Abstract
Piwi proteins and Piwi-interacting RNAs (piRNAs) are essential for germ cell development, but analysis of the molecular mechanisms of these ribonucleoproteins remains challenging in most animal germ cells. To address this challenge, we systematically characterized Xiwi, a Xenopus Piwi homologue, and piRNAs from Xenopus eggs and oocytes. We used the large size of Xenopus eggs to analyze small RNAs at the single cell level, and find abundant piRNAs and large piRNA clusters in the Xenopus tropicalis genome, some of which resemble the Drosophila piRNA-generating flamenco locus. Although most piRNA clusters are expressed simultaneously in an egg, individual frogs show distinct profiles of cluster expression. Xiwi is associated with microtubules and the meiotic spindle, and is localized to the germ plasm—a cytoplasmic determinant of germ cell formation. Xiwi associates with translational regulators in an RNA-dependent manner, but Xenopus tudor interacts with Xiwi independently of RNA. Our study adds insight to piRNA transcription regulation by showing that individual animals can have differential piRNA expression profiles. We suggest that in addition to regulating transposable elements, Xiwi may function in specifying RNA localization in vertebrate oocytes.
Keywords: piRNA, Piwi, Xenopus
Introduction
Proper germ cell development is critical for fertility, and thus for the survival of an organism's lineage. Recently, the RNA interference (RNAi) pathway has emerged as an important component of germ cell development. RNAi is a transcript and chromatin regulatory pathway in eukaryotes controlled by Argonaute and Piwi proteins, and the small regulatory RNAs bound by these proteins. In animals, Argonaute proteins are present in all cell types and bind microRNAs (miRNAs) and endogenous small interfering RNAs (endo siRNAs)(Okamura and Lai, 2008). In vertebrates and ecdysozoan invertebrates, Piwi proteins are expressed in germ line tissue, and bind a separate class of small RNAs defined by their interaction with Piwi (piRNAs) (Aravin et al, 2006; Girard et al, 2006; Lau et al, 2006; Saito et al, 2006; Vagin et al, 2006; Brennecke et al, 2007).
Fruit flies, mice, and fish with mutated Piwi genes lack piRNAs and are sterile, with some mutants inducing apoptosis of developing germ cells (Cox et al, 1998; Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004; Carmell et al, 2007; Houwing et al, 2007; Unhavaithaya et al, 2008). Genetic studies of Piwi-pathway mutants implicate activation of transposable elements (TEs), which might cause genome damage, as a potential cause of apoptosis (Vagin et al, 2006; Brennecke et al, 2007; Aravin et al, 2007b; Klattenhoff and Theurkauf, 2008). Although the mechanism for Piwi proteins silencing TEs is not fully understood, it is hypothesized to involve an RNA cleavage event (Lau et al, 2006; Saito et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007) or interactions with chromatin that can result in DNA methylation (Carmell et al, 2007; Aravin et al, 2008; Kuramochi-Miyagawa et al, 2008) (mice) or covalent histone modifications (Klenov et al, 2007; Yin and Lin, 2007)(fruit flies).
Key to dissecting the mechanistic function of Piwi proteins and piRNAs is a biochemically tractable model system. However, in vitro culture systems that are amenable to biochemistry have not yet been developed for germline tissues. The African clawed frogs, Xenopus laevis and Xenopus tropicalis, are prominent model organisms for studying developmental and biochemical processes because their eggs are large and plentiful, and egg extracts are noted for their ability to recapitulate many cell-cycle processes in vitro (Desai et al, 1999). We reasoned that Xenopus eggs would express Piwi proteins and piRNAs, and explored the feasibility of using Xenopus as a system to analyze Piwi and piRNA function.
Here we report a systematic study of Xenopus Piwi (Xiwi) and X. tropicalis egg small RNAs. We examine Xiwi abundance, localization, and association with new proteins, messenger RNAs, and cytoskeleton components. A fundamental question in piRNA biology concerns the uniformity of piRNA production in different cells. The large size of Xenopus eggs allowed us to carry out deep sequencing analysis from single eggs, thereby allowing a comparison of a single cell population to an overall population, and showing that single cells differ in their potential to produce piRNAs. Our data also suggest possible roles for Xiwi in translation regulation and RNA transport, thus expanding Piwi protein function beyond TE silencing.
Results
Identification of a piRNA-binding protein expressed in Xenopus germ cells
We identified three Xenopus members of the Piwi family through gene models present in the X. tropicalis genome in the Ensembl database version 52 (see Materials and methods) and raised antibodies against Xiwi, the protein most closely related to Drosophila Piwi. On carrying out western blots of egg extracts prepared from mature X. laevis and X. tropicalis eggs, this antibody recognized a single protein of the predicted molecular weight of 100 kDa (Figure 1A) and shows negligible cross reactivity with the closely related Xili protein. A recent study (Wilczynska et al, 2009) indicated that there are two paralogues of Xiwi (1a and 1b). RT–PCR analysis showed that both Xiwis are expressed throughout oogenesis (data not shown), and given the high sequence identity (79%) between these two proteins, we expect that our antibody will react with both proteins. Radioactive end-labeling of Xiwi-associated small RNAs revealed a range of 24–31 nt small RNAs from immunoprecipitations (IPs) with this antibody from egg extracts, confirming that this antibody recognized Xiwi (Supplementary Figure 1A).
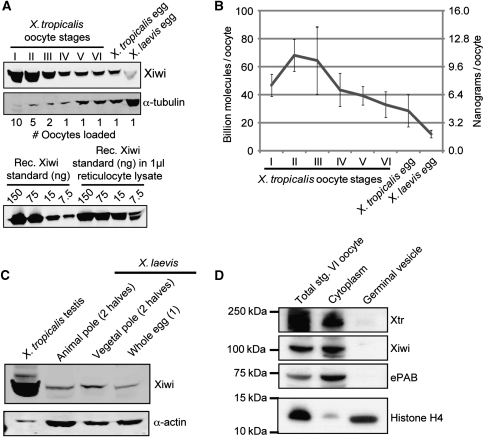
Figure 1.
Xiwi expression during Xenopus oogenesis. (A) Quantitative western blot of Xiwi in staged X. tropicalis oocytes and X. tropicalis and X. laevis eggs. Each lane contains the indicated number of oocytes or eggs. Expression levels were calculated from standard curves of the recombinant Xiwi fragment used to generate the antibody. Our antibody shows lower affinity for Xiwi from X. laevis (data not shown), which explains the apparent lower levels of Xiwi in X. laevis egg extract. (B) Quantification of Xiwi expression levels from quantitative western blots carried out in triplicate. (C) Xiwi was detected in X. tropicalis testes extracts or X. laevis eggs that were manually dissected into animal and vegetal halves. Xiwi concentration in testes extracts was comparable with that observed in stage I oocytes, and approximately equal amounts of Xiwi were observed in the animal and vegetal halves of a X. laevis egg. (D) Stage VI X. laevis oocytes were manually dissected to separate the germinal vesicle from the cytoplasm and the resulting fractions were probed for Xiwi, Xtr, ePAB (as a control for cytoplasm), and histone H4 (as a control for nuclear protein).
Using quantitative western blots, we determined that Xiwi is most abundantly expressed in testes and early stage oocytes (Figure 1), whereas the concentration of piRNAs continues to increase throughout oogenesis (Supplementary Figure S7). In X. tropicalis egg extracts Xiwi concentration is ∼300 nM. In fact, X. tropicalis oocytes contain >20–40 billion Xiwi molecules, which is consistent with a recent report of Xiwi expression in X. laevis oocytes (Wilczynska et al, 2009). This shows that Xiwi (∼6 ng per stage VI oocyte) is a moderately abundant RNA-binding protein compared with known RNA-binding proteins ePAB (50 ng per stage VI oocyte) (Voeltz et al, 2001) and Staufen (2 ng per stage VI oocyte) (Allison et al, 2004).
In Drosophila, Piwi is localized to the nucleus during oogenesis (Cox et al, 1998; Saito et al, 2006; Brennecke et al, 2007). To determine whether Xiwi is localized to the germinal vesicle (GV or nucleus) during Xenopus oogenesis, we manually separated the GV and cytoplasm, and examined Xiwi distribution by western blotting. In contrast to the nuclear localization of Drosophila Piwi, Xiwi is predominately localized to the cytoplasm, similar to embryonic poly-A-binding protein (ePAB), whereas a known nuclear protein—histone H4—is found exclusively in the nuclear fraction (Figure 1D). We conclude that Xiwi is expressed in both male and female germ cells and that Xiwi is localized to the cytoplasm during oogenesis.
Xiwi is a microtubule-associated protein that localizes to the germ plasm
To examine the localization of Xiwi, we carried out whole mount immunofluorescence and confocal microscopy on oocytes, eggs, and early embryos (see Ferrell, 1999; King et al, 2005) for descriptions on the stages of Xenopus oogenesis). In stage I oocytes, Xiwi was distributed diffusely throughout the cytoplasm but co-localized with Grp94 in the Balbiani body (Chang et al, 2004), an optically dense organelle containing mitochondria, endoplasmic reticulum, and many mRNAs, (Kloc and Etkin, 2005) (Figure 2A). In stage III–VI oocytes, Xiwi condenses into a thin line at the vegetal cortex of the oocyte (Figure 2B). Mature eggs show two distinct localizations of Xiwi: first, it is localized to punctate granules at the vegetal cortex of the egg and, second, Xiwi localized to the meiotic spindle in the animal cortex of the egg (Figure 2C) similar to the localization of Seawi in sea urchin embryos (Rodriguez et al, 2005). Consistent with this observed localization to both poles of the egg, we found that there were equal amounts of Xiwi present in manually hemisected eggs (Figure 1C). Finally, in early embryos Xiwi staining is lost from the mitotic spindles and localizes exclusively to germ plasm islands that become restricted to a few blastomeres during the course of development (Figure 2E). Our data suggest that Xiwi and, potentially, piRNAs are additional components of the germ plasm, consistent with the localization of Piwi proteins in other organisms (Klattenhoff and Theurkauf, 2008).
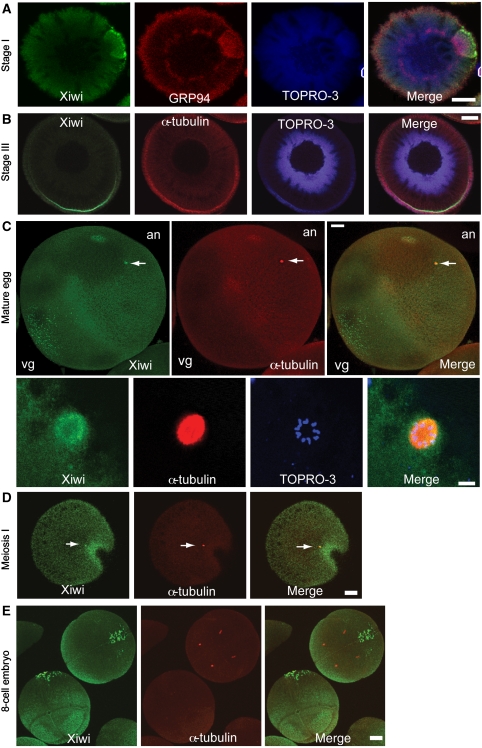
Figure 2.
Xiwi localizes to germ plasm and microtubules throughout Xenopus oogenesis. (A, B) Xiwi was detected by immunofluorescence in X. laevis stage I (A) or stage III (B) eggs, and was compared with the localization of α-tubulin or GRP-94, a marker for the Balbiani body. (C) Xiwi was detected by immunofluorescence in mature meiosis II-arrested X. laevis eggs and observed to localize to the granules in the vegetal (Vg) pole of the egg and to co-localize with α-tubulin at the meiosis II spindle in the animal pole (An) of the egg. Higher magnification view of Xiwi localization to the meiotic spindle is shown. (D) Stage VI X. laevis eggs were matured in vitro using progesterone and Xiwi localization was examined during meiosis I. (E) X. tropicalis 8-cell stage embryos were stained for Xiwi and α-tubulin. Xiwi was found to localize predominately to germ plasm islands. TOPRO-3 was used to stain nucleic acids. Scale bars are 100 μm in all panels except (C) lower panel, which is 10 μm. Arrows point specifically to localization of Xiwi and α-tubulin in the meiotic spindle.
To determine whether Xiwi associated with both the meiosis I and meiosis II spindles, we matured oocytes in vitro with progesterone (which initiates GV breakdown and spindle assembly), and observed Xiwi localization to the meiosis I spindle (Figure 2D). We also detected significant amounts of Xiwi (∼3% of the cellular total) specifically associating with microtubules purified from egg extracts (Supplementary Figure 2). We did not, however, observe any effect on in vitro mitotic spindle assembly following Xiwi immunodepletion (Supplementary Figure S2), which suggests Xiwi may be a passive cargo on microtubules as observed for mRNAs localized to the germ plasm during oogenesis.
piRNAs are the predominant class of small RNAs in Xenopus eggs and oocytes
We employed deep sequencing technology to identify small RNAs from: (1) egg extract, (2) purified microtubules and (3) immunoprecipitations (IP) of Xiwi from both X. laevis and X. tropicalis egg extracts; and (4) single eggs laid by two different female X. tropicalis. After quality filtering (see Materials and methods), these libraries ranged from 5215–702 590 filtered reads (Supplementary Table S1). The size distribution of RNAs from X. tropicalis and X. laevis egg extracts, microtubules, and Xiwi IPs were unimodal over a principal range of 26–31 nt (Figure 3A). This distribution is consistent with piRNA lengths from other animals, and indicates that the predominant class of small RNAs in Xenopus egg extract and associated with microtubules is piRNA.
Figure 3.
Xenopus piRNA genomics. (A) Size distribution of small RNA reads from Xiwi IP libraries sequenced on the Illumina GA-II (left), and microtubule and egg extract libraries from Xenopus laevis and Xenopus tropicalis sequenced on the 454 Life Science GA-FLX (right). (B) Statistics and annotations of three select small RNA libraries. The complete list of statistics and annotations are detailed in Supplementary Table S1. (C) X. tropicalis piRNA partial cluster maps from the three scaffolds with the greatest number of uniquely mapping reads. Normalized reads are superimposed, and positive and negative reads refer to plus strand and minus strand matches, respectively. Gap, gene and TE annotations were selected from the UCSC genome browser for X. tropicalis genome build 4.1, using the Xenrep5 database to plot the TE annotations (Hinrichs et al, 2006).
X. tropicalis small RNAs were further analyzed by mapping perfect matches to the genome and to specific X. tropicalis databases of: (1) TEs from Repbase (Jurka et al, 2005), (2) mRNAs and ESTs from the UniGene collection (Wheeler et al, 2005), and (3) miRNAs from miRBASE (Griffiths-Jones et al, 2008). On average, ∼64% of filtered reads could be mapped to the genome, ∼18% of filtered reads mapped to TEs, whereas mRNAs/ESTs matches comprised ∼17% of reads (Figure 3B and Supplementary Table S1). Only the X. tropicalis genome has been sequenced (JGI, 2004), so we analyzed X. laevis small RNAs in specific X. laevis databases similar to the ones described above.
The annotation metrics of these frog egg small RNA libraries are similar to rodent testes small RNA libraries (Girard et al, 2006; Lau et al, 2006), and yet X. tropicalis eggs contain relatively few miRNAs and endo siRNAs, which contrasts with the small RNA composition in mammalian eggs (Tam et al, 2008; Watanabe et al, 2008). Our small RNA isolation and sequencing procedure should capture miRNAs (Lau et al, 2006), and yet miRNAs represent, at most, 1.5% of total reads in the X. laevis egg extract or averaging ∼0.37% of total reads amongst the Xenopus libraries (Supplementary Table S4). Mouse and Drosophila oocytes contain a significant population of endo siRNAs, which are 21–22 nt long and derive from an endogenous dsRNA intermediate (Okamura and Lai, 2008). Genomic mapping and annotation of the Xenopus small RNAs also failed to reveal strong examples of endo siRNAs (data not shown). From this analysis, we conclude that piRNAs are the most abundant population of small RNAs in Xenopus female germ cells.
Similar to piRNAs from mammals, fish, and fruit flies (Lau et al, 2006; Houwing et al, 2007; Yin and Lin, 2007; Brennecke et al, 2008), retrotranposons are the major TE class targeted by Xenopus TE-associated piRNAs (>59%), and these piRNAs show a pronounced antisense match bias against TEs (>80%, Supplementary Table S2). This antisense bias is evident in Drosophila piRNAs (Saito et al, 2006; Vagin et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007), and has also been observed in zebrafish and mammalian piRNAs targeting retrotransposons (Girard et al, 2006; Grivna et al, 2006; Lau et al, 2006; Houwing et al, 2007; Aravin et al, 2007b). DNA transposons comprise a minority of the genome's TEs in fruit flies and mammals, but are more abundantly represented in Xenopus and zebrafish genomes (Jurka et al, 2005). Accordingly, the proportion of Xenopus TE-associated piRNAs targeting DNA transposons is greater than adult mammalian TE-associated piRNAs (37 versus 5%, Supplementary Table S2).
DNA transposons are thought to transpose through a ‘cut-and-paste' mechanism that does not rely on an RNA intermediate like retrotransposons (Jurka et al, 2007). Xenopus piRNAs may also be suppressing DNA transposons at the post-transcriptional level, because we observe similar numbers of piRNAs antisense to a DNA transposon, such as the Polinton-1 element, compared with a non-LTR retrotransposon, L1–55 (∼19K reads against L1–55, ∼12K reads against Polinton-1) (Supplementary Figure S3). Intriguingly, one DNA transposon, the Harbinger-2 element, contains a significant number of sense reads (Supplementary Table 3), but these reads are actually complementary to a Myb-like DNA-binding protein transcript coded by the antisense strand of the TE (Supplementary Figure S3C). The piRNAs mapping antisense to Harbinger-2 may target the tranposase transcript, and do not seem to overlap with the sense piRNAs targeting the Myb-like DNA-binding protein transcript. Thus, the Xenopus Piwi pathway may have the capacity to target the mRNAs arising from DNA transposons, but how the pathway discriminates TE-associated mRNAs from the bulk of other mRNAs is unclear.
Out of the 159 DNA transposons and 95 non-LTR retrotransposon families that have been defined in the Xenopus genome (Jurka et al, 2005), only specific elements are highly represented in piRNAs (Supplementary Table S3), reflecting either greater activity of these elements or propensity of relics of these TEs to reside in piRNA cluster transcription units. As a class, mammalian adult piRNAs are under-represented in TE annotations compared with the total TE content of mammalian genomes, whereas the composition of TE-derived piRNAs in zebrafish is over-represented (O'Donnell and Boeke, 2007). In this spectrum, Xenopus piRNAs as a class are similar to mammals in terms of TE annotation, however, the majority of Xenopus piRNAs (∼70%) map to multiple genomic loci whereas two-thirds of mammalian piRNAs map uniquely (Supplementary Table S1).
The ‘ping-pong' model of piRNA biogenesis is denoted by congruent pairs of piRNAs with 10 basepairs of homology in the 5′ portion of the piRNAs and a sequence composition biased for uridine at the first nucleotide of one piRNA (5′U), followed by adenine at the 10th nt of the other piRNA (A10) (Brennecke et al, 2007; Gunawardane et al, 2007). To search for ping-pong model signatures in the Xenopus egg small RNA libraries, we examined the base compositions of reads apportioned according to individual lengths between 18–31 nt, in groups of 18–23 nt and 24–31 nt, and in groups of reads that either starts with a 5′U or some other base (Supplementary Figure S4). As expected from studies in other animals, the strongest base bias observed in Xenopus libraries is a 5′U. The A10 bias was more apparent in X. laevis libraries than X. tropicalis, especially among reads that do not begin with U; only microtubule-associated small RNAs in X. tropicalis showed a modest A10 bias. Indeed, when examining X. tropicalis reads mapped to the genome, the microtubule-associated small RNA library had the highest number of read pairs that overlap at 10 nt, whereas ‘ping-pong' signatures were notably absent from the Xiwi IP from X. tropicalis (see Supplementary Figure S5 and Materials and methods section for details on ping-pong analysis). In summary, we detect evidence for ‘ping-pong' piRNA amplification in Xenopus egg small RNA libraries, and perhaps the molecules involved in this mechanism are enriched on microtubules.
Some X. tropicalis piRNA clusters resemble the Drosophila flamenco locus
As the majority of X. tropicalis piRNAs map to multiple genomic loci, we defined piRNA clusters by scanning for regions containing at least 10 uniquely mapping piRNA reads within 10 kb. This algorithm yielded 645 loci that we consider as ‘partial' piRNA clusters (Supplementary Table S5), because the X. tropicalis genome is unassembled beyond scaffolds. The publicly available build 4.1 consists of 19 758 scaffolds that range from 2 kb to 7.8 Mb in length. The majority of the ‘partial' piRNA clusters extend to the end of a scaffold (Figure 3C), and as scaffolds are not ordered against a physical map, it is likely that several of these partial clusters comprise single larger clusters. Although the piRNA cluster on scaffold 166 is limited on one side by the end of the scaffold, this cluster still spans 420 kb, making it one of the largest piRNA clusters identified. Nearly all the piRNAs from scaffold 166 arise from the plus strand, though a minority map to the minus strand. At least 10 Xenopus partial piRNA clusters with over 1000 uniquely mapping reads span 90 kb or more.
Neither extensive strand polarity switching, as observed in zebrafish, nor bi-directional cluster configurations similar to mammals were observed in Xenopus, as practically all clusters were composed of piRNAs mapping to one strand. Only one partial cluster on scaffold 34 shows possible strand switching based on normalized reads, but the uniquely mapping reads are strongly biased for one strand (Supplementary Figure S3D). Perhaps future assembly of scaffolds onto ordered X. tropicalis chromosomes will reveal bi-directional cluster configurations; however, the configuration of piRNAs mapped to the Harbinger-2 element can be interpreted as a possible bi-directional cluster (Supplementary Figure S3C).
We examined the composition of TEs in X. tropicalis piRNA clusters and found that TEs were not over-represented in piRNA clusters compared with the rest of the genome (data not shown). However, we noticed a strand bias for TE orientation in many of the clusters with the most abundant piRNAs. For example, TE strand orientation was biased for the minus strand in the regions of the piRNA clusters on scaffolds 166 and 871 (Figure 3C), but was more evenly distributed on both genomic strands outside the piRNA clusters. As the piRNAs seem to derive predominantly from the plus strand, the minus-strand bias of TEs residing in these clusters could explain the propensity of piRNAs to be complementary to TEs. This configuration of TEs in a vertebrate piRNA cluster is highly reminiscent of the flamenco locus in Drosophilids, which produce abundant primary piRNAs in a strand-biased manner, and is rife with TEs that are predominantly oriented in the genomic strand that is complementary to the piRNAs (Brennecke et al, 2007; Ghildiyal et al, 2008; Malone et al, 2009). All together, ∼60% of the uniquely mapping X. tropicalis piRNAs and ∼30% of clusters reside in loci with pronounced TE-strand bias, whereas other piRNAs and clusters tend to reside in unannotated, intergenic regions.
Single cell analysis of Xenopus egg small RNAs
How are the numerous piRNA clusters transcriptionally regulated, given that a large diversity of piRNAs can be generated from any single cluster? One model could be that piRNA cluster expression is static in any single germ cell but stochastic between germ cells, which would appear as a diversity of piRNA clusters being expressed within a cell population. An alternative model is that germ cells transcribe many clusters simultaneously so that all cells contain the same piRNA content. This is an interesting biological question because animal cells are known to express single genes from multi-copy gene families, such as odorant receptors or immunoglobulins, and little is known with regard to how piRNA expression is controlled. As clonal expansion of germ cells is not yet possible, and it is difficult to isolate and clone small RNAs from single cells, Xenopus eggs are a unique system to examine total small RNA content at a single cell level because of their ease of isolation and abundance of maternal RNAs. To accomplish this, we constructed small RNA libraries from four eggs obtained from two individual X. tropicalis, and sequenced all four libraries together in a multiplexed flow cell lane, along with one library in a single flow cell lane (Figure 4A). Bar codes allowed us to parse out the individual libraries, which generally showed similar read size distributions (Figure 4B).
Figure 4.
Single cell small RNA deep sequencing reveals differential piRNA cluster expression (A). Experimental scheme for multiplex sequencing of small RNA libraries from single X. tropicalis eggs. (B) Size distribution of reads from single-egg small RNA libraries. (C) Pairwise correlation comparison of genome-wide small RNA profiles in which the larger library is normalized to the size of the smaller library. Small RNA profiles measured the polarity and normalized number of uniquely mapping reads in 2 kb windows (see Materials and methods). (D) Partial clusters with similar or different expression between single frog eggs. The cluster on scaffold 1403 has similar normalized expression levels between eggs of frog A (marked with arrows) and frog B. Clusters on scaffold 2356 and 5393 are more highly expressed in eggs from frog B, whereas clusters on scaffold 199 and 166 are more highly expressed in frog A.
Although our sequencing depth and overlap in reads was modest for these X. tropicalis libraries (Supplementary Table S1 and Figure S5B), we were still able to compare the similarity of libraries by considering all uniquely mapping reads and their read frequencies normalized to the smallest size library. By analyzing in aggregate, the read frequencies matched to 2 kb window genomic coordinates, we were able to compute a Pearson correlation coefficient for pairs of libraries. This method allowed us to assess the robustness of multiplexed small RNA sequencing and technical replicates. We observed a strong correlation (R2=0.98) between the genome-wide profiles of reads from frog egg B1 that were sequenced in one flow cell lane and in the multiplexed pool (Figure 4C); analysis of partial piRNA cluster representation indicated all clusters from the smaller library were present in the larger library (Supplementary Figure S5C). Therefore, we conclude that these libraries and methods allow for comparison of the genome-wide profile of piRNA content at a single cell level.
To assess the small RNA content of individual cells from different females, we compared the genome-wide profiles of piRNA content between pairs of small RNA libraries. Pairwise comparisons indicated that eggs from an individual were much more similar than eggs from different individuals (e.g. egg B1 versus egg B2: R2=0.96; egg B1 versus egg A3: R2=0.37). We extended this analysis to our multiple-egg libraries to determine whether this correlation held true. As the Xiwi IP was obtained from egg extract derived from several different frogs, its correlation with other libraries was lower, though a high correlation was observed between the microtubule and egg extract libraries, likely due to the fact that the same extract was used to prepare both libraries (Figure 4C). Despite lower correlation between small RNA libraries coming from different frogs, all X. tropicalis libraries share expression of several partial piRNA clusters (Supplementary Figure S5C). For example, the numbers of uniquely mapping reads from the partial piRNA cluster on scaffold 1403 are similar in all four egg libraries. However, there are marked differences in uniquely mapping read frequencies at other clusters between eggs of frog A versus frog B (Figure 4D). Although these frogs are clearly non-isogenic, we hypothesize that the variation in cluster expression levels is due to transcription or processing regulatory differences rather than gross genomic DNA diversity, because differentially low expressing clusters are not devoid of reads. In summary, our data argues that germ cells in an individual will express multiple piRNA clusters concurrently, but germ cells of distinct individuals can show differential expression of certain piRNA clusters.
Xiwi associates with a Tudor protein and factors involved in translation and mRNA metabolism
Genetics and piRNA genomics have implicated Piwi proteins in transposon silencing (Aravin et al, 2007a), however, most piRNAs in adult vertebrates do not seem to target transposons, implying that additional Piwi functions outside of TE regulation might exist. To explore this idea, we IPed Xiwi from egg extracts to identify interacting factors. We pursued two general approaches to identify Xiwi interacting proteins: use of a native IP (Figure 5A), and IP after mild formaldehyde cross-linking (0.1%) to stabilize protein–protein and protein–RNA interactions (Figure 5B). Several proteins consistently co-IPed with Xiwi using both approaches. From the native IP, we identified ePAB, Vera/Vg1RBP, Rap55, the RNA-binding protein/transcription factor CBTF122, and numerous heat shock proteins. From cross-linked extracts, we identified the RNA-binding protein symplekin, small ribosomal subunits, and the Xenopus Tudor protein, Xtr (Hiyoshi et al, 2005).
Figure 5.
Xiwi associates with proteins involved in mRNA translation. (A) Xiwi was immunoprecipitated (IPed) from X. laevis egg extracts and separated by SDS–PAGE. Associated proteins were excised from the gel and identified by mass spectrometry. Protein identities of the top hits are indicated next to the excised band. (B) X. laevis egg extract was cross-linked by the addition of formaldehyde. Xiwi was then IPed, and associated proteins were identified by mass spectrometry. The top hits are indicated next to each band. (C) Xiwi-associated proteins were IPed from X. laevis egg extracts and blotted for the presence of Xiwi. For the ePAB experiment, recombinant GFP–ePAB was added to extracts and immunoprecipitated using anti-GFP antibodies. Anti-Xiwi antibodies cross react with both GFP–ePAB and GFP because the Xiwi antigen containing sequence present in the pET30 expression vector that is present in GFP–ePAB and GFP. (D) RNA was purified from Xiwi IPs and separated on an agarose gel. Xiwi IPs were compared with control IgG IPs. Size markers indicated are DNA markers. (E) Xiwi was IPed from X. laevis egg extracts and the IPs were washed with or without RNaseA to digest associated RNAs, then blotted for associated proteins.
To validate observed interactions, we carried out reciprocal native co-IPs with antibodies against Xtr, symplekin, and GFP–ePAB. Indeed, Xiwi specifically co-IPed with each of these factors (Figure 5C). Many of the Xiwi-associated proteins we identified are mRNA-binding proteins. To determine whether Xiwi IPs contain RNAs other than piRNAs, we purified and resolved the RNAs from an IP of Xiwi on an agarose gel. Long RNAs (>100 bp) co-precipitate with Xiwi, but not with control IgG (Figure 5D). We generated a random primed cDNA library from Xiwi immunopreciptated long RNAs from X. laevis and X. tropicalis extracts. Our small scale sequencing of cDNA clones yielded 87 and 72 matches to sequence databases from X. laevis and X. tropicalis, respectively, and 89% and 75% of those respective matches were to mRNAs (Supplementary Table S6), suggesting that mRNA association with xPiwi is a conserved property.
To examine the relationship between Xenopus piRNAs and mRNAs, we looked for perfect matches between sequenced piRNAs, and mRNAs and ESTs in the UniGene databases. Among the 562 X. tropicalis transcripts with ⩾100 mapping piRNAs, 245 and 217 transcripts showed a pronounced sense or antisense bias in piRNAs, respectively. The nature of piRNAs deriving from the sense strand of mRNAs is not clear, as these piRNAs were not detected to be obvious partners in ‘ping-pong' amplification (data not shown). However, mRNAs with the antisense piRNA matches might possibly be regulatory targets, such as a RAB11 family-interacting protein-4 gene and an arachidonate 5-lipoxygenase-activating protein in X. tropicalis; or the Xenopus posterior (Xpo) gene and a mesendoderm nuclear factor (menF) gene from X. laevis, which are annotated genes with the most number of antisense piRNA matches. Out of the mRNAs that might be targeted by piRNAs, 6.5% (37/562) are conserved between the two Xenopus species, although this is likely to be an underestimate because of the unfinished state of the genome sequence in both species. Interestingly, one of these conserved putative targets is a homologue of an antigen of the CD81 antibody, which inhibits proliferation. Curiously, cloned mRNAs did not significantly surface amongst mRNAs with >100 perfect piRNA matches, suggesting that rules governing mRNA and Piwi protein association is not apparent.
To test whether some proteins are interacting with Xiwi through a shared interaction with mRNAs, we treated Xiwi IPs with RNaseA to digest associated RNAs and probed these IPs for ePAB, symplekin, CBTF122, and Xtr. We found that the interaction of Xiwi with ePAB, symplekin, and CBTF122 required RNA, whereas the Xiwi interaction with Xtr was RNA independent (Figure 5E). This shows that Xiwi is present in a complex with RNA-binding proteins and mRNAs, and suggests that Xiwi may regulate the translation of a subset of mRNAs in the egg.
Xiwi and piRNAs do not appear to regulate translation in meiosis II-arrested CSF extracts
The interaction of Xiwi with many proteins known to regulate translation, and the suggestion that Miwi and Mili modulate the translation of mRNAs in spermatogonia (Deng and Lin, 2002; Grivna et al, 2006; Unhavaithaya et al, 2008) suggested that Xiwi might regulate translation in egg extracts. To test whether Xiwi regulated mRNA translation, we designed a series of luciferase reporter constructs containing piRNA-binding sites in the 3′UTR of Renilla luciferase. We designed two types of constructs, first we cloned pieces of the MenF cDNA—which generates hundreds of piRNAs in both sense and antisense orientation—and second, synthetic piRNA-binding sites for the same piRNA and corresponding mismatch mutant reporters. We incubated the mRNAs in CSF extract with firefly luciferase mRNA as a normalization control. mRNAs containing piRNA-binding sites were always expressed at a higher level than mRNAs containing miRNA-binding sites, or no binding sites (Supplementary Figure S6A).
To determine whether Xiwi was responsible for the observed translational upregulation of piRNA reporters, we immunodepleted Xiwi from CSF extracts and measured luciferase activity of each of the reporters compared with control immunodepetions. We found that there was a very modest, but statistically insignificant, upregulation of piRNA reporter constructs in the absence of Xiwi (Supplementary Figure S6B). The lack of effect of Xiwi depletion on luciferase reporter activity could show that Xiwi does not regulate the translation of target mRNAs or that there are redundant factors (such as Xili or xPiwi3) that are also required for translational regulation.
To determine whether Xiwi regulates the translation of proteins from endogenous mRNAs, we immunodepleted Xiwi from CSF extracts and labelled translation using biotin lysine tRNA. We found that Xiwi depletion had no global effect on the translation of endogenous proteins in CSF extract (Supplementary Figure 6D) or on the translation of a specific mRNA, XMAP215, which we found in Xiwi IPs (Supplementary Figure 6E). From these results, we conclude that piRNA-binding sites in the 3′UTR of an mRNA does not confer translational repression as seen for miRNAs, and that Xiwi does not globally regulate translation in CSF extracts.
Figure 6.
Xiwi co-localizes with Xtr in the germ plasm. Xiwi localization was examined during X. laevis oogenesis and compared with the localization of the xPiwi-associated protein Xtr. (A) In stage I oocytes, Xiwi was observed to localize to the Balbiani body at which it co-localized with Xtr. (B) In stage III oocytes Xiwi localization coalesced into a thin ribbon at the vegetal cortex of the oocytes in which it also co-localized with Xtr. In stage VI and mature eggs, Xiwi localized to granular structures present in the vegetal pole of the egg, reminiscent of the germ plasm. At no stage during oogenesis did we observe Xiwi localization to the nucleus as judged by a lack of co-staining with histone H4. Scale bars are 100 μm in A and B and 10 μm in (C).
A previous study indicated that Miwi is able to cleave a target complementary to an abundant piRNA (Lau et al, 2006), and sequence signatures of piRNAs suggest that RNA cleavage by piwi proteins is important for piRNA generation. To determine whether Xenopus egg extracts are capable of recapitulating piRNA-mediated RNA cleavage, we incubated 32P body-labelled luciferase RNA constructs in control or Xiwi-depleted extracts. We found that Renilla luciferase and Renilla luciferase with a non-specific Xenopus 3′UTR were stable and that their stability was unaffected by Xiwi depletion (Supplementary Figure 6C). In contrast, we found that both Renilla constructs containing piRNA-binding sites from MenF were cleaved to an RNA species of approximately the size of Renilla luciferase alone. Interestingly, we found that the cleavage of Renilla MenFas construct was dependent on the presence of Xiwi, as cleavage was reduced ∼50% in the absence of Xiwi. Renilla MenFs also showed a weak, but reproducible Xiwi dependence for cleavage (Supplementary Figure 6C). We found that Renilla MenFas RNA bound strongly to Xiwi, Renilla MenFs RNA bound weakly and we could detect no interaction with Xiwi and Renilla (Supplementary Figure 6F). We were unable to detect any piRNAs loaded into Xiwi from these cleavage reactions (data not shown). We conclude that Xenopus egg extracts recapitulate some aspects of piRNA-mediated RNA cleavage, but that cleavage of reporter RNAs does not affect their translational activity. Clearly, much more work will be required to investigate the structure of the 5′ and 3′ ends generated by Xiwi cleavage and to determine how cleavage events lead to loading of piRNAs into Piwi proteins.
Xiwi and Xtr co-localize throughout oogenesis
The observation that Xiwi and Xtr interact in an mRNA-independent manner suggested that they act together in a complex. To investigate the Xtr and Xiwi interaction in situ, we stained staged oocytes for Xiwi and Xtr, and examined the localization using confocal microscopy. We found that Xiwi co-localized with Xtr in the vegetal pole and germ plasm of all stages of oocytes and eggs (Figure 6A–C). Furthermore, we found that ∼5% of Xtr co-purifies with microtubules in egg extract, similar to Xiwi (Supplementary Figure 2B) and that Xtr is exclusively localized to the cytoplasm in stage VI oocytes (Figure 1D). We conclude that Xiwi and Xtr form a complex that localizes to the germ plasm in which it may regulate the localization or translation of maternal mRNAs. Our results are consistent with two recent studies of Tudor domain proteins interacting with mouse Piwi homologues (Vasileva et al, 2009; Wang et al, 2009) and earlier observations that Drosophila Tudor interacts with Aubergine, a Piwi homologue (Thomson et al, 2008).
Discussion
Xenopus oocytes provide an important system for understanding piRNA biogenesis and molecular function. The large size of the oocyte allowed the sequencing of piRNAs from individual cells, yielding results that suggest differential piRNA profiles can exist in eggs from different individual vertebrates. We find that Xiwi associates with the meiotic microtubules, raising the possibility for a functional role in RNA transport or meiosis in addition to previously proposed roles in TE regulation. These results establish the usefulness of Xenopus as a model system for cell biological and biochemical analysis of the Piwi pathway in vertebrates.
Could Xiwi modulate mRNA function through localization to the germ plasm?
In many animals, germ cells are specified during oogenesis through the localization of maternal proteins and mRNAs to a specialized cytoplasm, termed the germ plasm (Saffman and Lasko, 1999). Our observation that Xiwi localizes to the germ plasm in Xenopus oocytes, eggs, and embryos is consistent with observations of Piwi proteins in germ cell organelles like nuage in fruit flies and nematodes (Brennecke et al, 2007; Batista et al, 2008) or the chromatoid body in mice (Kotaja et al, 2006).
Many germ plasm-localized mRNAs move to the vegetal cortex in a microtubule-dependent manner during oogenesis (Kloc and Etkin, 2005). Our observation that Xiwi and piRNAs associate with purified microtubules and the meiotic spindle, coupled with a lack of effect of Xiwi depletion on spindle assembly, suggests that Xiwi is transported on microtubules where it may regulate piRNA processing or mRNA localization. Several recent studies have identified interactions between small RNA gene regulation and intracellular transport on the cytoskeleton (Rodriguez et al, 2005; Kotaja et al, 2006; Parry et al, 2007; Brodersen et al, 2008), suggesting that there is dynamic interplay between the microtubule cytoskeleton, and small RNA processing and regulation. A role for the microtubule cytoskeleton in piRNA generation is supported by the ping-pong piRNA signatures detected from purified microtubules. Perhaps the Xenopus homologue of Drosophila AGO3 is enriched on microtubules where it may provide a platform for concentrating piRNAs coming from the sense strand of transposons, which are vastly outnumbered by antisense piRNAs (Vagin et al, 2006; Brennecke et al, 2007).
Finally, our study along with two recent mouse studies (Vasileva et al, 2009; Wang et al, 2009), shows an evolutionarily conserved interaction of Piwi proteins with Tudor-domain proteins. Tudor was first discovered in Drosophila as a posterior group gene important for germ plasm formation and germ cell development (Boswell and Mahowald, 1985), and has been shown to interact with Aubergine, a Piwi family protein (Thomson et al, 2008). Xtr is homologous to Drosophila Tudor, both of which are large (>250 kDa) proteins consisting of multiple tudor domains. The biochemical function of Xtr and Tudor is unclear, but the tudor domain is common in RNA helicases in vertebrates and fruit flies, such as spindle-E, a gene with a known role in piRNA biogenesis (Vagin et al, 2006; Klenov et al, 2007). A recent study showed that Xiwi contains symmetrically dimethylated arginine (Kirino et al, 2009), which may mediate the interaction between Xiwi and Xtr (Sprangers et al, 2003). Interestingly, mutations in mouse tudor domain-containing proteins, Tdrd1 and Tdrd6, result in a loss of germ cells without affecting piRNA levels or activation of TEs, and thus further suggesting a role of Piwi outside TE silencing (Vasileva et al, 2009; Wang et al, 2009).
Evolutionary and epigenetic implications of piRNAs
In addition to the wide sequence diversity of piRNAs, another striking feature is that clusters of piRNAs seem to derive from multi-kb loci with pronounced strand bias. In Xenopus, piRNA clusters seems to be bigger than fish and mammalian clusters, and TE-associated piRNAs may target mRNAs arising from DNA transposons, a TE class not thought to rely on an RNA intermediate for transposition (Jurka et al, 2007). As the X. tropicalis genome is incomplete, we neither observe nor can we rule out bi-directional clusters in Xenopus. The general structure of Xenopus piRNA clusters and the fact that only ∼20% of Xenopus piRNAs map to TEs suggest that these non-coding RNA genes are similar to mammals, but the TE strand bias in many large piRNA clusters harkens to the flamenco piRNA locus in Drosophila. Finally, our study illustrates a pressing need to complete the fragmented X. tropicalis genome to allow complete mapping of this important class of non-coding RNAs.
By using genome-wide piRNA cluster representation as a proxy for overall small RNA content, our single cell analysis indicates that single eggs express multiple piRNA clusters simultaneously, and that eggs from the same mother have highly correlated small RNA content whereas the small RNA content between different individuals is less similar. This raises several interesting questions. What promoter motif is present at piRNA clusters that allows for simultaneous activation by an unknown transcription factor? How critical is any given piRNA cluster to germ cell development? How might the health of progeny be affected by distinct piRNA profiles?
The idea that piRNAs are agents of epigenetic inheritance has recently been examined in incompatible hybrid strains of Drosophila melanogaster (Brennecke et al, 2008), which shows that sterile hybrids are born when the mother fails to deposit a particular class of TE-associated piRNAs to an embryo whose paternal genome contains the active TE. Can hybrid dysgenesis also be explained by piRNA-based mechanisms in vertebrates? Can non-transposon-derived piRNAs contribute to hybrid dysgenesis? Future experiments using Xenopus as a model organism might address these question, as studies of hybrid incompatibility using Xenopus are possible (Michalak and Malone, 2008), Xiwi and piRNAs are deposited into the developing Xenopus embryo, and certain TE-associated piRNA populations are moderately abundant in one frog and absent in another (data not shown). These experiments will be able to explore the role of maternally deposited piRNAs in the health of vertebrates and determine whether a role for maternal piRNAs is conserved from fruit flies to frogs.
Materials and methods
Generation of anti-Xiwi antibodies
Xenopus Piwi gene models were identified in the Ensembl v52 database for X. tropicalis (model ENSXETG00000012572: Xiwi, model ENSXETG00000010533: Xili, and model ENSXETG00000006411: xPiwi3). Amino acids 51–334 were amplified by PCR from IMAGE cDNA clone 7686608, which is homologous to human piwi-like 1, and cloned into pET30a (pMB192). Protein was expressed at 22°C and purified using Ni–NTA resin (Qiagen). Recombinant Xiwi was used to generate antibodies in a rabbit (Covance). Antibodies were affinity purified before use.
Xenopus egg extract experiments
CSF arrested egg extracts were prepared from laid eggs from X. laevis and X. tropicalis as described by Brown et al (2007) with one modification, packed eggs were centrifuged for 15 min at 17 000 g and cytoplasm was extracted, the crude cytoplasmic extract was further centrifuged for 10 min at 17 000 g and cleared cytoplasm was extracted again away from the cloudy pellet.
To immunodeplete Xiwi, 25 μg of anti-Xiwi antibodies were pre-bound to 100 μl of protein A Dynabeads and incubated with 50 μl of CSF extract for 1 h on ice.
‘Cycled' or CSF spindles were prepared using Xiwi- or IgG-depleted extracts as described. Extracts were fixed in solution by dilution into 1 × BrB80 (80 mM PIPES, (pH 6.8), 1 mM MgCl2, 1 mM EGTA), 30% glycerol, 4% paraformaldehyde, 0.05% glutaraldehyde, and 0.5% Triton X-100. Extracts were incubated using fixative for 5 min at room temperature then centrifuged onto coverslips through a glycerol cushion as described. RanGTP asters were prepared by the addition of RanQ69L to 25 μM in Xiwi- or IgG-depleted extracts and centrifuged onto coverslips as described.
Xiwi immunofluorescence was carried out on spindles fixed as described above. Anti-Xiwi antibody was used at a concentration of 10 μg/ml.
Renilla luciferase (pMB341) and firefly luciferase (pMB342) (Promega) were PCR amplified and TOPO cloned into PCR2.1TOPO. 4 × perfect mir-16 sites were cloned 3′ Renilla luciferase using the SpeI site to generate pMB378 or seed mismatches to generate pMB380. 4 × perfect piRNA-binding sites for piR-XL2 (TACTGAAGGATGGCGTACTATTCCTGCCA) were cloned into the SpeI site to generate pMB380 or seed mismatches to generate pMB383. The 1872–3047 bp stretch of IMAGE clone 7296684 from X. laevis MenF was cloned into the SpeI site 3′ to Renilla luciferase in the sense orientation to generate pMB343, or in antisense orientation to generate pMB344. Templates for in vitro transcription were amplified using M13F and M13R PCR and used to transcribe capped mRNA in vitro using T7 RNA polymerase. Each transcript was diluted in water to generate a 20 nM stock solution, aliquoted, and stored at −20°C. Transcripts were added to CSF-arrested extracts at a concentration of 1 nM Renilla construct in combination with 1 nM firefly construct. mRNAs were incubated in extract for 12 h at 19°C and sequentially assayed for firefly and renilla activity using the Stop-and-Glow kit (Promega). Luciferase activity was read using a Trilux from Perkin Emer, and all readings were taken in duplicate. For immunodepletion reactions, Xiwi-depleted extract was compared with mock IgG-depleted extracts. For RNA stability assays in vitro transcription reactions contained 32P UTP. RNAs were added to Xiwi- or IgG-depleted extracts at a concentration of 1 nM and incubated for 2 h at 19°C. Total RNA was purified using TRIzol, separated on a formaldehyde agarose gel, transferred to nylon membrane, and exposed to a phosphor screen. Screens were read using a Typhoon imager and band intensities were quantified using ImageQuant.
Mass spectrometry identification and validation of Xiwi-interacting proteins
A total of 250 μg of anti-Xiwi antibodies were used to immunoprecipitate Xiwi from 500 μl of CSF extract for 1 h on ice. Dynabeads were collected using a magnet and washed 5 times with 1 ml PBS over the course of ∼5 min. Proteins were eluted from the beads using 1% SDS and separated by SDS–PAGE and stained with Coomassie Blue. Bands were excised from the gel and identified by mass spectrometry at the Harvard Taplin Biological Mass Spec facility.
Antibodies against Xenopus Tudor were a kind gift from Kazefumi Takamune, antibodies against Xenopus CBTF122 were a kind gift from Brenda Bass, antibodies against Xenopus ePAB were a kind gift from Joan Steitz, and antibodies against symplekin were obtained from Novus. Each of the above antibodies was pre-complexed with protein A or G Dynabeads and used to precipitate the relevant protein from CSF extracts as described for Xiwi. Immunoprecipitations were separated by SDS–PAGE and blotted for both Xiwi and the immunoprecipitated protein. For RNase treatment of IPs, antibody complexes were split after the initial retrieval on a magnet and were washed with either PBS containing 1 μg/ml RNaseA or PBS.
GFP–ePAB was expressed in BL21 bacteria and purified using Ni–NTA resin. GFP or GFP–ePAB was added to CSF extracts at a concentration of 1 μM and incubated for 1 h on ice. GFP-tagged proteins were immunoprecipitated using anti-GFP antibodies (Covance), and beads were washed and eluted as described above for Xiwi immunoprecipitations.
Oocyte, egg, and embryo cell biology
Oocytes were liberated from dissected ovaries in 0.8 U/ml of Liberase Blendzyme 3 (Roche) in 1 × modified Barth's saline (MBS) for ∼2–3 h at room temperature. Eggs and oocytes were homogenized with a pestle in embryo lysis buffer (100 mM NaCl, 20 mM NaF, 50 mM Tris (pH 7.5), 5 mM EDTA, 0.2% (v/v) NP40 (IGEPAL), 0.2% (w/v) Na deoxycholate, and 1:50 EDTA-free complete protease inhibitor (Roche)), and centrifuged to separate pigments and lipids. Embyros from a natural mating, eggs, and oocytes were fixed in Dent's fix (20% DMSO in methanol), bleached with 10% hydrogen peroxide, permeabilized, and blocked with 0.5% Triton X-100 in 5% milk/TBS. Samples were probed overnight at 4°C with a 1:1000 dilution of primary antibodies in 5% milk/TBS, >4 h with 1:1000 dilution of Alexa fluor-labelled secondary antibodies, and then mounted in a 1:2 solution of benzyl alcohol:benzyl benzoate (BABB). Images were obtained and analyzed on a Leica SP2 confocal microscope system. To accomplish double staining of Xiwi and Xtr with rabbit polyclonal antibodies, we first stained oocytes and eggs with antibodies against Xtr and histone H4 (Abcam, monoclonal antibody) followed by Alexa-fluor 488 and 647 secondary antibodies. These samples were then extensively washed, and finally triple stained with a Xiwi antibody that was directly labelled using the FluoReporter Tetramethylrhodamine Protein Labeling Kit (Invitrogen) before mounting in BABB.
Deep sequencing and bioinformatics
Small RNA libraries from Xenopus microtubules and egg extracts were prepared for sequencing on the Genome Sequencer FLX system as previously described by Lau et al (2006). To minimize sample loss in small RNA library construction from single eggs, we modified the library construction procedure as follows. Total RNAs were extracted from single eggs using TRI Reagent (∼0.1–1 μg), and a pre-adenylated 3′ linker sequence designed for an Illumina GA-II sequencer flowcell was ligated to the total RNA using T4 RNA ligase. The 3′ linker ligation reactions were then resolved on a 15% denaturing polyacrylamide gel alongside 18 nt and 31 nt RNA markers that were also marked with the 3′ linker. A 5′linker was ligated to purified 3′ ligation products overnight at 16°C using T4 RNA ligase. The 5′ ligation products were then directly subjected to reverse transcription and limited (<15) cycles of PCR, and the PCR products representing the small RNA population was gel purified, and this sample was further amplified 12 more cycles before final preparation for sequencing on the Illumina GAII.
To achieve multiplex deep sequencing, we designed four-base barcodes (AAAA, TTAA, CCAA, GGAA) at the 3′ end of the 5′ linker sequence. This barcode becomes the first 4 bases sequenced on the Illumina GA-II flowcell, and thus retain the highest sequence fidelity. Although individual libraries for each egg or IP sample were constructed separately, library DNAs were quantified by Nanodrop and equal mass amounts for XTIP and the XLIP libraries or the four single egg libraries were combined before loading onto the flowcell. We then carried out 40-cycle sequencing runs on the Illumina GA-II instrument using 4 pM of template, and base calls were calculated using the manufacturer's software.
Libraries were parsed according to barcode, and then scanned for, at least, 5 bp matching to the 5′ end of the 3′ linker. The barcode and a matching sequence for the 3′ linker were trimmed from each sequence, though ∼11% of reads failed to show a match to the 3′ linker, and were counted as 36 bp reads, but these reads often failed to map to the genome due the degenerated 3′ linker sequence. Reads that were 16–36 nt long were then queried against a database containing Xenopus and select microbial ribosomal RNAs and structural RNAs that include transfer RNAs, spliceosomal RNAs, RNAseP, and telomerase RNAs. Perfect matches to this query were removed, which was most significant for libraries from single eggs in which >60% of reads were ribosomal/structural RNA matches. Interestingly, vast majority of these ribosomal/structural RNAs were 22, 23, and 25 nt long.
Reads were collapsed into unique sequences, which were then used in various database alignments to accomplish the annotation. For miRNA annotation, we applied BLAST with parameters that the expectation value is ⩽0.00005 to all mature and stem-loop entries of miRNAs from X. laevis, X. tropicalis, and Mus musculus, from miRBase release 12.0. For alignment to the Xenrep5 TE database provided by Vladimir Kapitonov, the X. laevis and X. tropicalis UniGene database, or to the X. tropicalis genome, we used the ShortQueryLookup alignment program that is part of the Broad Institute's Arachne package. Counts of annotation hits for TEs, miRNAs, and mRNAs were maintained using Microsoft Excel and Access.
To identify piRNA clusters, we first counted the number of uniquely mapping reads or normalized read totals in 2 kb windows, as the smallest scaffolds are 2 kb long. Normalized read count is the sequencing frequency of a sequence divided by the number of genomic loci with perfect matches to the read sequence. We defined a ‘partial' piRNA cluster as scanning in 10 kb windows for uniquely mapping reads and considering the only a genomic domain that contains at least 10 uniquely mapping reads. As many domains terminate at the end of a scaffold, we consider the defined piRNA clusters as ‘partial' in their characterization.
For pairwise comparison of piRNA profiles between libraries, we first normalized the read frequencies of uniquely mapping (UM) reads by the size of the smallest library. For example, consider a loci with 10 UM reads in library A and 2 UM reads in library B, but library A had 140 000 filtered reads whereas library B had 70 000 reads. To compare library A with B, we normalized UM read count to 5 in library A and 2 in library B. By counting such UM read counts across all 2 kb windows in the X. tropicalis genome, and taking into account whether the reads map to the plus or minus strand, we then calculated a Pearson correlation coefficient.
Venn analysis of read sequence overlap was carried out by simple sequence matching. To determine overlap in piRNA cluster expression, we generated partial cluster lists for each library using the criteria described above, and then employed SQL queries to find matching numbers of clusters. Base composition was plotted using the Weblogos application. Plotting of read frequencies from 2 kb windows was carried out using Kaleidagraph v4.0. Ping-pong pairing analysis was carried out on uniquefied sequences, where sequences are first mapped to the genome, then checked whether another sequence in the library maps on the opposite strand. The overlap is counted and tallied in bins to yield a distribution, which is highest at position 10 in the microtubule small RNA libraries.
Supplementary Material
Supplementary Figures
Supplementary Tables
Supplemental Figures and Table Legends
Review Process File
Acknowledgments
We thank David Bartel, Clotilde Perbost and 454 Life Sciences for pilot sequencing runs, Lee Lim and Tomas Babak for advice on informatics, and Vladimir Kapitonov for providing data on Xenopus TEs. We are grateful to Blake Riggs, Rebecca Heald, Dianne Schwarz, Judith Sharp, Kazufumi Takamune, Mathew Guille, Brenda Bass, and Joan Steitz for reagents and experimental assistance. We thank Dianne Schwarz, Judith Sharp, and Matt Simon for comments on the paper. This study was supported by funds from Whitney foundation fellowship and a K99 award from the NIH (to NCL); grants from the NIH (to REK and MDB).
Author Contributions: NCL and MDB designed and carried out the experiments, TO and MB provided informatics and sequencing analysis, and NCL, MDB, and REK wrote the paper. The small RNA sequencing data from this paper are deposited in the Gene Expression Omnibus under the series accession number GSE15556.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N (2004) Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA 10: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J (2007a) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc′his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007b) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC (2008) PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP (1985) tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43: 97–104 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R (2007) Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol 176: 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc′his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML (2004) Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15: 4669–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE (1999) The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol 61: 385–412 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (1999) Xenopus oocyte maturation: new lessons from a good egg. Bioessays 21: 833–842 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H (2006) MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A et al. (2006) The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi M, Nakajo N, Abe S, Takamune K (2005) Involvement of Xtr (Xenopus tudor repeat) in microtubule assembly around nucleus and karyokinesis during cleavage in Xenopus laevis. Dev Growth Differ 47: 109–117 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- JGI (2004) Xenopus tropicalis genome, build 4.1 (http://genome.jgi-psf.org/Xentr4/Xentr4.home.html)
- Jurka J, Kapitonov VV, Kohany O, Jurka MV (2007) Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet 8: 241–259 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL (2005) Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97: 19–33 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z (2009) Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W (2008) Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Etkin LD (2005) RNA localization mechanisms in oocytes. J Cell Sci 118: 269–282 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P (2006) Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 119: 2819–2825 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T (2008) DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P, Malone JH (2008) Testis-derived microRNA profiles of African clawed frogs (Xenopus) and their sterile hybrids. Genomics 91: 158–164 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD (2007) Mighty Piwis defend the germline against genome intruders. Cell 129: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC (2008) Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G (2007) A whole-genome RNAi screen for C. elegans miRNA pathway genes. Curr Biol 17: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, M DIC, Suprenant KA, Bonder EM (2005) Seawi—a sea urchin piwi/argonaute family member is a component of MT–RNP complexes. RNA 11: 646–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman EE, Lasko P (1999) Germline development in vertebrates and invertebrates. Cell Mol Life Sci 55: 1141–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M (2003) High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 327: 507–520 [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P (2008) Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H (2008) MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. J Biol Chem 284: 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Müller-Reichert T, Jessberger R (2009) Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA (2001) A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 15: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, H. L (2009) Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pontius JU, Pruitt KD, Schuler GD, Schriml LM et al. (2005) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 33: D39–D45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N (2009) Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA 15: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H (2007) An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables
Supplemental Figures and Table Legends
Review Process File