Abstract
While probing the role of RNA for the function of SET1C/COMPASS histone methyltransferase, we identified SET1RC (SET1 mRNA-associated complex), a complex that contains SET1 mRNA and Set1, Swd1, Spp1 and Shg1, four of the eight polypeptides that constitute SET1C. Characterization of SET1RC showed that SET1 mRNA binding did not require associated Swd1, Spp1 and Shg1 proteins or RNA recognition motifs present in Set1. RNA binding was not observed when Set1 protein and SET1 mRNA were derived from independent genes or when SET1 transcripts were restricted to the nucleus. Importantly, the protein–RNA interaction was sensitive to EDTA, to the translation elongation inhibitor puromycin and to the inhibition of translation initiation in prt1-1 mutants. Taken together, our results support the idea that SET1 mRNA binding was dependent on translation and that SET1RC assembled on nascent Set1 in a cotranslational manner. Moreover, we show that cellular accumulation of Set1 is limited by the availability of certain SET1C components, such as Swd1 and Swd3, and suggest that cotranslational protein interactions may exert an effect in the protection of nascent Set1 from degradation.
Keywords: COMPASS, complex assembly, Saccharomyces cerevisiae, SET1C complex, translation
Introduction
Most proteins in a cell are integral components of heterogeneous multi-subunit complexes. Assembly pathways for such complexes have been studied, for example, for ribonucleoproteins that form on small nuclear or small nucleolar RNAs (Filipowicz and Pogacic, 2002; Paushkin et al, 2002; Staley and Woolford, 2009) and for membrane-bound complexes in organelles (Choquet and Vallon, 2000; Ackerman and Tzagoloff, 2005). The manner in which large structures such as ribosomal subunits are built poses another huge challenge for biologists (Fatica and Tollervey, 2002); moreover, very little is known about the formation of a large number of diverse multi-protein complexes in the cytosol. In particular, the mechanisms that ensure specific association of a certain set of interaction partners remain largely obscure.
Chromatin modification enzymes such as histone methyltransferases (HMTases) are often contained in multi-protein assemblies (Kouzarides, 2007). In yeast, the Set1 protein is required for all methylation of histone 3 lysine 4 (H3K4) (Briggs et al, 2001; Roguev et al, 2001; for a recent review see Dehe and Geli, 2006). The protein stably associates with seven other subunits, which have been given multiple denominations: Swd1/Cps50p/Saf49p, Swd2/Cps35p/Saf35p, Swd3/Cps30p/Saf35p, Spp1/Cps40p/Saf41p, Bre2/Cps60p, Sdc1/Cps25p/Saf19p and Shg1 (Miller et al, 2001; Roguev et al, 2001; Nagy et al, 2002). The resulting SET1 complex is also known as COMPASS (Miller et al, 2001). For simplicity, we will use the standard nomenclature of the proteins as indicated by the Saccharomyces Genome Database (http://www.yeastgenome.org/). Only the Swd2 subunit of the complex is essential for cell viability, which is probably because of an independent role of the protein as a component of the cleavage and polyadenylation factor (CPF) complex (Cheng et al, 2004; Dichtl et al, 2004).
H3K4 HMTases from yeast to humans carry, in addition to variable enzyme-specific subunits, a common set of associated proteins: the so-called ‘MLL core complex'. This complex consists of a catalytic SET domain containing subunit and WDR5, RpBP5 and Ash2L proteins, which are homologous to yeast Swd3, Swd1 and Bre2 (reviewed in Ruthenburg et al, 2007). Analysis of these core components in vivo and in vitro showed a central role for WDR5 in complex integrity and substrate interaction (Wysocka et al, 2005; Dou et al, 2006; Steward et al, 2006). It is noteworthy that the requirement for certain core components to maintain complex integrity seems to be conserved in yeast, as a mutation of the essential WD-repeat protein Swd2 (Dichtl et al, 2004) or a deletion of the genes encoding the non-essential Swd1 and Swd3 subunits resulted in a severe under-accumulation of Set1 (Dehe et al, 2006; Steward et al, 2006). Although a reconstitution of an MLL core enzyme was recently achieved with heterologously expressed proteins in vitro (Dou et al, 2006), little is known about the manner by which HMTase complexes are formed in vivo.
Set1 harbours an evolutionarily conserved RNA-binding domain in its amino-terminal half, comprising two RNA recognition motifs (RRM1 and RRM2) (Tresaugues et al, 2006). We recently showed that, although this domain can bind RNA in vitro, mutations within RRM2 do not interfere with the HMTase activity of SET1C in vivo (Tresaugues et al, 2006). However, removal of the entire RRM domain or point mutations within RRM1 resulted in a specific loss of tri-methylation, consistent with the idea that RNA may have a role in regulating SET1C (Schlichter and Cairns, 2005; Tresaugues et al, 2006). Moreover, it was suggested that the catalytic SET domain and associated n-SET and post-SET domains can bind single-stranded DNA and RNA in vitro (Krajewski et al, 2005). Nevertheless, the physiological implications of nucleic acid binding by SET1C and related complexes remain unclear.
This study was initiated by a search for RNA molecules that bind to SET1C. Our analyses showed that SET1 mRNA was associated with a SET1C sub-complex, which we refer to as SET1RC. Our experiments provide evidence that SET1RC assembles on nascent Set1 during translation. We propose that this cotranslational assembly couples the synthesis and accumulation of Set1, the catalytic subunit of the complex, with the formation of SET1C. Cotranslational processes, as described here, may be of wider relevance for the assembly of multi-protein complexes.
Results
Specific enrichment of SET1 mRNA in Shg1–TAP purifications
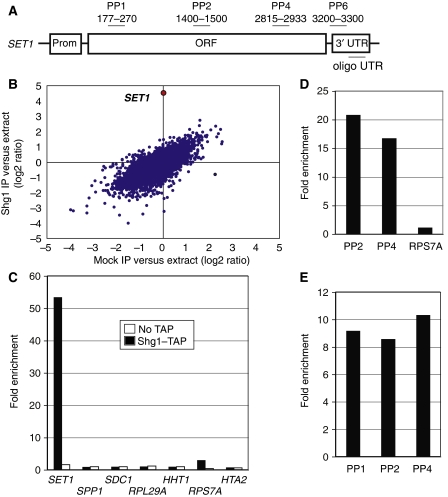
To test for RNA associated with SET1C, we partially purified the complex from Shg1–TAP extracts; a non-tagged isogenic wild-type strain served as control. RNA isolated from extracts and from purified Shg1–TAP material was used to generate cDNA probes labelled with different fluorescent dyes, which were then mixed and hybridized to Saccharomyces cerevisiae oligonucleotide microarrays (Gerber et al, 2004; Supplementary data). The scatter plot shown in Figure 1B illustrates that SET1 RNA was highly enriched in Shg1–TAP affinity isolates (average enrichment ∼23-fold; P-value was <0.0004, false discovery rate (FDR) approximated to 0%). Six additional RNAs were enriched; however, they had FDRs of >50% and hence were not likely to represent true targets. The microarray results were confirmed by real-time PCR (Figure 1C): SET1 mRNA was found to be approximately 50-fold enriched compared with ACT1 mRNA in Shg1–TAP isolates; transcripts encoding other SET1C subunits (SPP1, SDC1), ribosomal proteins (RPL29A and RPS7A) and histones (HHT1 and HTA2) were not enriched. Moreover, Figure 1D shows that a SET1 signal was obtained when cDNA synthesis was primed with oligo(dT) instead of random hexamers (which were used in Figure 1C), suggesting that the RNA contained poly(A). To exclude the fact that we detected short polyadenylated fragments, we used for cDNA synthesis a primer that anneals in the 3′ UTR (oligo UTR; see Figure 1A). We observed similar signal intensities for primer pairs PP1, PP2 and PP4, indicating that RNAs extended from the 3′ UTR to the 5′ end of the transcript (Figure 1E). Taken together, our analyses suggested that bona fide, full-length SET1 mRNA was associated with Shg1–TAP.
Figure 1.
Specific association of SET1 mRNA with Shg1–TAP. (A) Schematic presentation of the SET1 gene. Oligonucleotides (primer pairs PP1–PP6) used for qPCR analysis or reverse transcription (oligo UTR) and their location relative to the translational start codon (+1) are indicated. (B) Average RNA enrichment from four independent microarray hybridizations analysing Shg1–TAP affinity isolates (y-axis) and mock controls (x-axis). Data are shown for 6253 features. SET1 data are depicted in red. (C) Quantitative RT–PCR analysis of the indicated target genes on RNA co-purifying with partially purified Shg1–TAP (solid black bars) or from a control extract (open white bars). Fold enrichment was determined as described in ‘Materials and methods'. (D) Quantitative RT–PCR analysis as described in (C) with the indicated primer pairs, with cDNA that was obtained after oligo(dT)-primed reverse transcription. (E) Quantitative RT–PCR analysis as described in (C) with the indicated primer pairs with cDNA that was obtained after reverse transcription that was primed with oligo UTR.
SET1 mRNA is associated with a SET1C sub-complex
Most of cellular Shg1 is associated with SET1C (Roguev et al, 2001). To test whether other SET1C subunits were also bound to SET1 mRNA, we affinity purified all eight SET1C subunits individually from extracts. An immunoblot analysis confirmed the correct expression of TAP-fusion proteins and enrichment of proteins after IgG-Sepharose adsorption and elution by TEV-protease cleavage (Supplementary Figure S1A). We observed an enrichment of SET1 mRNA with TAP–Set1, Spp1–TAP, Swd1–TAP and Shg1–TAP, but not with Bre2–TAP, Sdc1–TAP, Swd2–TAP and a non-tagged wild-type strain (Figure 2A). Swd3–TAP gave a signal that was slightly above background, indicating that it may either associate loosely with the complex or that this protein does not support efficient purification because of other limitations. No enrichment of RPS7A mRNA took place in any case. To evaluate whether the TAP-tag compromised the functionality of the fusion proteins and potentially influenced the efficiency of SET1 mRNA co-purification, we tested H3K4 methylation by western blot. Supplementary Figure S1B shows that all analysed strains had similar levels of H3K4 di- and tri-methylation, suggesting that TAP-fusion proteins were functional.
Figure 2.
SET1 mRNA is associated with a SET1C sub-complex. (A) Affinity purification was carried out with extracts obtained from the indicated strains. After RNA extraction and cDNA synthesis, qPCR analysis was performed to detect SET1 RNA (PP4; solid black bars) or a control RNA (RPS7A; open white bars). Values represent the mean of three to five comparable experiments for each indicated strain and error bars indicate s.d. (B) The diagram (top) depicts the domain structure of Set1 (RRM1, RRM2, n-SET, SET and post-SET; not in scale) and numbers delineate the interaction domains for Shg1, Spp1 and Bre2, as derived from yeast two-hybrid results. The region of interaction (aa 1–900; Dehe et al, 2006) of the Swd1–Swd3 heterodimer is indicated. Sdc1 forms a heterodimer with Bre2 and does not directly bind to Set1; no interaction domain is known for Swd2. The bottom panels show GST pull-down assays with GST–Set1 and in vitro translated 35S-methionine-labelled Swd1, Spp1 and Shg1 proteins. Input lanes show 10% of radioactive protein included in the binding reactions. The GST lane shows background binding. (C) Shg1–TAP and associated material were partially purified by affinity chromatography on IgG-Sepharose and the obtained material was then applied on a Superdex 200 column. The upper graph displays the obtained protein elution profile. The indicated fractions were then analysed by RT–PCR for the presence of SET1 mRNA (lower graph). The western blot at the bottom of the figure shows the distribution of Shg1 in the Superdex 200 fractions. The arrow indicates the void volume of the column.
Next, we addressed whether Set1, Spp1, Swd1 and Shg1 associate with SET1 mRNA as constituents of the same complex. Results from yeast two-hybrid screening identified domains within Set1 that mediate interactions with Shg1 (amino acid (aa) 462–560) and Spp1 (aa 762–794) (BD, unpublished data; Figure 2B). Furthermore, Bre2 interacted with a Set1 domain encompassing aa 900–1081, consistent with previous results (Dehe et al, 2006). No interaction domains on Set1 are known for Swd1, Swd2 and Swd3, but the Swd1–Swd3 heterodimer does not require the C-terminal SET domain (aa 901–1081) for association with SET1C (Dehe et al, 2006). In pull-down assays, Swd1, Spp1 and Shg1 efficiently bound to GST–Set1 but not to GST alone (Figure 2B), consistent with the view that the three proteins directly associate with Set1. Furthermore, a Superdex 200 gel-filtration analysis of material associated with Shg1–TAP after IgG purification suggested that SET1 mRNA was associated with a large molecular weight complex that eluted with the void volume of the column (Figure 2C). Taken together, our results showed the existence of a SET1 mRNA containing complex that we will henceforth refer to as SET1RC (SET1 mRNA-associated complex).
Swd1, Spp1 and Shg1 are dispensable for SET1 mRNA binding
To determine the requirements for SET1RC formation and stability, we initially tested SET1 mRNA association to TAP-tagged SET1RC subunits in the absence of other SET1RC proteins. As shown in Figure 2A, SET1 mRNA association was observed with TAP–Set1 in Δswd1, Δspp1 and Δshg1 backgrounds, with Spp1–TAP in Δswd1 and Δshg1 backgrounds, with Shg1–TAP in Δswd1 and Δspp1 backgrounds and with Swd1–TAP in the Δshg1 background. The signal obtained with the TAP–Set1 Δspp1 strain was elevated in this analysis, but remained within the limits of variation that we observed in numerous such experiments. The only combination that gave no signal was Swd1–TAP in Δspp1, indicating that the absence of Spp1 weakens the binding of Swd1–TAP to the complex. The absence of Bre2 and Sdc1, which we found not to associate with SET1RC, was tested in Spp1–TAP and Shg1–TAP backgrounds, respectively, and did not interfere with the binding of SET1 mRNA.
We, along with others, previously found that inactivation of Swd1, Swd2 and Swd3 resulted in a severe under-accumulation of Set1 (Dichtl et al, 2004; Dehe et al, 2006; Nedea et al, 2008), and western blot analysis confirmed strongly reduced levels of TAP–Set1 in Δswd1 and Δswd3 cells (Figure 6A). Remarkably, co-purification of SET1 mRNA with TAP–Set1 occurred with the same efficiency, irrespective of the presence or absence of either Swd1 or Swd3 (Figure 2A). This indicated that the amount of TAP–Set1 associated with SET1RC was not significantly reduced, despite strongly reduced cellular levels of the protein. In summary, we found that SET1RC formation can occur in the absence of Swd1, Spp1 and Shg1, suggesting that only Set1 is required for SET1 mRNA binding.
mRNA binding is not associated with a specific Set1 domain
Set1 carries RRM1- and RRM2 RNA-binding motifs in the N-terminal half and the catalytic SET domain at its C-terminus (Figure 3A). To test which domains are required for SET1 mRNA binding we fused Set1 fragments to ProteinA and assayed whether encoding mRNAs co-purified. The data in Figure 3B illustrate that both the N- and the C-terminal half of the protein (residues 1–571 and 572–1081) bound their encoding mRNA with an efficiency similar to full-length protein (residues 1–1081). Thus, SET1 mRNA binding occurred both with a Set1 protein, which carried the RRM domains, and the C-terminal part of the protein, which lacked RRMs. Moreover, proteins carrying mutations in RRM1 (1–571YF/AA) or RRM2 (1–571H/A), which strongly reduced RNA binding in vitro (Tresaugues et al, 2006), retained the ability to interact with cognate mRNA (Figure 3B). Thus, RNA binding of the N-terminal domain did not depend on intact RRMs, consistent with the observation that the C-terminal fragment, which completely lacked these motifs, also bound RNA.
Figure 3.
Characteristics of SET1 mRNA association. (A) Schematic presentation of Set1 (not in scale); numbers indicate approximate amino-acid positions of RRM1, RRM2 and SET domains. Also indicated are fragments that were expressed as plasmid-encoded ProteinA fusions in Δset1 strains. A strain carrying the empty ProteinA-fusion vector was analysed for control. The 1-571 YT/AA fragment carries two point mutations in RRM1, and 1–571 H/A carries a single point mutation in RRM2 (asterisks indicate the presence of point mutations; Tresaugues et al, 2006). (B) RT–PCR analysis of RNA isolated from IgG purifications of the indicated ProteinA-fusion proteins. PP2 and PP4 were used to detect SET1 mRNA or fragments thereof (primer locations are indicated in panel C). RPS7A served as control. Note that PP2 will only detect full-length and N-terminal fragments and PP4 will detect only full-length and C-terminal fragments. (C) Schematic representation of N- and C-terminal halves of Set1, which were expressed as ProteinA fusions from plasmids in strains carrying a chromosomal SET1-MYC gene (1–1081-Myc). The location of primer pairs used for qPCR is also shown. Wavy lines represent mRNAs that correspond to Set1 proteins as indicated. Double-headed arrows indicate interactions between proteins and RNA that were tested. The crossed-out double-headed arrows indicate interactions that were not observed. (D) RT–PCR analysis of RNA associated with the indicated ProteinA-fusion proteins. The 1–571 mRNA was detected with PP1 and 572–1081 mRNA was detected with PP4. 1–1081-Myc mRNA was detected with PP6 in the 1–571 strain and with PP1 in the 572–1081-expressing strain, as indicated. (E) RNA analysis of SHG1-TAP strains carrying plasmids encoding the 5′ and 3′ halves of the SET1 mRNA, under control of a T7 RNA polymerase promoter. RT–PCR was carried out with primers specific for T7 RNAs (T7-5′ and T7-3′ primer pairs, respectively) to determine the fold-increase of T7 RNA compared with that of chromosomally encoded SET1 mRNA. (F) RT–PCR analysis of RNA co-purifying with Shg1–TAP strains as described in (E). Endogenous SET1 mRNA was detected with PP1 and PP4 (solid black bars; note that PP1 does not detect the T7-3′ fragment and that PP4 does not detect the T7-5′ fragment). The T7-derived RNAs were detected with T7-5′ and T7-3′ primer pairs (open white bars; note that T7 primer pairs specifically detect T7 transcripts and not endogenous SET1 mRNA). (G) Sub-cellular fractionation of Shg1–TAP cells. Total extract (XT), post-nuclear supernatant (PNS) and nuclear (Nc) fractions were analysed by western blot with the indicated antibodies. H3K4me3 is a nuclear-specific marker, whereas Pgk1 is cytoplasmic.
Next, we asked whether the interaction may be restricted to mRNAs and to their derived proteins, or whether Set1 proteins were able to bind SET1 RNA derived from a second SET1 locus. For this purpose, we expressed the ProteinA fusions of the N- and C-terminal Set1 halves in a strain that also carries a chromosomal full-length SET1-MYC gene (Figure 3C). We found that both Set1 fragments efficiently co-purified their encoding mRNA, but neither of the fragments bound to SET1-MYC mRNA (Figure 3D). This suggested that binding was restricted to Set1 proteins and their encoding mRNAs.
To address the requirements of sub-cellular RNA localization for association within SET1RC, we asked whether nuclear-restricted SET1 RNAs were included in the complex. RNAs corresponding to the N- and C-terminal halves of SET1 were expressed in SHG1-TAP strains with nuclear localized T7 RNA polymerase (Dower and Rosbash, 2002). As termination of these transcripts was mediated by a T7 terminator, the RNAs will not be exported to the cytoplasm (Dower and Rosbash, 2002). Figure 3E shows that we achieved efficient transcription of T7-derived RNAs (approximately 180-fold relative to endogenous SET1 mRNA levels). Analysis of RNA co-purifying with Shg1–TAP showed, however, that neither of the T7 RNAs was enriched with this procedure (Figure 3F). In contrast, chromosomally encoded SET1 mRNA was bound, as expected (Figure 3F). These results suggested that nuclear-restricted RNAs do not qualify for association with SET1RC.
The above data are consistent with the possibility that SET1RC formation may rely on a cytoplasmic localization of its components. To test this hypothesis, we performed fluorescence microscopy on strains carrying GFP-tagged alleles of SHG1 and SET1. We observed nuclear localization of these SET1C components as expected (Supplementary Figure S4; Huh et al, 2003). However, we could not conclude about cytoplasmic protein levels, as the signal strength was very weak compared with auto-fluorescence obtained with a GFP-only control strain (Supplementary Figure S4). To circumvent this problem, we used Shg1–TAP-expressing cells and performed sub-cellular fractionation. Figure 3G shows that Shg1–TAP distributed to both nuclear and post-nuclear fractions. In contrast, histone H3 tri-methylated at lysine 4 (H3K4me3) was exclusively nuclear and the glycolysis enzyme 3-phosphoglycerate kinase (Pgk1) localized to the post-nuclear supernatant. We conclude that some cellular Shg1–TAP was associated with the cytoplasmic compartment.
SET1 mRNA binding is sensitive to EDTA and puromycin
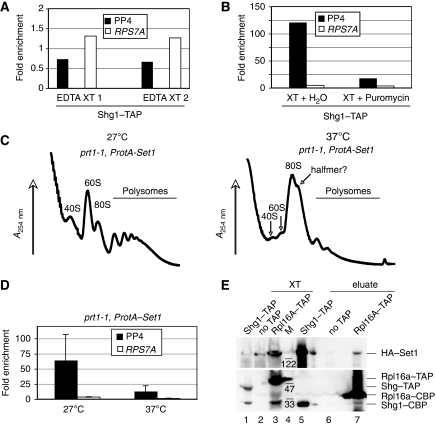
One of the simplest explanations to account for our observation in Figures 1, 2 and 3 is that the SET1 mRNA interactions depended on nascent Set1 peptides that were associated with their mRNAs during translation. Therefore, we wanted to test whether SET1RC formation was indeed translation dependent. For this purpose, we initially performed RNA co-purification experiments with Shg1–TAP extracts in the presence of EDTA. This treatment dissociates ribosomes and we found that SET1 mRNA binding was indeed abolished under these conditions (Figure 4A). However, EDTA could also disrupt binding independently of ribosome dissociation. Therefore, we performed affinity purification in the presence and absence of the translation inhibitor puromycin. Figure 4B shows that the drug reduced co-purification of SET1 mRNA by more than six-fold, suggesting that SET1 mRNA binding required translation.
Figure 4.
SET1 mRNA binding is translation dependent. (A) RT–PCR analysis of RNA associated with Shg1–TAP, which was purified in the presence of 10 mM EDTA. PCR was carried out with PP4 to detect SET1 mRNA; RPS7A served as control. Shown are results from two independent extracts (EDTA XT 1 and EDTA XT 2). (B) Shg1–TAP was purified from identical extracts on IgG-Sepharose in the absence (XT+H2O) or presence of 200 μg/ml puromycin (XT+puromycin). Shown is a representative RT–PCR using PP4 to detect SET1 mRNA; RPS7A served as control. (C) A254 recordings of sucrose-gradient fractionations of extracts from a prt1-1 strain expressing plasmid-born ProtA-SET1, after growth at 27°C or after a shift to 37°C for 35 min, as indicated. (D) RT–PCR analysis of RNA associated with ProtA–Set1 in extracts that were obtained as described in (C). PCR was carried out with PP4 to detect SET1 mRNA; RPS7A served as control. Values represent the mean of at least three independent extracts produced for each strain at the indicated conditions and error bars indicate s.d. (E) Co-immunoprecipitation experiment with extracts (XT) prepared from the indicated TAP-tagged strains that also carried a plasmid-expressing HA–Set1. After adsorption of material on IgG-Sepharose, bound material was eluted by TEV-protease cleavage (eluate). The obtained fractions were analysed by western blot using anti-HA (top) and anti-TAP (bottom) antibodies, respectively. Migration of molecular weight marker (M) and HA- and TAP-tagged proteins is indicated on the right.
To further scrutinize the relationship of SET1RC and translation, we initially tested the efficiency of SET1 mRNA binding when glucose was withdrawn from the medium, a condition that rapidly inhibits the initiation of protein synthesis (Ashe et al, 2000). Sucrose-gradient fractionation showed that most polysomes disappeared on glucose withdrawal, but a minor polysomal signal remained (we estimate less than 5%; Supplementary Figure S2A), in agreement with previous reports (Ashe et al, 2000). The bulk of RPL29 mRNA exited polysomal fractions (8–12) on glucose withdrawal and relocated to early fractions (1–5), whereas some mRNA remained associated with fractions 7–9, overlapping the 80S monosome peak (Supplementary Figure S2B). In contrast, SET1 mRNA distribution was very similar in the presence and absence of glucose (Supplementary Figure S2B). Consistent with this, we also found that binding of Shg1–TAP to SET1 mRNA was not sensitive to glucose withdrawal (Supplementary Figure S2C). We conclude that control of SET1 mRNA translation does not respond to glucose withdrawal and therefore occurs uncoupled from most cellular mRNAs.
To interfere with translation initiation using an alternative approach, we analysed SET1 mRNA binding in prt1-1 temperature-sensitive strains expressing plasmid encoded ProtA–Set1, as translation initiation was shown to be disrupted in these mutants (Nielsen et al, 2004). Figure 4C shows that polysomes associated with the mutant at 27°C were strongly reduced at a non-permissive temperature (Figure 4C). This decrease coincided with the accumulation of an 80S monosome peak and a prominent shoulder that may reflect a ribosomal halfmer, as suggested earlier (see Figure 4C; Nielsen et al, 2004). Most interestingly, we found that SET1 mRNA co-purified efficiently from the cells grown at 27°C but the signal was clearly reduced after growth at the non-permissive temperature (Figure 4D). This effect was not because of reduced SET1 mRNA levels, which remained stable at 37°C as determined by qPCR analysis (data not shown). These data show that the observed mRNA binding was indeed translation dependent.
Set1 interacts with the translational machinery
To obtain further evidence for the interaction between SET1C and SET1RC with the translational machinery, we examined a large high-confidence high-throughput protein–protein interaction data set, using a modified testing procedure (Collins et al, 2007; PK unpublished results). A systematic analysis of the available protein complexes and significant interactions between the various protein complexes defined within this data set support the idea that SET1C interacts with components of the translational machinery (Supplementary Figure S3). These include interactions with the large ribosomal subunit (purple nodes, P-value 4.6 × 10−4); the small ribosomal subunit (dark blue nodes, P-value 9.4 × 10−10); and the translation elongation factor 1 (EF1) complex (green nodes, P-value 1.3 × 10−4). The yellow (P-value 5.1 × 10−5) and red nodes (P-value 2.1 × 10−6) do not uniquely map to individually known protein complexes, but all proteins are involved in ribosome biogenesis and assembly, as well as in translation initiation, and together consist of various known sub-complexes involved in translation. Furthermore, the light blue nodes indicate the CPF complex (P-value 3.2 × 10−12), which is linked to SET1C through the common Swd2 subunit (Roguev et al, 2001). Taken together, these statistically significant protein interactions are consistent with the idea that assembly of the SET1C complex is linked to translation.
Alternatively, these in silico analyses may also point to an interaction between mature SET1C components and the ribosome. We tested this idea with co-immunoprecipitation (Co-IP) experiments and found that HA–Set1 indeed co-purified with the TAP-tagged large ribosomal subunit component Rpl16a (Figure 4E). No signal was obtained with the wild-type (no TAP) extract, whereas Shg1–TAP bound to HA–Set1 as expected. It is important to point out that these experiments detected the binding of full-length HA–Set1 to the ribosome. In contrast, nascent Set1, which we predict to assemble the SET1RC complex, is heterogenous in size and therefore unlikely to produce a detectable full-length signal in a Co-IP experiment. Irrespective of this, our observations suggest that mature Set1 or SET1C may serve a hitherto unrecognized function through an association with the translational machinery.
TAP–Set1 is targeted for degradation in the absence of Swd1 and Swd3
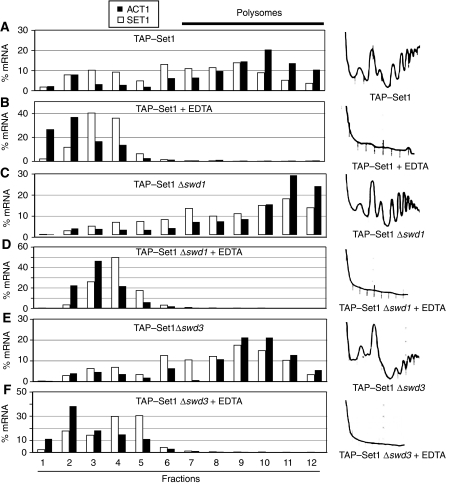
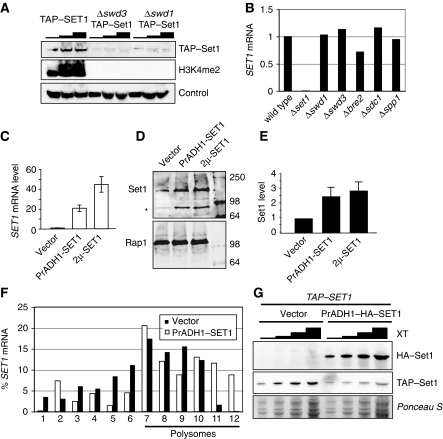
Our data support the idea that SET1RC forms on SET1 mRNA during translation. We reasoned that such an interaction may coordinate the assembly of SET1C with the synthesis of its catalytic component Set1. We described earlier that inactivation of Swd1, Swd2 and Swd3 resulted in a severe under-accumulation of Set1 (Dichtl et al, 2004; Dehe et al, 2006). Western blot analysis confirmed strongly reduced levels of TAP–Set1 in Δswd1 and Δswd3 mutant cells (Figure 6A). Furthermore, the lack of any di-methylated histone 3 lysine 4 (H3K4me2) signal indicated the absence of functional SET1C in these strains. This defect was not due to transcriptional downregulation, as strains lacking individual SET1C subunits had similar levels of SET1 mRNA (Figure 6B). Interestingly, the absence of either Swd1 or Swd3 did not abrogate SET1RC formation, as judged by the binding of TAP–Set1 to SET1 mRNA (Figure 2A). Therefore, we predicted that TAP–Set1 translation continued in Δswd1 and Δswd3 strains and that the protein was instead targeted for degradation. To this end, we analysed TAP-SET1 extracts that contained or lacked either Swd1 or Swd3 on sucrose gradients. Analysis of SET1 and ACT1 mRNA distributions showed an association of SET1 mRNA with polysome containing fractions 7–12 independent of Swd1 and Swd3 (Figure 5A, C and E). To ensure that fractions 7–12 indeed contained polysomes, we repeated the analysis in the presence of EDTA and found that no mRNA association took place anymore (Figure 5B, D and F). These data indicated that the under-accumulation of Set1 in the absence of Swd1 or Swd3 was not due to an inhibition of translation.
Figure 5.
Set1 translation continues in Δswd1 and Δswd3 strains. (A–F) Sucrose-gradient analysis of extracts prepared from strains expressing TAP–Set1 in the presence or absence of Swd1 and Swd3, as indicated. Extracts were prepared in the presence of 10 mM EDTA, as indicated, to disrupt ribosome association. RT–PCR analysis was carried out to detect ACT1 (filled black bars) and SET1 (open white bars) mRNAs. RNA levels were normalized to an unrelated control RNA (spike) that was added in equal amounts to individual fractions before RNA extraction. mRNA content is represented as % mRNA in relation to the total amount of mRNA present in all fractions. A254 nm absorption spectra obtained during collection of gradient fractions are shown on the right.
Set1 over-expression is limited in wild-type strains
Reduced Set1 levels in the absence of either Swd1 or Swd3 may indicate that the accumulation of proteins requires the presence of other complex components. Therefore, we hypothesized that Set1 over-expression may be limited in wild-type strains when the levels of other SET1C subunits remained constant. We initially tested this idea with two expression strategies: (1) driving SET1 transcription with the robust ADH1 promoter, and (2) expressing the SET1 gene from a multi-copy plasmid. We confirmed the over-expression of SET1 mRNA (20- and 45-fold; Figure 6C) and then determined Set1 protein levels using quantitative western analysis (Figure 6D and E). Importantly, a titration analysis of increasing amounts of respective extracts demonstrated that the measurements were obtained in the linear signal range (data not shown). We found that the strong increase in SET1 mRNA copy number resulted in a modest increase in Set1 protein levels of up to three-fold over wild-type levels (Figure 6E). Moreover, we found that SET1 mRNA, which was over-expressed with the ADH1 promoter, remained polysome associated, suggesting that it was engaged in translation (Figure 6F). These data suggest that a significant amount of newly synthesized Set1 was unstable and rapidly degraded.
Figure 6.
Over-expression of Set1 protein. (A) Western blot analysis of strains expressing TAP–Set1 in the absence of Swd1 and Swd3, respectively. Increasing amounts of total extract were separated by SDS–PAGE; protein detection was carried out with antibodies directed against the indicated epitopes. An unspecific band observed with the anti-H3K4me2 antibody served as loading control. (B) RT–PCR analysis of SET1 mRNA levels in strains lacking SET1C components. The signal obtained with wild-type cells served as reference (100%) for representation of relative SET1 levels measured in the mutants. (C) RT–PCR analysis of SET1 mRNA levels in wild-type strains carrying empty plasmid or over-expressing SET1 mRNA with the ADH1 promoter (PrADH-SET1) or on a multi-copy plasmid (2 μ-SET1), respectively. The shown mean values are relative to empty vector control and were derived from RNAs obtained from two independent cultures, which were analysed by two independent reverse-transcription reactions. Error bars indicate s.d. (D) Set1 (upper panel) and Rap1 (lower panel) expressions were detected by western blotting with monoclonal anti-Set1 antibodies and anti-Rap1 polyclonal antibodies, respectively. Infrared fluorescence-labelled secondary antibodies IRDye 800 anti-rabbit IgG and Alexa Fluor 680 anti-mouse IgG were used for direct infrared fluorescence detection of primary antibodies with an Odyssey imaging system. Asterisk indicates a Set1 degradation product. (E) Representation of signals obtained in (D) after normalization of Set1 levels relative to Rap1. Values were normalized to the empty vector control and represent the mean of three independent extracts. Error bars indicate s.d. (F) Analysis of SET1 mRNA distribution after sucrose-gradient fractionation of extracts as described in the legend of Figure 5. Extracts were obtained either from wild-type strains carrying an empty vector (black bars) or from the PrADH-SET1 over-expression construct (white bars). (G) Western blot analysis of TAP-SET1 strains carrying an empty vector or the PrADH-HA–SET1 construct. Increasing amounts of extract (XT) were resolved by SDS–PAGE and were examined with antibodies directed against the TAP-tag and HA-tag. Ponceau S staining served as loading control.
To further test this idea, we adopted yet another over-expression strategy. We transformed strains carrying a single chromosomal TAP-SET1 copy, either with an empty vector or with a vector over-expressing HA–Set1, with the ADH1 promoter. Interestingly, the presence of additional HA–Set1 resulted in a marked decrease in the levels of TAP–Set1 (Figure 6G), consistent with the idea that the availability of SET1C components restricted the accumulation of Set1. As a cellular surplus of Set1 is targeted for degradation, we propose that cotranslational protein interactions may stabilize newly synthesized Set1 (Figure 7).
Figure 7.
Model for cotranslational assembly of the SET1C complex. Model for the cotranslational formation of the SET1RC complex and maturation of SET1C in wild-type (upper panel) and Δswd1 strains (lower panel) (see text for details). Nascent Set1 protein is indicated by a red line, SET1 mRNA is indicated by a black line. Mature Set1 and other SET1C subunits are indicated as ovals, ribosomes are depicted in turquoise. Proteins found in SET1RC are indicated with bold letters. For reasons of simplicity, the representation does not consider known protein–protein interactions and stoichiometries of subunits associated within SET1C.
Discussion
In a search for RNA molecules co-purifying with SET1C, we found SET1 mRNA as the only transcript that stably bound to the Shg1 subunit under the conditions described in this study. This observation led to the identification of SET1RC, a complex including SET1 mRNA and four out of eight polypeptides, which also constitute SET1C. We propose that SET1RC assembles cotranslationally on nascent Set1 protein and that SET1RC has an effect as a transitional complex to coordinate Set1 expression and the assembly of SET1C.
A major conundrum of our observations was how the specific association of SET1 mRNA with SET1RC was established. Our analyses defined the SET1RC complex to minimally consist of SET1 mRNA in association with Set1, Swd1, Spp1 and Shg1. As Swd1, Spp1 and Shg1 can directly bind to GST–Set1 in pull-down assays, and as the four proteins are also components of SET1C, we consider it likely that they are also present in the same complex to form SET1RC. We show that Swd1, Spp1 and Shg1 were not essential for SET1 mRNA binding, suggesting that only Set1 was required. Unexpectedly, we found that RNA binding occurred independently of canonical Set1 RNA-binding domains and was also associated with the C-terminal half of the protein. Another key observation was that RNA binding was restricted to proteins and to their encoding transcript, and did not occur ‘in trans' when protein and target mRNA were derived from different genes. Moreover, SET1RC did not include RNAs that were nuclear restricted, arguing that association required cytoplasmic localization of the RNA. Consistent with this idea, we could detect Shg1–TAP in post-nuclear fractions. Each of these observations can be coherently accounted for if SET1 mRNA binding was mediated by a nascent Set1 protein that emerged from ribosomes during translation of its mRNA. Although it is trivial that a nascent protein interacts with its mRNA during translation, the exciting observation here was that other SET1C proteins (Shg1, Spp1 and Swd1) also bound to SET1 mRNA, suggesting that this association occurred in a cotranslational manner.
Importantly, we provide several independent lines of evidence supporting the idea that SET1RC is indeed a translation-dependent complex. We show that binding of Shg1–TAP to SET1 mRNA was sensitive to both EDTA and to the translation inhibitor puromycin. Furthermore, binding of SET1 mRNA by TAP–Set1 remained unchanged in strains lacking Swd1 or Swd3 (Figure 2A), although steady-state levels of TAP–Set1 were strongly reduced (Figure 6A). The observation that TAP–Set1 translation continues normally in the absence of Swd1 or Swd3 is fully consistent with the idea that binding of nascent TAP–Set1 to SET1 mRNA was detected in these experiments. Finally, we show that binding of ProtA–Set1 to its mRNA was reduced when translation initiation was inhibited in a prt1-1 mutant background at non-permissive temperature (Figure 4D).
The association of Swd1, Spp1 and Shg1 within SET1RC may define the early steps of the SET1C assembly pathway, which are initiated as soon as interaction domains of nascent Set1 emerge from the ribosome (Figure 7). The binding site for Swd1 on Set1 has not yet been mapped, but it does not include the C-terminal SET domain (aa 900–1081, Figure 2B; Dehe et al, 2006). Shg1 recognizes a central domain of Set1 within RRM2, and Spp1 binds in the vicinity of the n-SET region (Figure 2B). A further maturation of the complex is probably preceded by, or coincides with, the release of full-length Set1 and associated proteins from the ribosome. Therefore, certain steps of SET1C assembly probably occur post translationally. We expect that Swd3 joins the complex through an interaction with the Swd1 subunit, as the two proteins are known to form a heterodimer (Roguev et al, 2001; Dehe et al, 2006). The failure to detect an Swd3-SET1 mRNA association may indicate a post-translational association of Swd3 with Swd1. It seems possible that Sdc1 and Bre2 associate as a pre-assembled heterodimer (Roguev et al, 2001; Dehe et al, 2006). These proteins interact through a direct binding of Bre2 to the SET domain, which is located close to the C-terminal end of Set1 (Dehe et al, 2006; BD, unpublished results), providing a plausible explanation as to why they are not components of SET1RC. Currently, we cannot clearly define the entry point of Swd2 in this assembly pathway. The observation that Swd2 is required for a stable accumulation of Set1 (Dichtl et al, 2004; Nedea et al, 2008) may indicate that it will join the complex early. However, the protein was suggested to interact dynamically with SET1C on chromatin (Lee et al, 2007), opening the possibility that it joins the complex when it is bound to nucleosomes. The assembly pathway of SET1C may therefore have both cytoplasmic and nuclear phases or, alternatively, the composition of the nuclear form of the complex may be dynamic.
Most interestingly, the formation of certain cytoskeletal structures, including myosin and vimentin, has been suggested to involve cotranslational assembly processes (Fulton and L'Ecuyer, 1993). To the best of our knowledge, our experiments provide evidence for the first time that assembly of a soluble multi-protein complex is initiated on a nascent protein during translation in the cytosol. It seems likely that similar pathways are in place to assemble other multi-protein complexes. Research over the last decade showed a remarkable interdependence of cellular processes (Maniatis and Reed, 2002; Orphanides and Reinberg, 2002), but what is the significance and advantage of coupling assembly of a complex to translation? An obvious advantage would be to ensure an ordered assembly of interacting subunits once the respective binding sites emerge from the ribosome. It seems possible that the resulting interactions may have an effect on the folding of the nascent polypeptide chain or in the stabilization of folded domains. Notably, there are well-documented examples of chaperone proteins that aid the folding of nascent proteins both in prokaryotic and eukaryotic cells (Bukau et al, 2000; Frydman, 2001; Hartl and Hayer-Hartl, 2002). Considering the crowded environment of the cytosolic compartment, cotranslational complex formation may further monitor the association of the correct proteins at the right time in the assembly pathway. The availability of interaction partners may have an immediate effect on the functionality and fate of the translated protein or in the resulting assembled complex. This possibility is well exemplified here with Swd1 or Swd3 subunits of SET1C. The absence of either protein resulted in a severe under-accumulation of Set1, whereas SET1 mRNA transcription, SET1RC formation and Set1 translation seemed to function normally. These observations suggest that the maturation of the complex did not proceed after release from the ribosome and it seems likely that the newly translated Set1 was instead channelled into a turnover pathway. Similarly, over-expression of SET1 mRNA resulted only in a modest increase in protein levels and over-expression of HA–Set1 resulted in reduced TAP–Set1 levels in the same cells. This effect most likely reflects the limited availability of the remaining SET1C subunits and suggests that cotranslational protein interactions are required to stabilize newly synthesized Set1.
Although our experiments suggest that assembly of SET1C is initiated during translation, we cannot exclude the possibility that post-translational assembly will occur in a parallel pathway and it remains unclear whether cotranslational protein interactions are obligatory for successful Set1 synthesis. The answer to this question will likely depend on the mechanisms and kinetics of Set1 protein degradation, which have not been addressed in this study. Initial experiments showed, however, that treatment of Set1-over-expressing yeast cells with the inhibitor MG132 stabilized the degradation products of Set1, pointing to proteasomal involvement (AH and BD, unpublished results). Most interestingly, earlier studies indicate that, in S. cerevisiae, nascent proteins can be targeted for proteasomal degradation in significant amounts (50% of all nascent peptides) and that a kinetic competition may exist between protein folding and cotranslational degradation (Turner and Varshavsky, 2000). Therefore, it is tempting to speculate that cotranslational protein interactions may enhance the efficiency of complex formation by protecting nascent Set1 from degradation. Such a mechanism would be an efficient means to match the amount of newly synthesized Set1 with the availability of other SET1C subunits. The Swd3 homologue, WDR5, is known to contribute to the recognition of the H3K4 substrate and it is required for the formation of H3K4me3 on target genes and for normal metazoan development (Wysocka et al, 2005). Therefore, adverse effects on gene expression and cell growth seem likely when SET1C complexes are not assembled with the complete set of subunits at proper stoichiometry. The cell could escape such situations by coupling the synthesis and stable accumulation of Set1 to the assembly of the complex.
Materials and methods
Growth and construction of yeast strains
Growth and handling of yeast was carried out by standard techniques. Yeast was grown in a rich YPD medium (2% glucose, 2% bacto-tryptone, 1% yeast extract) or in a synthetic drop-out medium when selection for plasmid-dependent growth was required (2% glucose, 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 1 × aa). The strains used in this study are listed in Supplementary Table S1. Chromosomal targeting for tagging the SET1 gene with Myc-epitope was carried out as described (Longtine et al, 1998). Gene deletion was performed as described (Rothstein, 1991), by transferring kanMX4-disrupted alleles from the Euroscarf yeast gene deletion collection (Winzeler et al, 1999).
Oligonucleotides
Oligonucleotides used in this study for cloning, strain construction, reverse transcription and PCR are listed in Supplementary Table S2.
Constructs
ProteinA fusions of SET1 were produced by PCR amplification of DNA encoding the indicated aa regions and cloning of PCR products into the NcoI and BamHI or NcoI and SalI sites of pUN100-NOP1p-ProtA-TEV-ADH1t (Senger et al, 1998); the resulting plasmid constructs were as follows: pHZ724 (NcoI-XmaI; aa 1–1081), pHZ671 (NcoI-BamHI; aa 1–571), pHZ672 (NcoI-SalI; aa 572–1081), pHZ725 (NcoI-BamHI; aa 1–571 YF/AA) and pHZ726 (NcoI-BamHI; aa 1–571 H/A). For expression of SET1 fragments with T7 RNA polymerase, SET1 sequences were cloned into pRS426 with oligonucleotide-encoded HindIII and NotI restriction sites, generating plasmids pHZ692 (5′ half) and pHZ693 (3′ half). The plasmid encoding nuclear-targeted T7 RNA polymerase (pRS315 nlsT7RNAP) was a kind gift from M Rosbash (Brandeis, USA).
Isolation of RNA associated with TAP-tagged proteins
Purification of RNA bound to TAP-fusion proteins was carried out as described (Gerber et al, 2004), with modifications. One-and-a-half litres of yeast was grown in an appropriate medium to an OD600 nm of 0.6–0.8 and was collected in the presence of 0.1 mg/ml cycloheximide. Cell pellets were washed twice with 25 ml ice-cold buffer A (20 mM Tris–HCl pH 8.0, 140 mM KCl, 2 mM MgCl2, 0.1% NP-40, 0.2 mg/ml heparin, 0.1 mg/ml cycloheximide) and frozen as drops in liquid nitrogen. Cells were disrupted with three 4-min bursts in a Mixer Mill MM301 (Retsch, Haan, Germany) and the cell powder was re-suspended in cold buffer B to a final volume of 15 ml (buffer A supplemented with 0.5 mM DTT, 1 mM PMSF, 100 U/ml RNase OUT (Invitrogen), one tablet complete mini protease inhibitors EDTA free (Roche) and 20 U/ml RQ1 DNase (Promega)). The suspension was cleared by three successive centrifugation steps at 4°C with increasing r.p.m. (15′/4 kr.p.m., 15′/8 kr.p.m. and 40′/12 kr.p.m.). Buffer B was added to the supernatant to a final volume of 9 ml to produce an extract with a protein concentration of 10–25 mg/ml. This material served for total RNA extraction, for western blot analysis or for binding to IgG beads; 4 ml of the obtained extract was bound to 200 μl slurry of IgG-Sepharose 6 Fast Flow (Amersham Biosciences) for 2 h at 4°C. The beads were then washed twice for 15 min in 5 ml buffer B and thrice for 15 min in 10 ml buffer C (20 mM Tris–HCl pH 8.0, 140 mM KCl, 2 mM MgCl2, 10% glycerol, 0.5 mM DTT, 0.01% NP-40, 10 U/ml RNase OUT). The beads were then re-suspended in 200 μl buffer C with 0.25 U/μl AcTEV protease (Invitrogen) and incubated for 2 h at 15°C. The supernatant and one 200 μl wash with buffer C were collected and combined to obtain ∼0.4 ml of the final TEV eluate. The eluate was then cleaned with the RNeasy MicroElute Cleanup kit from Qiagen, according to the manufacturer's instructions.
RNA extraction, reverse transcription and qPCR
RNA extraction was carried out using the hot-phenol method as described earlier (Dichtl et al, 2004). All RNA samples that were analysed by reverse transcription were first treated with DNase. For this purpose, 1 μg of RNA was incubated with 1 U RQ1 DNase (Promega) at 37°C for 30 min with the supplied reaction buffer. The reaction was stopped by the addition of 1 μl stop solution and DNase was inactivated at 65°C for 10 min. Reverse transcription was performed using 25–150 ng of Dnase-treated RNA with the Reverse Transcriptase Core kit from Eurogentech, according to the manufacturer's instructions. qPCR analysis was carried out on an Applied Biosystems 7900HT fast real-time PCR system with the following temperature profile: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 1 min; 95°C for 15 s, 60°C for 15 s and 95°C for 15 s. The PCR reaction was based on the SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's instructions. In co-purification experiments, all ct values for SET1 mRNA or other mRNAs, such as the RPS7A control mRNA, were normalized to levels for ACT1 mRNA. This was carried out for RNA isolated from total extract (total) or from immunopurified material (IP). The ratio between the IP and the total resulted in the fold-enrichment factor that is indicated in the figures.
Polysome analysis
Polysome analysis was carried out essentially as described (Halbeisen and Gerber, 2009). Cell lysates were prepared from a 100 ml exponential growth culture (OD600 nm 0.6). At the time of collection, cycloheximide was added to a final concentration of 0.1 mg/ml in the presence of ice. Cells were pelleted by centrifugation at 4000 r.p.m. for 5 min at 4°C and washed twice in 2.5 ml of lysis buffer (20 mM Tris–HCl pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 0.1 mg/ml cycloheximide, 1 mg/ml heparin). For cell lysis, the pellet was re-suspended in 700 μl of lysis buffer. A 400 μl volume of glass beads was added and the reaction mixture was vortexed at full speed for 20 s, followed by incubation on ice for 100 s. This process was repeated four times. After cell lysis, excess cell debris and glass beads were removed in a two-step centrifugation performed for 5 min at 4700 r.p.m. and for 5 min at 9500 r.p.m. at 4°C. The obtained cell lysate was brought to a final volume of 1 ml with lysis buffer. Cell lysate of 0.8 ml was loaded on a 10–50% sucrose gradient that contained lysis buffer lacking Triton X-100. The samples were sedimented in a Beckman SW41 rotor at 4°C for 160 min at 35 000 r.p.m. and collected, whereas the A254 value was monitored using a continuous flow cell UV detector. RNA was extracted from polysome fractions (100 μl) with RNeasy Mini columns according to the manufacturer's instructions (Qiagen). A spike RNA (0.2 ng) was added to each fraction before RNA extraction. The obtained RNA was precipitated overnight with LiCl (1.5 M final concentration) to remove residual heparin.
Microarrays, sub-cellular fractionation and bioinformatic analysis
Microarray experiments, sub-cellular fractionation and bioinformatic analysis are described in Supplementary data. Microarray data were deposited at GEO (www.ncbi.nlm.nih.gov/geo; GEO accession: GSE15247).
Supplementary Material
Supplementary Data
Review Process File
Acknowledgments
We are grateful to T Scherrer for help with DNA microarray analysis and polysomal gradients, to Werner Boll for assistance in fluorescence microscopy and to Erica Bogan for improving the paper. We thank F Stewart, A Roguev and A Hinnebusch for yeast strains; P Nagy for anti-Set1 and D Shore for anti-Rap1 antibodies, as well as R Erdmann and R Rucktäschel for anti-Pgk1 antibodies; M Rosbash for T7 RNAP plasmids, E Hurt for the ProteinA-fusion vector and Mai Thuong Pham for the HA–Set1 construct. IG was supported by a Müller fellowship through the Zürich Molecular Life Sciences PhD Program. APG was supported by a Career Development Award from the International Human Frontier Science Program Organization (HFSP). The study conducted in V Géli's laboratory is supported by ‘La Ligue Nationale contre le Cancer' (équipe labellisée). This study was supported by the University of Zürich and by grants from the Swiss National Science Foundation to BD (PP00A-102941 and PP00A3_120490/1).
References
- Ackerman SH, Tzagoloff A (2005) Function, structure, and biogenesis of mitochondrial ATP synthase. Prog Nucleic Acid Res Mol Biol 80: 95–133 [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB (2000) Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Deuerling E, Pfund C, Craig EA (2000) Getting newly synthesized proteins into shape. Cell 101: 119–122 [DOI] [PubMed] [Google Scholar]
- Cheng H, He X, Moore C (2004) The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol 24: 2932–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Vallon O (2000) Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ (2007) Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics 6: 439–450 [DOI] [PubMed] [Google Scholar]
- Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, Shevchenko A, Rosaleny LE, Tordera V, Chavez S, Stewart AF, Geli V (2006) Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281: 35404–35412 [DOI] [PubMed] [Google Scholar]
- Dehe PM, Geli V (2006) The multiple faces of Set1. Biochem Cell Biol 84: 536–548 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Aasland R, Keller W (2004) Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA 10: 965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Dower K, Rosbash M (2002) T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14: 319–327 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Fulton AB, L'Ecuyer T (1993) Cotranslational assembly of some cytoskeletal proteins: implications and prospects. J Cell Sci 105 (Part 4): 867–871 [DOI] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Gerber AP (2009) Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol 7: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krajewski WA, Nakamura T, Mazo A, Canaani E (2005) A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol 25: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA 99: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL (2008) The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell 29: 577–587 [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG (2004) Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J 23: 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D (2002) A unified theory of gene expression. Cell 108: 439–451 [DOI] [PubMed] [Google Scholar]
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G (2002) The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol 14: 305–312 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194: 281–301 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Schlichter A, Cairns BR (2005) Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J 24: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J 17: 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Woolford JL Jr (2009) Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol 21: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13: 852–854 [DOI] [PubMed] [Google Scholar]
- Tresaugues L, Dehe PM, Guerois R, Rodriguez-Gil A, Varlet I, Salah P, Pamblanco M, Luciano P, Quevillon-Cheruel S, Sollier J, Leulliot N, Couprie J, Tordera V, Zinn-Justin S, Chavez S, van Tilbeurgh H, Geli V (2006) Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol 359: 1170–1181 [DOI] [PubMed] [Google Scholar]
- Turner GC, Varshavsky A (2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289: 2117–2120 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Review Process File