more than 70 tripartite motif genes in the human genome. Tripartite refers to proteins bearing a RING finger, a B-box and a coiled-coil domain. The two latter domains promote TRIM5 multimerization and are almost always required for anti-HIV-1 restriction activity [126,127]. The TRIM5 RING finger has E3 ubiquitin ligase activity in vitro and in vivo [128-130] and, while not strictly required for restriction activity, it clearly plays an important role in TRIM5-mediated restriction. This apparent paradox has been explained by the finding that TRIM5 blocks HIV-1 infection at more than one step, reverse transcription and nuclear import [101,131,132].
The CypA domain in the TRIM5-CypA fusions is clearly a CA-binding determinant [106,120]. A single amino acid change in the CypA domain distinguishes the owl monkey and macaque proteins which alters retroviral restriction specificity [122]. It has been much harder to demonstrate CA-binding activity for the PRY-SPRY domain (a domain present in the Dictystelium discoideum dual-specificity kinase and the PYRIN protein gene family) of the TRIM5alpha isoform [133-135], apparently because its interaction is much weaker than that of the CypA-CA [127]. It is allele specific differences in the PRY-SPRY domain that determine the differences in retroviral restriction specificity for different TRIM5alpha orthologues [136-139].
The most important question that remains unanswered about TRIM5 is how it restricts retroviral infection. Is it simply that a multimer of TRIM5 binds to incoming virion cores and prematurely uncoats them? Or is the mechanism more complicated, requiring accessory factors (see Figure 4) [107-109,140]? What, if any, is the role of ubiquitin in the restriction mechanism? What is the three-dimensional structure of TRIM5 and how does it recognize the virion core? TRIM5 forms cytoplasmic bodies but the significance of these remains unclear, especially since the endogenous protein has not been visualized.
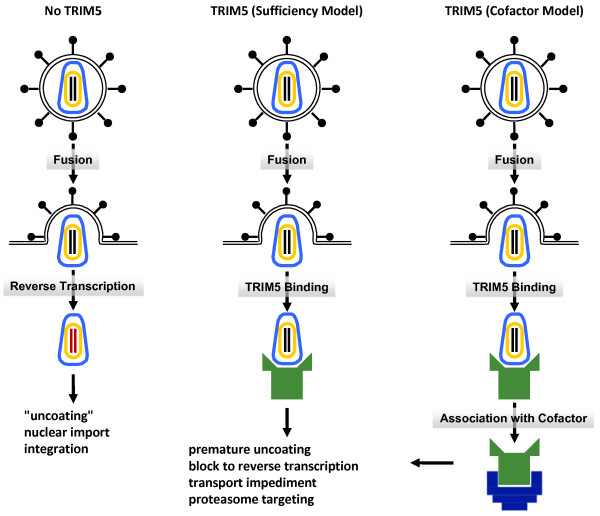
Figure 4.
The effect of TRIM5 on retroviral transduction. In the absence of a restrictive TRIM5α orthologue (No TRIM5), the retroviral genomic RNA is reverse transcribed, translocated to the nucleus and integrated. In the presence of a restrictive allele of TRIM5, retroviral infection is blocked at the level of reverse transcription and, probably, also at the level of nuclear import. According to some investigators, TRIM5 binding to the incoming virion capsid is sufficient to disrupt infection (sufficiency model). According to other investigators, the antiviral effects of TRIM5 require as yet to be identified, cellular cofactors (cofactor model).
Non-coding microRNAs and RNA-silencing
Although restriction factors have traditionally been thought of as proteins, recent findings suggest similar roles played by non-coding RNAs. This concept seems to make sense because, while less than 2% of the human genome encode for proteins, >70% of human DNA are transcribed into non-coding RNAs (ncRNAs) [141]. NcRNAs come in several forms including siRNA, miRNA, and piRNA with each being regulated differently [142-147]. Amongst these RNAs, the best studied is perhaps the ~21-23 nucleotide long microRNA (miRNA). The biogenesis of miRNAs has been reviewed in detail elsewhere [146] and currently more than 800 discrete members are listed in the human miRNAs database (see http://microrna.sanger.ac.uk).
For RNA interference (RNAi) activity, miRNAs assemble into RNA-induced silencing complexes (RISC) which contain an Argonaute protein with additional RNA-binding co-factors such as TRBP (TAR RNA-binding protein) [148-150]. An active miRNA-RISC silences a target mRNA via limited, and sometimes imperfect, base-pairing between the 5'-positioned 2-8 nucleotides of the miRNA (the 'seed' region) [151] and the mRNA. Currently, it is held that a miRNA primarily targets the 3' untranslated region (UTR) of a mRNA. However, coding sequences and the 5' UTR of mRNAs can serve also as miRNA-substrates [152-154]. Indeed, a single miRNA through 'imperfect' (wobbled) base-pairing with mRNA-targets could potentially recognize and regulate the expression of up to one hundred different mRNAs [155].
miRNA-restriction of HIV-1
MiRNAs are used by plants and lower eukaryotic cells to restrict infecting viruses [156]. In higher eukaryotes, the same functional complement of miRNA components and RNA-silencing machinery are conserved. However, it has been debated whether mammals might have extinguished this ncRNA-based antiviral strategy [157-160]. To date, experimental findings, as outlined below, support the idea that mammals continue to employ ncRNAs to 'silence' viruses. First, short interfering RNAs (siRNAs) [161], piwi-interacting RNAs [142,162] and Dicer-processed miRNAs [163] have been shown to suppress mammalian endogenous retroviruses in somatic, germ and mouse embryonic stem (ES) cells. Secondly, bioinformatics analyses have extended this notion of antiviral defense to a plethora of human miRNAs that can target many types of viruses [164,165]. Specifically, miRNA-regulation of HIV-1 infection has been experimentally verified and independently reported by four groups of investigators [166-169]. Thirdly, recent results show that human cells can process HIV-1 RNAs into small ncRNAs that could potentially interdigitate into the cellular RNAi pathways [170]. Collectively, these discrete findings converge to suggest that RNAi-restriction pathways in human T-lymphocytes and macrophages are operative for HIV-1 replication (Figure 5).
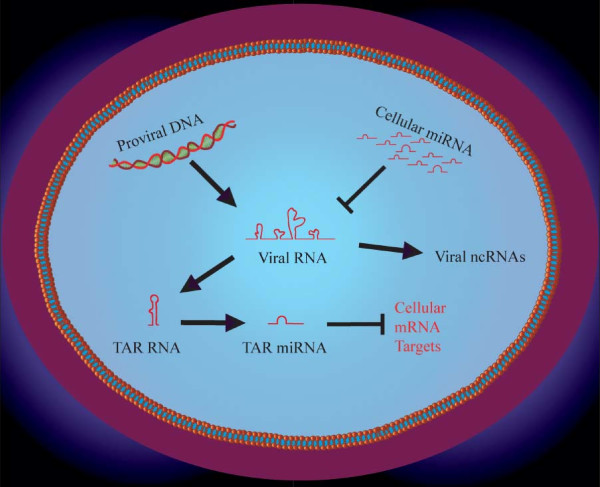
Figure 5.
Schematic representations of roles played by small non-coding RNAs in HIV-1 replication. Integrated HIV-1 provirus is transcribed into viral RNA which can be targeted by cellular miRNAs. HIV-1 transcripts can also be processed into small ncRNAs that could function in cellular RNAi pathway(s). The viral trans-activation responsive TAR RNA has been reported to be processed into a viral miRNA which could act to modulate the expression of cellular mRNAs.
Suppression of RNAi enhances HIV-1 replication
Whether or not RNAi is a physiological mechanism for modulating viral replication can be addressed by asking what happens to an HIV-1 infection when the infected cells are attenuated for their RNAi activity. The RNAi pathway has been deliberately disabled in human cells by the knock down of either the Dicer protein [171] or the RISC components [172] and such suppressions have enhanced HIV-1 replication in cells. Separately, when heterologous RNA-silencing suppressors (RSS), such as the Vaccinia virus E3L protein, influenza A virus NS1 protein, Ebola virus VP35 protein [173] or NS3 protein of Rice Hoja Blanca virus [174], were over expressed in human cells (Table 1) HIV-1 replication in RSS-expressing cells was also considerably enhanced over that of control cells. The most simple interpretation of these results is that the RSS proteins encoded by the viruses repressed a physiologically restrictive cellular RNAi on HIV-1 and that the neutralization of this RNAi directly increased viral replication. The generality of RNAi as a mammalian antiviral mechanism is illustrated by the finding that many viruses encode RSS proteins (Table 1) and that, when cellular RNAi is suppressed, the replication of vesicular stomatitis virus (VSV) [175] and influenza A virus [176] is also enhanced. Collectively, the extant observations are consistent with a conserved mammalian ncRNA/Dicer-RISC/RNAi innate restriction pathway(s) that guards human cells against endogenous and exogenous viruses.
Table 1.
Examples of RNA silencing suppressors encoded by several viruses.
| Virus | Viral Suppressors | References |
| Adenovirus | VA RNA | Lu 2004 [199]; Andersson 2005 [200]; Xu 2007 [201] |
| HCV | Core protein | Chen 2008 [202]; Wang 2006 [203] |
| Ebola | VP35 protein | Haasnoot 2007 [184] |
| Influenza A virus | NS1 protein | Li 2004 [204]; Delgadillo 2004 [205]; Bucher 2004 [206]; Haasnoot 2007 [184]; de Vries 2009 [186] |
| Nodamura Virus | NoV B2 protein | Sullivan 2005 [207] |
| Primate Foamy Virus | Tas protein | Lecellier 2005 [208] |
| HIV | Tat protein, TAR RNA | Bennasser 2005 [182]; Haasnoot 2007 [184]; Bennasser 2006 [183]; Qian 2009 [185] |
| Rice Hoja Blanca Virus | NS3 | Schnettler 2009 [174] |
[This table is modified after Grassmann and Jeang, Ref. [159]].
At what point of an infection might HIV-1 encounter cellular RNAi-restriction? Berkhout et al. reported that the infecting HIV-1 genome is initially sheathed [177] to repel RNAi surveillance. However, at a later stage the RNAi machinery can access and process HIV-1 RNA, as evidenced by the cellular production of virus-encoded TAR (trans-activation responsive RNA) miRNAs [178-180] (Figure 5). Nevertheless, despite the RNAi activity of the cell, in most settings, HIV-1 continues to replicate. This replication is partly explained by the action of the virus-encoded arginine-rich RNA-binding protein, Tat, which acts in an RNA-binding dependant manner [181] as an RNAi-suppressor [182] and by the viral TAR RNA which acts as an RNAi-decoy [183]. The function of HIV-1 Tat as an RNAi-suppressor has been characterized by several investigators and appears to be widely conserved in diverse experimental settings [182,184-186]. Finally, as already noted, RNAi-suppressors appear to be commonly conserved in many mammalian viruses (Table 1). Indeed, viruses such as herpes simplex do employ RNAi-suppression for their replication in human cells [187].
Cellular RNAi virus interaction has been speculated to reshape viral genome sequences overtime [157,188]. Accordingly, it serves viruses, such as HIV-1, well to evolve functions that can alter the miRNA expression profile of the cell to repress virus-noxious miRNAs and to enhance virus-propitious miRNAs. Evidence that such changes occur during viral infection has been reported [171,189,190], suggesting a dynamic strike-counterstrike interplay between the virus and the host [191,192]. Future investigations are needed in order to better understand how RNA-based antiviral restriction complements protein-based restriction.
Concluding remarks
We have raised several examples of host cell factors that can act to impede HIV-1 replication. The emerging scenario suggests that a number of different steps in HIV-1 replication (for example, uncoating, reverse transcription, transcription and virus release) can be targeted by APOBEC3G, cyclophilin, TRIM5α, BST-2 and RNAi. Remarkably, the cellular stratagems against the virus are apparently moderated by HIV-1 encoded countermeasures (for example, Vif, Vpu and Tat). Hence, intricate strike-counterstrike interactions apparently govern the ultimate outcome of HIV-1/cell infection. The overall picture is likely to be even more complex. Recent genome-wide screenings have revealed many hundreds of host cell proteins that appear to be needed in order to activate HIV-1 replication in human cells [193-196]. Additional genome-wide screens will undoubtedly uncover an equally numerous number of host cell factors that restrict HIV-1 replication. The limited examples of HIV-1 restriction factors presented here represent a prelude to the many more restriction entities that still await identification. The next challenge is to elucidate these entities and then apply them to the development of additionally useful interventions against HIV-1/AIDS. For example, the further discovery of cellular factors which obstruct HIV-1 integration [197] could serve to aid the development of novel adjunct therapies that synergize with extant integrase inhibitors [198]. Exciting progress in antivirals lies ahead.
Abbreviations
CA: caspid protein; CML: calcium modulating cyclophilin; CypA: cyclophilin A; Env: envelope protein; FIV: feline immunodeficiency virus; Gag: group specific antigen; GPI: glycosyl-phosphatidylinositol; HMM: high molecular mass; Itk: inducible T-cell kinase; LMM: low molecular mass; miRNA: microRNA; ncRNA: non-coding RNA; Nef: negative factor; piRNA: PIWI interacting RNA; Pol: polymerase; Rev: regulator of viral RNA expression; RISC: RNA-induced silencing complexes; RNA: ribonucleic acid; RNAi: RNA interference; shRNA: short hairpin RNA; siRNA: short interfering RNA; RSS: RNA-silencing suppressors; SHIV: simian-human immunodeficiency virus; SIV: simian immunodeficiency virus; TAR: trans-activation responsive RNA; Tat: transcriptional activator of transcription; TM: transmembrane; TRBP: TAR RNA-binding protein; UTR: untranslated region; Vif: viral infectivity factor; Vpr: viral protein R; Vpu: viral protein U; Zprl: zinc finger protein 1.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KS, JL and KTJ participated equally in the writing of this review article.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
Work in the laboratories of KS and KTJ was supported by intramural funding from NIAID, NIH and the Intramural AIDS Targeted Antiviral Program (IATAP) of the office of the Director NIH. Work in the JL laboratory was supported by the National Institutes of Health grant RO1AI36199 and the Swiss National Science Foundation grant 3100A0-113558. We are grateful to D Schmidt for his assistance with Figure 5.
Contributor Information
Klaus Strebel, Email: Kstrebel@NIH.gov.
Jeremy Luban, Email: Jeremy.Luban@unige.ch.
Kuan-Teh Jeang, Email: Kjeang@NIH.gov.
References
- Wainberg MA, Jeang KT. 25 years of HIV-1 research - progress and perspectives. BMC Med. 2008;6:31. doi: 10.1186/1741-7015-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever AM, Jeang KT. Replication of human immunodeficiency virus type 1 from entry to exit. Int J Hematol. 2006;84:23–30. doi: 10.1532/IJH97.06112. [DOI] [PubMed] [Google Scholar]
- Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Jeang KT. Insights into cellular microRNAs and human immunodeficiency virus type 1 (HIV-1) J Cell Physiol. 2008;216:327–331. doi: 10.1002/jcp.21488. [DOI] [PubMed] [Google Scholar]
- Ross SR. Are viruses inhibited by APOBEC3 molecules from their host species? PLoS Pathog. 2009;5:e1000347. doi: 10.1371/journal.ppat.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, et al. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Rosler C, Kock J, Kann M, Malim MH, Blum HE, Baumert TF, et al. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, Gummuluru S, Hu J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol. 2007;81:4465–4472. doi: 10.1128/JVI.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, et al. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei YC, Tian YJ, Ding HH, Wang BJ, Yang Y, Hao YH, et al. N-terminal and C-terminal cytosine deaminase domain of APOBEC3G inhibit hepatitis B virus replication. World J Gastroenterol. 2006;12:7488–7496. doi: 10.3748/wjg.v12.i46.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chertova E, Chen J, Ott DE, Roser JD, Hu WS, et al. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Goila-Gaur R, Miyagi E, Khan MA, Opi S, Takeuchi H, et al. Production of infectious virus and degradation of APOBEC3G are separable functional properties of human immunodeficiency virus type 1 Vif. Virology. 2007;369:329–339. doi: 10.1016/j.virol.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Khan MA, Miyagi E, Strebel K. Differential sensitivity of "old" versus "new" APOBEC3G to human immunodeficiency virus type 1 vif. J Virol. 2009;83:1156–1160. doi: 10.1128/JVI.01734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, et al. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J Virol. 2007;81:13235–13241. doi: 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pery E, Rajendran KS, Brazier AJ, Gabuzda D. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N-terminus of HIV and SIV Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–52. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, He Z, Wang T, Xu R, Yu XF. A patch of positively charged amino acids surrounding the HIV-1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J Virol. 2009;83:8674–82. doi: 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Saadatmand J, Li X, Guo F, Niu M, Jiang J, et al. Function analysis of sequences in human APOBEC3G involved in Vif-mediated degradation. Virology. 2008;370:113–121. doi: 10.1016/j.virol.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, et al. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J Mol Biol. 2009;389:819–832. doi: 10.1016/j.jmb.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A, Nagata T, Matsugami A, Habu Y, Sugiyama R, Hayashi F, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Takeuchi H, Kao S, Khan MA, Miyagi E, Goila-Gaur R, et al. Monomeric APOBEC3G is catalytically active and has antiviral activity. J Virol. 2006;80:4673–4682. doi: 10.1128/JVI.80.10.4673-4682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friew YN, Boyko V, Hu WS, Pathak VK. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Kamata M, Nagaoka Y, Chen IS. Reassessing the role of APOBEC3G in human immunodeficiency virus type 1 infection of quiescent CD4+ T-cells. PLoS Pathog. 2009;5:e1000342. doi: 10.1371/journal.ppat.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Schubert U, Peden K, Strebel K. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J Virol. 1996;70:820–829. doi: 10.1128/jvi.70.2.820-829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J Virol. 1996;70:8285–8300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter GD, Jr, Yamshchikov G, Cohen SJ, Mulligan MJ. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J Virol. 1996;70:2669–2673. doi: 10.1128/jvi.70.4.2669-2673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abada P, Noble B, Cannon PM. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J Virol. 2005;79:3627–3638. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V, Smith RM, Bour SP, Strebel K, Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc Natl Acad Sci USA. 2003;100:15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan MA, Handley MA, Lee YH, Talbot KJ, Harper JW, Panganiban AT. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family. J Virol. 1998;72:5189–5197. doi: 10.1128/jvi.72.6.5189-5197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K, Seharaseyon J, Dong P, Bour S, Marban E. Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Mol Cell. 2004;14:259–267. doi: 10.1016/s1097-2765(04)00183-2. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Van DN, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V, Heimann-Nichols E, Smith RM, Sun Y, Bram RJ, Ali S, et al. Identification of calcium-modulating cyclophilin ligand as a human host restriction to HIV-1 release overcome by Vpu. Nat Med. 2008;14:641–647. doi: 10.1038/nm1778. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bartee E, McCormack A, Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2:e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Andrew AJ, Kao S, Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci USA. 2009;106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa J, Kaisho T, Tomizawa H, Lee BO, Kobune Y, Inazawa J, et al. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics. 1995;26:527–534. doi: 10.1016/0888-7543(95)80171-h. [DOI] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, et al. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol. 2009;83:7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, et al. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Hue S, Schaller T, Verschoor E, Pillay D, Towers GJ. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Inbody SH, Mulcahy ER, et al. Scrambling of the amino acids within the transmembrane domain of Vpu results in a simian-human immunodeficiency virus (SHIVTM) that is less pathogenic for pig-tailed macaques. Virology. 2005;339:56–69. doi: 10.1016/j.virol.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Paul M, Mazumder S, Raja N, Jabbar MA. Mutational analysis of the human immunodeficiency virus type 1 Vpu transmembrane domain that promotes the enhanced release of virus-like particles from the plasma membrane of mammalian cells. J Virol. 1998;72:1270–1279. doi: 10.1128/jvi.72.2.1270-1279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Yamamoto SP, Misawa N, Yoshida T, Miyazawa T, Koyanagi Y. Comparative study on the effect of human BST-2/Tetherin on HIV-1 release in cells of various species. Retrovirology. 2009;6:53. doi: 10.1186/1742-4690-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, et al. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, et al. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J Virol. 2009;83:4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Gers-Huber GLMZMLJPV. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Strebel K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Clouse KA, Strebel K. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol. 1995;69:7699–7711. doi: 10.1128/jvi.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Francica JR, grawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S, Akari H, Miyagi E, Strebel K. Naturally occurring amino acid substitutions in the HIV-2 ROD envelope glycoprotein regulate its ability to augment viral particle release. Virology. 2003;309:85–98. doi: 10.1016/s0042-6822(02)00128-9. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Zhao LJ, Mukherjee S, Leung K, Sheffer D, Joag SV, et al. Chronology of genetic changes in the vpu, env, and Nef genes of chimeric simian-human immunodeficiency virus (strain HXB2) during acquisition of virulence for pig-tailed macaques. Virology. 1998;248:275–283. doi: 10.1006/viro.1998.9300. [DOI] [PubMed] [Google Scholar]
- Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- Terwilliger EF, Cohen EA, Lu YC, Sodroski JG, Haseltine WA. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- Friborg J, Ladha A, Gottlinger H, Haseltine WA, Cohen EA. Functional analysis of the phosphorylation sites on the human immunodeficiency virus type 1 Vpu protein. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:10–22. [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, et al. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Vandegraaff N, Li Y, Gee-Estrada K, Stremlau M, Welikala S, et al. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006;351:404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Lin TY, Emerman M. Determinants of cyclophilin A-dependent TRIM5 alpha restriction against HIV-1. Virology. 2008;379:335–341. doi: 10.1016/j.virol.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari H, Greco G, Luban J. Cyclophilin A peptidyl-prolyl isomerase activity promotes ZPR1 nuclear export. Mol Cell Biol. 2002;22:6993–7003. doi: 10.1128/MCB.22.20.6993-7003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, et al. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity. 2004;21:189–201. doi: 10.1016/j.immuni.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Colgan J, Asmal M, Yu B, Luban J. Cyclophilin A-deficient mice are resistant to immunosuppression by cyclosporine. J Immunol. 2005;174:6030–6038. doi: 10.4049/jimmunol.174.10.6030. [DOI] [PubMed] [Google Scholar]
- Braaten D, Franke EK, Luban J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci USA. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Sayah DM, Luban J. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Sebastian S, Sokolskaja E, Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci USA. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Yang R, Aiken C. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J Virol. 2008;82:12001–12008. doi: 10.1128/JVI.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Sebastian S, Sokolskaja E, Luban J. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J Virol. 2004;78:11739–11750. doi: 10.1128/JVI.78.21.11739-11750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, et al. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Luban J. Selection for loss of Ref1 activity in human cells releases human immunodeficiency virus type 1 from cyclophilin A dependence during infection. J Virol. 2004;78:12066–12070. doi: 10.1128/JVI.78.21.12066-12070.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. J Virol. 2006;80:2855–2862. doi: 10.1128/JVI.80.6.2855-2862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Aiken C. Analysis of human cell heterokaryons demonstrates that target cell restriction of cyclosporine-resistant human immunodeficiency virus type 1 mutants is genetically dominant. J Virol. 2007;81:11946–11956. doi: 10.1128/JVI.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisiak R, Horban A, Gallay P, Bobardt M, Selvarajah S, Wiercinska-Drapalo A, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, et al. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Si Z, Vandegraaff N, O'huigin C, Song B, Yuan W, Xu C, et al. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci USA. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen LM, Keckesova Z, Webb BL, Gifford RJ, Smith TP, Towers GJ. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J Virol. 2006;80:7332–7338. doi: 10.1128/JVI.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J Virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan WA, Schaller T, Ylinen LM, Hosie MJ, Towers GJ, Willett BJ. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J Virol. 2009;83:8270–8275. doi: 10.1128/JVI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro IP, Menezes AN, Moreira MA, Bonvicino CR, Seuanez HN, Soares MA. Evolution of cyclophilin A and TRIMCyp retrotransposition in New World primates. J Virol. 2005;79:14998–15003. doi: 10.1128/JVI.79.23.14998-15003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SJ, Webb BL, Ylinen LM, Verschoor E, Heeney JL, Towers GJ. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoux L, Sebastian S, Sayah DM, Luban J. Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues. J Virol. 2005;79:7883–7888. doi: 10.1128/JVI.79.12.7883-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, Kaul S, et al. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5 alpha and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Dodding MP, Stoye JP. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J Virol. 2006;80:4061–4067. doi: 10.1128/JVI.80.8.4061-4067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Grutter C, de Castillia CS, Pertel T, Olivari S, Grutter MG, et al. An invariant surface patch on the TRIM5alpha PRYSPRY domain is required for retroviral restriction but dispensable for capsid binding. J Virol. 2009;83:3365–3373. doi: 10.1128/JVI.00432-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama EE, Miyoshi H, Nagai Y, Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J Virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube J, Bouchard A, Berthoux L. Both TRIM5alpha and TRIMCyp have only weak antiviral activity in canine D17 cells. Retrovirology. 2007;4:68. doi: 10.1186/1742-4690-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2007;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, et al. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development. 2009;136:965–973. doi: 10.1242/dev.029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Benkirane M, Jeang KT. Small non-coding RNAs, mammalian cells, and viruses: regulatory interactions? Retrovirology. 2007;4:74. doi: 10.1186/1742-4690-4-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- terBrake O, Haasnoot J, Kurreck J, Berkhout B. ESF-EMBO symposium: antiviral applications of RNA interference. Retrovirology. 2008;5:81. doi: 10.1186/1742-4690-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, Kant HJ van de, Bourc'his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kishi A, Yachie N, Kanai A, Tomita M. Computational analysis of microRNA-mediated antiviral defense in humans. FEBS Lett. 2007;581:4603–4610. doi: 10.1016/j.febslet.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4(+) T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L JK. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, et al. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W, Haasnoot J, van der Velden J, van Montfort T, Zorgdrager F, Paxton W, et al. Increased virus replication in mammalian cells by blocking intracellular innate defense responses. Gene Ther. 2008;15:545–552. doi: 10.1038/gt.2008.12. [DOI] [PubMed] [Google Scholar]
- Schnettler E, de Vries W, Hemmes H, Haasnoot J, Kormelink R, Goldbach R, et al. The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep. 2009;10:258–263. doi: 10.1038/embor.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol. 2007;88:2627–2635. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- Westerhout EM, ter BO, Berkhout B. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006;3:57. doi: 10.1186/1742-4690-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de HP, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Zhong X, Yu L, Ding B, de HP, Boris-Lawrie K. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc Natl Acad Sci USA. 2009;106:605–610. doi: 10.1073/pnas.0806822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W, Haasnoot J, Fouchier R, de HP, Berkhout B. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J Gen Virol. 2009;90:1916–1922. doi: 10.1099/vir.0.008284-0. [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhu Y, Bisaro DM, Parris DS. Herpes simplex virus type 1 suppresses RNA-induced gene silencing in mammalian cells. J Virol. 2009;83:6652–6663. doi: 10.1128/JVI.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Yeung ML, de L V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delelis O, Carayon K, Saib A, Deprez E, Mouscadet JF. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A. In-Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology. 2007;4:21. doi: 10.1186/1742-4690-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, et al. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res. 2008;133:250–258. doi: 10.1016/j.virusres.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, et al. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- Bucher E, Hemmes H, de HP, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]