Abstract
We report the nucleotide sequence of two novel cryptic plasmids (4357 and 14 662 base pairs) carried by a Yersinia enterocolitica biotype 1A strain isolated from pork. As distinguished from most biotype 1A strains, this isolate, designated 07-04449, exhibited adherence to eukaryotic cells. The smaller plasmid pYe4449-1 carries five attributable open reading frames (ORFs) encoding the first CcdA/CcdB-like antitoxin/toxin system described for a Yersinia plasmid, a RepA-like replication initiation protein, and mobilizing factors MobA and MobC. The deduced amino acid sequences showed highest similarity to proteins described in Salmonella (CcdA/B), Klebsiella (RepA), and Plesiomonas (MobA/C) indicating genomic fluidity among members of the Enterobacteriaceae. One additional ORF with unknown function, termed ORF5, was identified with an ancestry distinct from the rest of the plasmid. While the C+G content of ORF5 is 38.3%, the rest of pYe4449-1 shows a C+G content of 55.7%. The C+G content of the larger plasmid pYe4449-2 (54.9%) was similar to that of pYe4449-1 (53.7%) and differed from that of the Y. enterocolitica genome (47.3%). Of the 14 ORFs identified on pYe4449-2, only six ORFs showed significant similarity to database entries. For three of these ORFs likely functions could be ascribed: a TnpR-like resolvase and a phage replication protein, localized each on a low C+G island, and DNA primase TraC. Two ORFs of pYe4449-2, ORF3 and ORF7, seem to encode secretable proteins. Epitope-tagging of ORF3 revealed protein expression at 4°C but not at or above 27°C suggesting adaptation to a habitat outside swine. The hypothetical protein encoded by ORF7 is the member of a novel repeat protein family sharing the DxxGN(x)nDxxGN motif. Our findings illustrate the exceptional gene pool diversity within the species Y. enterocolitica driven by horizontal gene transfer events.
1. Introduction
Pathogenicity within the genus Yersinia typically relies on the presence of the virulence plasmid pYV encoding a type 3 secretion system (T3SS) [1]. The enteropathogenic species Yersinia enterocolitica is subdivided into nonpathogenic (biotype 1A), high-pathogenic (biotype 1B) and low-pathogenic (biotypes 2–5) representatives, based on their virulence potential in the oral mouse infection model [2]. Isolates of Y. enterocolitica biotype 1A typically lack the virulence plasmid and other known chromosomally encoded virulence factors and are, therefore, considered avirulent in general. However, several lines of evidence indicate that at least some biotype 1A strains are pathogenic to human provoking gastrointestinal symptoms indistinguishable from those caused by Y. enterocolitica strains harbouring the virulence plasmid [3, 4]. Biotype 1A strains frequently harbour plasmids of various size [5] that could carry novel virulence determinants to explain the apparent pathogenicity of certain strains. A major animal reservoir of Y. enterocolitica is swine resulting in contaminated pork as important transmission route [6].
Among 20 arbitrarily selected strains of Y. enterocolitica biotype 1A isolated from patients with diarrhoea, of animal source or from environmental sampling, a single isolate, designated 07-04449, exhibited significant adherence to several eukaryotic cell lines. This strain, isolated from pork, harbours two novel plasmids which we have sequenced in the search of novel virulence determinants.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
Y . enterocolitica biotype 1A isolates were selected from the strain collection of the National Reference Centre for Salmonella and other enteric pathogens, Robert Koch Institute, Wernigerode, Germany. All strains were routinely cultured in Luria-Bertani (LB) broth. To avoid possible stimulation of virulence gene expression, all strains were cultured at 27°C for standard procedures except for infection studies. Where appropriate, antibiotics were supplemented as follows: kanamycin at 30 μg/mL, streptomycin at 30 μg/mL.
2.2. Isolation of Plasmids
Plasmids carried by strain Yersinia enterocolitica 07-04449 were isolated by using a plasmid preparation kit (Qiagen). Plasmids were resolved on 0.7% agarose gels. Plasmids pYe4449-1 and pYe4449-2 were cut out from the agarose gel and were purified with the QIAquick gel extraction kit (Qiagen).
2.3. Cell Adhesion Analyses
Culturing of eukaryotic cell lines and analysis of adhesion of yersiniae to cell lines Int-407, HeLa, Hep-2, and IEC-6 were performed essentially as described [7]. Int-407, Hep-2, and HeLa cells were cultured in EMEM medium (Cell Concepts) supplemented with 1% nonessential amino acids, 2 mM glutamine, and 5% heat-inactivated fetal calf serum (FCS), and IEC-6 cells were cultured in DMEM (Cell Concepts) supplemented with 2 mM glutamine, 5% heat-inactivated FCS, and 0.1 IU/mL insulin. For infection experiments, eukaryotic cells were cultivated in SonicSeal slide wells (Nunc). After infection with yersiniae (multiplicity of infection of 50), cells were incubated for 3 hours at 37°C in the presence of 5% CO2, then washed five times with PBS, fixed in methanol for 5 minutes, stained with Accustain modified Giemsa stain (Sigma), and examined under the microscope.
2.4. Sequencing and Assembly
Plasmid pYe4449-1 was incubated with the EZ-Tn5 <KAN-2> Tnp Transposome kit (Epicentre Biotechnologies) in vitro, transformed into E. coli Top10 (Invitrogen) and selected for kanamycin resistant clones. Plasmids from four colonies were isolated, and sequencing reactions were performed with the transposon-specific primers provided with the transposome kit. Gaps of pYe4449-1 were filled by primer walking sequencing. Plasmid pYe4449-2 was digested with restriction endonuclease BamHI. One restriction fragment of approximately 0.8 kbp was cut out from an agarose gel, purified, and inserted into vector pMOSBlue (pMOSBlue Blunt Ended Cloning Kit, Amersham Biosciences). The vector was transformed into E. coli Top10. Grown colonies were picked and plasmids were isolated as described earlier. The cloned pYe4449-2 fragment was sequenced with pMOSBlue-specific primers. Subsequently, the rest of the plasmid was sequenced by primer walking. Sequence coverage was approximately 4-fold for both plasmids. The sequences were assembled with DNASTAR Lasergene v7.2 SeqMan Pro. The nucleotide sequences of plasmids pYe4449-1 and pYe4449-2 have been deposited in the GenBank database (accession numbers FJ696405 and FJ696406, resp.).
2.5. Sequence Analyses
Potential coding regions were determined by softberry (http://www.softberry.com/) tools and confirmed applying Glimmer (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). Comparison of nucleotide sequences of pYe4449-1 and pYe4449-2 and deduced protein sequences with sequences available in databases was accomplished using various NCBI (http://www.ncbi.nlm.nih.gov/) tools. However, C+G islands, direct and inverted repeats were identified with programs provided by the Mobyle bioinformatics portal (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py). Superhelically induced duplex destabilization (SIDD) sites were identified applying WebSIDD (http://www.genomecenter.ucdavis.edu/benham/sidd/websidd.php; [8]). Search for putative protein secretion signals was performed with the SignalP server (http://www.cbs.dtu.dk/services/SignalP/; [9]). Possible phosphorylation sites were identified using the NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/). Protein secondary structure analyses were performed with different ExPASy tools (http://www.expasy.ch/).
2.6. Plasmid Copy Number Determination
Copy number of plasmids was estimated as described by Singh and Banerjee [10] by comparing total amount of chromosomal DNA and plasmid DNA using the assumption that the size of the chromosome of Y. enterocolitica 07-04449 is similar to that of sequenced strain Y. enterocolitica 8081, which is 4.62 Mbp [11].
2.7. Plasmid Curation
Attempts to cure plasmids pYe4449-1 and pYe4449-2 from Y. enterocolitica 07-04449 were accomplished following standard procedures [12–14]. In brief, curing agents ethidium bromide (500 μg/mL), acridine orange (450 μg/mL), or promethazine (120 μg/mL) were added to overnight cultures diluted 100-fold in LB broth and subsequently incubated for 24 hours at 37°C. Dilution series were plated on LB agar and incubated overnight at 37°C. Loss of plasmids was checked by PCR and plasmid isolation.
2.8. Mating Experiments
Plasmids pYe4449-1 and pYe4449-2 each were tagged with a kanamycin resistance cassette (KmR) by in vitro transposon mutagenesis applying the EZ-Tn5 <KAN-2> Tnp Transposome kit (Epicentre Biotechnologies). Donor strain Y. enterocolitica 07-04449 was electroporated with EZ-Tn5 <KAN-2> transposon-tagged plasmid pYe4449-1 and pYe4449-2, respectively, and mated with recipient E. coli TOP10 beforehand selected for spontaneous streptomycin resistance. Overnight cultures of donor and recipient strains grown at 27°C were diluted 1:40 in 2xYT medium supplemented with selecting antibiotics and incubated for 2 hours at 27°C. Also, 1 mL of donor and recipient cultures were washed twice and resuspended in 500 l μL 2xYT medium. Donor and the recipient were subsequently mixed and centrifuged at 3000 g for 2 minutes, resuspended in 1 mL 2xYT medium and were plated on a nutrient broth agar plate. After five hours of incubation at 27°C, the bacteria were washed from the plate, the suspension was centrifuged at 3000 × g for 2 minutes, resuspended in 200 l μL 2xYT medium, and plated on selective agar plates containing kanamycin and streptomycin. For comparison, occurrence of spontaneous streptomycin resistance of donor strain Y. enterocolitica 07-04449 was determined in parallel.
2.9. Plasmid Constructions
For epitope-tagging of ORF3 and ORF7 products, the respective genes were amplified by PCR including the putative promoter regions as predicted by the softberry package (http://www.softberry.com/). To amplify ORF3, primers 5′-GCTGAAAGCTTCTACGTGAGCACGCTTCTGCG-3′ and 5′-CGCAGTCGACTCAATGGTGATGGTGATGGTGCGATCCTCTCCAACCCTGCCCGCTCCAG-3′ were used thereby introducing HindIII and SalI restriction sites as indicated (underlined) and a sequence encoding the Arg-Gly-Ser-(His)6 epitope (doubly underlined). Analogously, primers 5′-GCTGAAAGCTTGTGGCATTCACCAGACATCGC-3′ and 5′-CGCAGTCGACTCAATGGTGATGGTGATGGTGCGATCCTCTATTACAGTAAGTATTTCCCAGGCT-3′ were used to amplify ORF7. PCR products were restricted with HindIII and SalI and ligated into plasmid pACYC184 (New England Biolabs) which was also cut with HindIII and SalI. Resulting plasmids termed pACYC184/ORF3-RGS(H)6 and pACYC184/ORF7-RGS(H)6 were transformed into insertional mutant strains carrying pYe4449-2/ORF3::Tn5 and pYe4449-2/ORF7::Tn5, respectively.
2.10. TCA Precipitation of Culture Supernatant: SDS-PAGE and Western Blotting
Precipitation of proteins from culture supernatant with trichloroacetic acid (TCA), SDS-PAGE and Western blotting, has been recently described [15]. Monoclonal antibody raised against the RGS(H)4 epitope was purchased from Qiagen.
3. Results and Discussion
3.1. The Adhesive Phenotype of Yersinia enterocolitica 07-04449
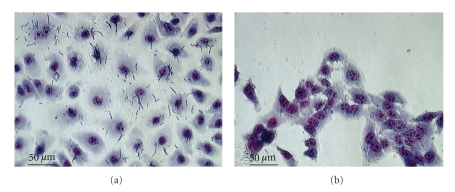
We screened 20 isolates of Y. enterocolitica biotype 1A from Germany for their potential to adhere to HeLa, Int-407, Hep-2, and IEC-6 eukaryotic cell lines as a first indicator of pathogenicity. Only one strain, termed 07-04449, which was obtained from pork, showed significant adherence to eukaryotic cells. On average, 4.2 +/− 0.5 bacteria of strain 07-04449 were found attached to each Int-407 cell, whereas only 0.2 +/− 0.2 bacteria of other strains were found attached to each Int-407 cell. Most pronounced adherence of the 07-04449 isolate was found in interaction with rat intestinal cell line IEC6 (Figure 1(a)). Interestingly, we observed that 0704449 yersiniae attaching to IEC-6 cells were markedly elongated compared to planktonic ones or compared to 07-04449 cells attached to HeLa, Int-407 or Hep-2 cells, respectively (cf. Figure 1(a) and Figure 1(b)). These findings stimulated us to search for possible plasmid-encoded adherence- and pathogenicity-related factors. We identified two plasmids, termed pYe4449-1 and pYe4449-2 that were completely sequenced.
Figure 1.
Yersinia enterocolitica biotype 1A strain 07-04449 adhering to eukaryotic cells. Rat intestinal IEC-6 cells (a) and human intestinal Int-407 cells (b) were infected with strain 07-04449 at a multiplicity of infection (MOI) of 50, stained with Giemsa and subjected to microscopy.
3.2. Characterisation of pYe4449-1 (4357 Base Pairs)
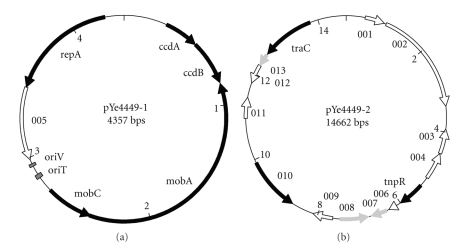
The smaller plasmid, pYe4449-1, consists of 4357 base pairs (bps) and a C+G content of 53.7%. For comparison, the C+G content of the Y. enterocolitica genome is 47.3% [11] and the C+G content of the recently sequenced 95.5 kbp plasmid pYE854 from another Y. enterocolitica biotype 1A strain was 42.3% [16]. Sequence analysis revealed six open reading frames (ORFs) with a minimum coding capacity of 50 amino acids (Figure 2(a) and Table 1). Plasmid pYe44491 encodes an antitoxin/toxin system, likely conducive to plasmid maintenance, with the highest similarity to the Salmonella enterica subsp. enterica CcdA/B system (70% and 68% identity, resp.). This is the first report of a CcdA/B family antitoxin/toxin system associated with a Yersinia plasmid. The chromosomally encoded CcdA/B-like system of the high-pathogenicity Y. enterocolitica strain 8081 is more divergent with 53% and 48% identity, respectively. Interestingly, several attempts to cure pYe44491 following different protocols failed, indicating that the CcdA/B system is functional.
Figure 2.
Maps of plasmids pYe4449-1 and pYe4449-2 . Open reading frames (ORFs) with attributable functions and e-values below e-20 are indicated by black arrows, ORFs with significant homology to database entries (e-values below e-10) but unclear functions are indicated by grey arrows, and ORFs without significant homology (e-values above 0.1) to database entries are indicated by white arrows. Details about identified ORFs are given in Tables 1 and 2.
Table 1.
ORFs identified on plasmid pYe4449-1.
| Name1 | Position (bp) | Strand | Size (aa) | MW2 (kDa) | pI2 | Identities | Expect | Best BlastP match3/function | Accession number |
|---|---|---|---|---|---|---|---|---|---|
| ccdA | 298–537 | + | 79 | 9 | 10.13 | 56/79 (70%) | 2e-26 | CdcA-like protein (Salmonella enterica subsp. enterica serovar Derby) | gb∣AAV53396.1∣ |
| ccdB | 537–845 | + | 102 | 11.06 | 5.02 | 70/102 (68%) | 5e-36 | cytotoxic protein CcdB(Salmonella enterica subsp. enterica serovar Schwarzengrund str. SL480) | ref∣ZP_02659678.1∣ |
| mobA | 848–2419 | − | 523 | 59.2 | 8.42 | 289/514 (56%) | 3e-133 | putative relaxase/mobilization nuclease MobA (Plesiomonas shigelloides) | emb∣CAB56514.1∣ |
| mobC | 2409–2735 | − | 108 | 12.5 | 10.18 | 84/109 (77%) | 4e-37 | putative mobilization protein (Plesiomonas shigelloides) | emb∣CAB56514.1∣ |
| ORF 5 | 2981–3481 | − | 166 | 18.7 | 6.41 | hypothetical protein | |||
| repA | 3481–4227 | − | 248 | 28.2 | 8.83 | 184/244 (75%) | 9e-103 | replication initiation protein A (Klebsiella sp. KCL-2) | ref∣NP_620615.1∣ |
1Gene names according to putative conserved protein domains if available.
2Molecular weights and isoelectric points are calculated using the algorithm from ExPASy's Compute pI/Mw program.
3Only significant BlastP matches with e-values below e-10 are listed; all others are indicated as “hypothetical protein” and show e-values above 0.1.
Directly adjacent to ccdA/B but in the opposite orientation, the mobilizing factors MobA and MobC are encoded. They show the highest similarity to Plesiomonas shigelloides proteins (45% and 77% identity, resp.). Further, an ORF encoding a RepA-like replication initiation protein was identified on pYe4449-1, sharing 75% identity with Klebsiella sp. KCL-2 RepA. Finally, a hypothetical protein without significant similarity to proteins in the databases is encoded by ORF5 (no hits with e-values below 0.1).
We analysed the C+G content of the plasmid in sections to identify possible horizontal gene transfer events and identified one low C+G island. Interestingly, this island colocalised with ORF5. While the C+G content of ORF5 is 38.3%, the rest of pYe4449-1 shows a C+G content of 55.7% suggesting a descent differing from its plasmid background. Downstream of ORF5, a palindromic sequence was identified capable of forming the 11-bp stem of a stem-loop hairpin likely involved in Rho-independent termination of the transcript associated with ORF5 [17].
Based on the finding of two mob genes with close homology to Plesiomonas shigelloides plasmid pUB6060 [18] (see Table 1), we compared the oriV and oriT origins described for pUB6060 [18, 19] to our pYe4449-1 sequence. We identified an oriV identical to that of pUB6060, positioned 2953CCTATATCAGATAACAGCCC2934 on pYe4449-1, and belonging to the ColE2-like replicons [18]. Also, an oriT could be identified in close proximity, positioned 2844GTAGAGGGGACGCTGTGTGTACTGGCTTA2872 (see Figure 2(a)). The oriT sequence only slightly deviates from that of pUB6060 (underlined bases are “C” in pUB6060). A recent phylogenetic analysis on conjugative relaxases allowed to classify MOB families [20]. According to this classification, MobA of pYe4449-1 together with MobA of pUB6060 belongs to the MOBHEN family of the MOBP5 clade.
Plasmid pYe4449-1 with the EZ-Tn5 <Kan-2> transposon insert was transformed into Y. enterocolitica 07-04449 to perform mating experiments. The tagged plasmid was efficiently transferred to E . coli Top10 cells indicating that the donor strain harbours a conjugation system. The estimated copy number for pYe4449-1 was 30–40 copies per cell in Yersinia and 10–20 copies per cell in E . coli.
3.3. Characterisation of pYe4449-2 (14662 base pairs)
The C+G content of the larger plasmid pYe4449-2 (54.9%) was similar to that of pYe4449-1 (53.7%). On the nucleotide level, the segment from bp position 12260 to 125 is 81% identical to bp segment 36316–34658 of a Klebsiella pneumoniae plasmid harbouring several antibiotic resistance genes (gb∣FJ223605.1∣). By contrast, the segment from bp position 126 to 5370 did not exhibit any significant similarity to deposited nucleotide sequences even when search options were optimized for “somewhat similar sequences.” We identified 14 ORFs with a minimal coding sequence length of 50 amino acids (Figure 2(b) and Table 2). Of the 14 ORFs identified, only six were significantly identical to sequences in the data bases (e-value below e-10). However, only 3 ORFs could be assigned to proteins with known functions, a resolvase, a phage protein, and a DNA primase.
Table 2.
ORFs identified on plasmid pYe4449-2.
| Name1 | Position (bp) | Strand | Size (aa) | MW2 (kDa) | pI2 | Identities | Expect | Best BlastP match3/function | Accession |
|---|---|---|---|---|---|---|---|---|---|
| ORF 1 | 446–913 | + | 155 | 16.8 | 5.3 | hypothetical protein | |||
| ORF 2 | 957–3410 | + | 817 | 86.4 | 9.9 | hypothetical protein | |||
| ORF 3 (yltA) | 3933–4511 | − | 192 | 20.9 | 6.2 | protein with unknown function expressed at low temperature | |||
| ORF 4 | 4511–5146 | − | 211 | 23.2 | 5.4 | hypothetical protein | |||
| tnpR | 5371–5997 | + | 208 | 23.3 | 9.6 | 92/208 (44%) | 2e-46 | Resolvase, N-terminal domain (Marinobacter aquaeolei VT8); TnpR-like protein | ref∣YP_957859.1∣ |
| ORF 6 | 6107–6301 | + | 64 | 7.1 | 10.8 | hypothetical protein | |||
| ORF 7 (ysrA) | 6361–6744 | + | 127 | 13.5 | 4.2 | 42/113 (37%) | 3e-10 | hypothetical protein BACOVA_04330 (Bacteroides ovatus ATCC 8483); putative repeat protein | ref∣ZP_02067325.1∣ |
| ORF8 | 6795–7478 | − | 227 | 25.3 | 5.3 | 52/127 (40%) | 8e-18 | hypothetical protein (Aeromonas hydrophila) | gb∣AAR06625.1∣ |
| ORF 9 | 7643–8119 | + | 158 | 17.6 | 10.2 | hypothetical protein | |||
| ORF 10 | 8637–9917 | − | 426 | 48.6 | 8 | 157/386 (40%) | 2e-80 | predicted P-loop ATPase (Bacteriophage APSE-3) | gb∣ACJ10118.1∣ |
| ORF 11 | 10971–11462 | + | 163 | 18.2 | 6.1 | hypothetical protein | |||
| ORF 12 | 11798–12250 | − | 150 | 16.5 | 5.8 | hypothetical protein | |||
| ORF 13 | 12247–12534 | − | 95 | 9.9 | 4.5 | 72/95 (75%) | 1e-28 | hypothetical protein pKlebpneu12_p49 (Klebsiella pneumoniae) | ref∣YP_002286974.1∣ |
| traC | 12592–13803 | − | 403 | 44.8 | 6.3 | 345/401 (86%) | 0 | hypothetical protein pKlebpneu12_p48 (Klebsiella pneumoniae) | ref∣YP_002286973.1∣ |
1Gene names according to putative conserved protein domains if available.
2Molecular weights and isoelectric points are calculated using the algorithm from ExPASy's Compute pI/Mw program.
3Only significant BlastP matches with e-values below e-10 are listed; all others show e-values above 0.1 and are either indicated as “hypothetical protein” or experimental confirmation is specified.
We identified a TnpR-like resolvase gene as typically associated with transposons. The hypothetical protein showed highest similarity to a Marinobacter aquaeolei TnpR resolvase (Table 2). The tnpR gene of pYe4449-2 is localised on a low C+G island together with ORF6 and ORF7 (Figure 3(b)), suggesting their ancient collective transfer to the plasmid via transposition. While the role of ORF6 is completely unclear, the features of ORF7, sharing homology with a few uncharacterised proteins, are detailed in Section 3.4.
Figure 3.
ORF7 of plasmid pYe4449-2 encodes a putative new repeat protein with a high content of phosphorylatable amino acids. (a) Scheme of the arrangement of the ORF7-encoded hypothetical protein. The nonameric repeat module “RTDSLGNTY” occurring twice in each repeat is indicated by dark grey boxes. (b) Alignment of the protein repeat segments encoded by ORF7, residues identical in all three repeats are highlighted against a dark grey background and residues identical in two of the three repeats are highlighted against a light grey background. The repeat module “RTDSLGNTY” occurring twice in each repeat is indicated by a bar below the consensus sequence. Below these bars, the interspecies repeat motif “DxxGN(x)nDxxGN” is given as deduced from the closest homologues of ORF 7 (BACOVA_04330; NE2449; MPG2080_02246). Serine and threonine residues most likely to be phosphorylated in a eukaryotic background according to NetPhos 2.0, server predictions (score 0.968 and above) were indicated by circles.
ORF10, which is situated in another low C+G region (Figure 3(b)) encodes a hypothetical protein 40% identical to a P-loop ATPase of APSE bacteriophages. APSE phages are lambda-like phages associated with the genomes of facultative endosymbionts such as Hamiltonella defensa that are closely related to Yersinia and found in aphids like Acyrthosiphon pisum and other sap-feeding insects [21, 22]. It is interesting to note that lysogenic APSE phages have been implicated in the carriage of virulence genes such as Shiga-like toxin, cytolethal distending toxin, and YD-repeat toxin genes [21]. This suggests that APSE-like phages might have contributed to pathogenicity of certain Y. enterocolitica biotype 1A strains by a carry-over of virulence determinants. Moreover, this finding lends support on the hypothesis that a regular part of the life cycle of Y. enterocolitica could be the colonization and infection of insects where genetic exchange with the facultative insect endosymbionts would be favoured. This hypothesis is substantiated by the endowment of Y. enterocolitica with insecticidal genes [23, 24].
As mentioned earlier, the segment from position 12 260 to 125 is highly related to a Klebsiella plasmid and, expectedly, ORFs 13 and 14 which are localised within this region exhibit highest similarity to Klebsiella proteins. While the role of ORF13 is unclear, ORF14 encodes DNA primase TraC suggesting that the plasmid is transferable. This was confirmed by tagging the plasmid with the EZ-Tn5 <Kan-2> transposon, retransformation into Y. enterocolitica 07-04449, and mating with E . coli Top10.
As described earlier for pYe4449-1, we have been also unsuccessful in curing pYe4449-2 following several protocols (see Section 2). This exceptional segregational stability indicates an important trait of both plasmids und suggests their usefulness to engineer novel cloning vectors.
No sequences homologous to known origins of replication could be identified on pYe4449-2. Superhelically induced duplex destabilization (SIDD; [8]) of pYe4449-2 was analysed to search for regulatory sequences that might indicate the positioning of the origin of replication [25]. SIDD analysis of the plasmid revealed a single highly significant site (8333-8463) representing a possible origin. Copy number of pYe4449-2 was approximately 15–20 in Y. enterocolitica 07-04449 and half of it in E . coli.
3.4. A Novel Secretable and Phosphorylatable Repeat Protein
Retrieval for the presence of repeats within the nucleotide sequences of both plasmids applying “etandem” of the Mobyle toolbox revealed a 108 nucleotide long triple tandem repeat sequence within ORF7 which is localized on a low C+G island of pYe4449-2. Subsequent analysis of the ORF7-deduced protein sequence identified three repeats, each 36 amino acids in length (Figure 3). Each repeat contains two nonameric repeat modules of the sequence RTDSLGNTY. The triple tandem organisation identified on the nucleotide and the translational level suggests that the likely functional unit contains two of these nonameric repeat modules. The closest homologues of the ORF7 product, Bacteroides ovatus BACOVA_04330, Nitrosomonas europaea NE2449, and MPG2080_02246 of a marine γ-proteobacterium are all hypothetical proteins with unknown functions. Multiple sequence analyses revealed that all these proteins are repeat proteins. From these repeats, we condensed the common repeat motif DxxGN(x)nDxxGN with a variable number of residues between two DxxGN sequences (Figure 3). BLAST searches revealed the existence of repeated arrangements of DxxGN(x)nDxxGN motifs in several predicted proteins (Table 3). The highest number of repeats, twenty of the form DxxGN(x)7DxxGN, was found in Pseudomonas aeruginosa hypothetical protein PLES_34491.
Table 3.
BLAST search for proteins containing the DxxGN(x)nDxxGN repeat motif.
| DxxGN(x)nDxxGN with n = | Number of repeats‡ | Reference, gene product, organism |
|---|---|---|
| 0 | 3 | CAQ39838.1, hypothetical protein, conserved in Plasmodium species, Plasmodium knowlesi strain H |
| 1 | 3 | ZP_03280828.1, hypothetical protein ENTCAN_00574, Enterobacter cancerogenus ATCC 35316 |
| 2 | 8 | YP_001743821.1, putative autotransporter, Escherichia coli SMS-3-5 |
| 8 | XP_001554189.1, predicted protein, Botryotinia fuckeliana B05.10 | |
| 3 | 4 | XP_002046888.1, GJ13133, Drosophila virilis |
| 3 | ZP_03508148.1, hypothetical protein RetlB5_24044, Rhizobium etli Brasil 5 | |
| 4 | 5§ | XP_002224392.1, hypothetical protein BRAFLDRAFT_86726, Branchiostoma floridae |
| 4* | XP_002206131.1, hypothetical protein BRAFLDRAFT_68704, Branchiostoma floridae | |
| 5 | 7 | ZP_03166503.1, hypothetical protein RUMLAC_00154 Ruminococcus lactaris ATCC 29176 |
| 6 | 7079543 LMHCC_2776, YD repeat protein, Listeria monocytogenes HCC23 | |
| 6 | 7 | ZP_02067325.1, hypothetical protein BACOVA_04330, Bacteroides ovatus ATCC 8483 |
| 6 | 7079543 LMHCC_2776, YD repeat protein, Listeria monocytogenes HCC23 | |
| 7 | 20 | YP_002441035.1, hypothetical protein PLES_34491, Pseudomonas aeruginosa LESB58 |
| 5 | XP_002226734.1, hypothetical protein BRAFLDRAFT_89165, Branchiostoma floridae | |
| 11¶ | 2 | XP_001635831.1, predicted protein, Nematostella vectensis |
‡BLAST hits with highest number of repeats were selected.
§Precisely, there are 11 consecutive DxxGN(x)4 repeats.
*Three of the four repeats are part of the triple tandem DxxGN(x)4DxxGN(x)4DxxGN.
¶For spacer regions (x)n with n > 7, a more specific search strategy is required to obtain significant hits; the repeat used for the Nematostella hit is DxLGN(x)11DxLGN.
Secondary structure prediction analyses of the ORF7 hypothetical protein indicate that it completely lacks α-helical elements whereas for almost 80% of the sequence, a random coil is predicted. Similar results were obtained for the ORF7 homologues and repeat-containing regions of proteins listed in Table 3. This suggests that the DxxGN(x)nDxxGN repeat motif triggers a specific conformation of functional importance. Due to the lack of significant sequence similarity to structurally resolved proteins, no three-dimensional prediction was possible using the ESyPred3D server [21].
The hypothetical protein encoded by ORF7 was further checked for a secretion signal using the SignalP server [9]. A highly significant signal sequence was identified (signal peptide probability: 0.998) and the most likely cleavage site predicted was within peptide 19VQA↓ST23 (cleavage site probability: 0.979) immediately before the first repeat domain. The protein is thus likely a secreted protein.
Moreover, the protein sequence was found to be extremely rich in serine, threonine, and tyrosine residues. Among the 106 residues to be secreted, there are 16 serine, 18 threonine, and 8 tyrosine residues constituting almost 40% of the protein altogether. This prompted us to speculate about a possible virulence function interfering with eukaryotic cell signalling. Consequently, we searched for putative phosphorylation sites applying the NetPhos 2.0 Server. In support of our hypothesis, 10 serine and 2 threonine residues were predicted as probable phosphorylation sites with a score of 0.917 and higher while the threshold is 0.5. Of these sites, four were predicted with scores above 0.99. The ten most likely phosphorylation sites with highly significant scores of 0.968 and above are marked in Figure 3(b). Interestingly, the serine residues localised within the nonameric repeat modules are all significant targets of phosphorylation. Thus, it seems very likely that this repeat protein, if entering the host cell, would interfere with host cell signalling pathways, possibly by trapping serine/threonine kinases and/or phosphatases.
Taken together, the hypothetical protein encoded by ORF7 is likely to be a secreted repeat protein, a member of a novel repeat protein family possibly sharing a conserved structural motif determined by DxxGN(x)nDxxGN repeats. We provisionally term it YsrA for Yersinia secretable repeat protein A. However, a possible role of YsrA as virulence factor is supported by its secretability, phosphorylatability, and the existence of DxxGN(x)nDxxGN repeats in eukaryotic proteins. Several bacterial toxins interfering with host cellular signalling pathways are suspected of being adopted from eukaryotes [26].
3.5. Expression of ORF3 and ORF7 Products and Relation to the Adhesive Phenotype
All hypothetical proteins encoded by pYe4449-1 and pYe4449-2 were checked for a putative secretion signal using the SignalP server [9]. Apart from ORF7 (see Section 3.4.), only ORF3 was identified to likely encode a secreted protein (signal peptide probability: 0.996). In an attempt to investigate a possible role of ORF3 and ORF7 in adhesion of strain 07-04449 to eukaryotic host cells, we screened for EZ-Tn5 <Kan-2> transposon insertions into these ORFs. We identified insertions into ORF3 and ORF7 after base pair positions 4303 and 6387, respectively. Plasmid profiles and PCR analyses confirmed that pYe4449-2 was completely substituted for the respective mutated plasmid in the host strain. The mutants were compared to the parental strain with respect to cellular adhesion as described earlier. However, both were indistinguishable from the wild-type strain suggesting that neither ORF3 nor ORF7 contributes to adhesion.
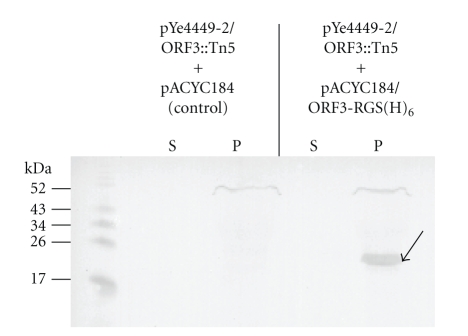
Next, we constructed plasmids to complement the ORF3 and ORF7 mutant strains using plasmid pACYC184 as backbone. In doing so, we introduced an Arg-Gly-Ser-(His)6 epitope-encoding sequence at the 3′-end of each ORF construct to allow detection of the respective protein by Western blotting. In each case, the likely promoter region according to Softberry analysis (http://www.softberry.com/) was included. The resulting plasmids pACYC184/ORF3-RGS(H)6 and pACYC184/ORF7-RGS(H)6 were transformed into insertional mutant strains carrying pYe4449-2/ORF3::Tn5 and pYe4449-2/ORF7::Tn5, respectively. Cultures were analysed after growth at 37°C, 27°C and 4°C. Figure 4 shows a Western blot analysis of the expression of epitope-tagged ORF3 product ORF3-RGS(H)6 after incubation at 4°C over night. Detection with an RGS(H)4-specific antibody revealed a band of 20–22 kDa in the bacterial pellet fraction (P) of the complemented ORF3 mutant but not in the corresponding supernatant (S). Since the calculated molecular mass of the tagged ORF3 product is 22.1 kDa for the unprocessed protein and 19.7 kDa after processing of the predicted secretion signal, it remains to be determined whether ORF3-RGS(H)6 was cytosolic or associated with the bacterial envelope after sec-dependent translocation. Strikingly, no expression was detected after incubation at 27°C or 37°C (data not shown). The ORF3 gene product was, therefore, termed Yersinia low temperature protein A (YltA) and the gene name abbreviated yltA accordingly. By contrast, no epitope-tagged ORF7 product could be detected in an analogous experiment (data not shown). We attribute this to specific characteristics of the ORF7 product for the reason that we could successfully produce a recombinant glutathione S-transferase (GST)-ORF7 fusion protein lacking the putative secretion signal (data not shown). However, after cleavage of the GST fusion with thrombin, only released GST could be visualized by Coomassie or silver stain on denaturing gels, but not the ORF7 fusion part. We have ruled out intrinsic thrombin cleavage sites, both by in silico analysis and by kinetic analyses of the cleavage reaction. We thus conclude that the lack of evidence of ORF7 expression could be due to aberrant behaviour of the protein during preparation.
Figure 4.
Detection of epitope-tagged ORF3 expressed at 4°C. A transposon mutant derivative of isolate 07-04449 carrying plasmid pYe4449-2/ORF3::Tn5 was transformed with plasmid pACYC184/ORF3-RGS(H)6 and pACYC184, respectively, and cultured over night at 4°C in LB broth. Bacterial pellet fractions “P” and TCA-precipitated supernatant fractions “S” were subjected to denaturing SDS-PAGE. Subsequently, the gel was Western-blotted followed by chromogenic detection using a monoclonal antibody directed against the RGS(H)4 epitope. The arrow indicates the ORF3-RGS(H)6 product.
It is interesting to note that Bresolin et al. [27, 28] recently demonstrated the enhanced expression of several genes, including insecticidal toxin genes, in a virulence plasmid-carrying Y. enterocolitica strain at 10°C suggesting that invertebrate hosts are part of their life cycle. Evidence presented on that ancestors of strain 07-04449 were in genetic exchange with endosymbionts of insects, and the finding of enhanced expression of YltA at low temperature fits well with this hypothesis.
3.6. Detection of Virulence-Associated Heat-Stable Enterotoxin B Gene (ystB) and Prevalence of pYe4449 Plasmids
To indicate a possible virulence potential of isolate 07-04449, we checked for the presence of virulence-associated ystB encoding the heat-stable enterotoxin B [29]. A recent study by Bhagat and Virdi [30] described the correlation of virulence-associated genes of biotype 1A strains such as ystB with clonal groups rather than with source of isolation (that is clinical or nonclinical). Among the 20 isolates we had initially screened for adherence, four isolates were of nonclinical origin (two from pork including 07-04449, one from lettuce, one from cured trout). All 20 isolates were ystB-positive (data not shown) and, therefore, likely belong to clonal group A which is believed to be better equipped for virulence compared to clonal group B [30]. This analysis may further indicate that clonal group B is underrepresented among isolates from Germany. However, a more comprehensive study is required to substantiate this trend.
Further, data on the prevalence of plasmids pYe4449-1 and pYe4449-2 were collected. To this end, we have analysed plasmid profiles from 54 Y. enterocolitica biotype 1A isolates and from 348 Y. enterocolitica isolates belonging to pYV carrying biotypes 2–5. Of the 54 biotype 1A strains analysed, 18 strains harboured plasmids with a size similar to pYe4449-1 and none was found with a size similar to that of pYe4449-2. The 18 strains with small plasmids were analysed by PCR with a pair of primers binding to the repA gene and to the mobA gene, respectively, designed to amplify the ccdA/B region inbetween. Two strains gave positive results, with amplicons similar in length to those obtained from the pYe4449-1 control, indicating homologous backbones. However, none of these plasmids was positive in an ORF5-specific PCR. Among the 348 pYV-carrying isolates, 21 harboured additional plasmids but none was similar in size to either pYe4449-1 or pYe4449-2. Taken together, we found both of the plasmids described here to be unique among the isolates tested.
3.7. Concluding Remarks
Howard et al. [31] have recently determined the core gene pool of 94 representatives of the species Y. enterocolitica. Strikingly, only 20.8% of the genes were shared by all strains illustrating the exceptional heterogeneity of this species. Our description of two novel plasmids adds to the diversity of the Y. enterocolitica gene pool. Interestingly, not a single ORF showed highest similarity to Yersinia sequences suggesting that this species was or still is in intensive genetic exchange with bacteria including genera that are not in the focus of any research yet. Consequently, efforts in understanding the life cycle of Y. enterocolitica will help to understand these genetic fluxes and vice versa. Studying plasmid biology of yersiniae will be a main pillar of such endeavours.
Acknowledgments
The authors thank the members of the sequencing core facility at the Robert Koch Institute for DNA sequencing. This research was supported with “Sonderforschungsmittel” provided by the Robert Koch-Institute.
References
- 1.Cornelis GR, Boland A, Boyd AP, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiology and Molecular Biology Reviews. 1998;62(4):1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottone EJ. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes and Infection. 1999;1(4):323–333. doi: 10.1016/s1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 3.Tennant SM, Grant TH, Robins-Browne RM. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunology & Medical Microbiology. 2003;38(2):127–137. doi: 10.1016/S0928-8244(03)00180-9. [DOI] [PubMed] [Google Scholar]
- 4.McNally A, Cheasty T, Fearnley C, et al. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999-2000. Letters in Applied Microbiology. 2004;39(1):103–108. doi: 10.1111/j.1472-765X.2004.01548.x. [DOI] [PubMed] [Google Scholar]
- 5.Lewin A, Strauch E, Hertwig S, Hoffmann B, Nattermann H, Appel B. Comparison of plasmids of strains of Yersinia enterocolitica biovar 1A with the virulence plasmid of a pathogenic Y. enterocolitica Strain. Zentralblatt für Bakteriologie. 1996;285(1):52–63. doi: 10.1016/s0934-8840(96)80022-3. [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson-Ahomaa M, Bucher M, Hank C, Stolle A, Korkeala H. High prevalence of Yersinia enterocolitica 4:O3 on pig offal in Southern Germany: a slaughtering technique problem. Systematic and Applied Microbiology. 2001;24(3):457–463. doi: 10.1078/0723-2020-00055. [DOI] [PubMed] [Google Scholar]
- 7.McNally A, La Ragione RM, Best A, Manning G, Newell DG. An aflagellate mutant Yersinia enterocolitica biotype 1A strain displays altered invasion of epithelial cells, persistence in macrophages, and cytokine secretion profiles in vitro. Microbiology. 2007;153(5):1339–1349. doi: 10.1099/mic.0.2006/000919-0. [DOI] [PubMed] [Google Scholar]
- 8.Bi C-P, Benham CJ. WebSIDD: server for prediction of the stress-induced duplex destabilized sites in superhelical DNA. Bioinformatics. 2004;20:1477–1479. doi: 10.1093/bioinformatics/bth304. [DOI] [PubMed] [Google Scholar]
- 9.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: signalP 3.0. Journal of Molecular Biology. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Singh SK, Banerjee PC. Nucleotide sequence analysis of cryptic plasmid pAM5 from Acidiphilium multivorum. Plasmid. 2007;58(2):101–114. doi: 10.1016/j.plasmid.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Thomson NR, Howard S, Wren BW, et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS genetics. 2006;2(12, article e206) doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande NM, Dhakephalkar PK, Kanekar PP. Plasmid-mediated dimethoate degradation in pseudomonas aeruginosa MCMB-427. Letters in Applied Microbiology. 2001;33(4):275–279. doi: 10.1046/j.1472-765x.2001.00995.x. [DOI] [PubMed] [Google Scholar]
- 13.Molnár A, Amaral L, Molnár J. Antiplasmid effect of promethazine in mixed bacterial cultures. International Journal of Antimicrobial Agents. 2003;22(3):217–222. doi: 10.1016/s0924-8579(03)00206-1. [DOI] [PubMed] [Google Scholar]
- 14.Ruan L, Xu X. Sequence analysis and characterizations of two novel plasmids isolated from Thermus sp. 4C. Plasmid. 2007;58(1):84–87. doi: 10.1016/j.plasmid.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Trček J, Wilharm G, Jacobi CA, Heesemann J. Yersinia enterocolitica YopQ: strain-dependent cytosolic accumulation and post-translational secretion. Microbiology. 2002;148(5):1457–1465. doi: 10.1099/00221287-148-5-1457. [DOI] [PubMed] [Google Scholar]
- 16.Hammerl JA, Klein I, Lanka E, Appel B, Hertwig S. Genetic and functional properties of the self-transmissible Yersinia enterocolitica plasmid pYE854, which mobilizes the virulence plasmid pYV. Journal of Bacteriology. 2008;190(3):991–1010. doi: 10.1128/JB.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biology. 2007;8(2, article R22) doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avison MB, Walsh TR, Bennett PM. pUB6060: a broad-host-range, DNA polymerase-I-independent ColE2-like plasmid. Plasmid. 2001;45(2):88–100. doi: 10.1006/plas.2000.1511. [DOI] [PubMed] [Google Scholar]
- 19.Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiology Reviews. 2004;28(1):79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiology Reviews. 2009;33(3):657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Applied and Environmental Microbiology. 2008;74(21):6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(22):9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs TM, Bresolin G, Marcinowski L, Schachtner J, Scherer S. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiology. 2008;8, article 214 doi: 10.1186/1471-2180-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heermann R, Fuchs TM. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics. 2008;9, article 40 doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaleski P, Wolinowska R, Strzezek K, Lakomy A, Plucienniczak A. The complete sequence and segregational stability analysis of a new cryptic plasmid pIGWZ12 from a clinical strain of Escherichia coli. Plasmid. 2006;56(3):228–232. doi: 10.1016/j.plasmid.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Stebbins CE, Galán JE. Structural mimicry in bacterial virulence. Nature. 2001;412(6848):701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 27.Bresolin G, Morgan JAW, Ilgen D, Scherer S, Fuchs TM. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Molecular Microbiology. 2006;59(2):503–512. doi: 10.1111/j.1365-2958.2005.04916.x. [DOI] [PubMed] [Google Scholar]
- 28.Bresolin G, Neuhaus K, Scherer S, Fuchs TM. Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. Journal of Bacteriology. 2006;188(8):2945–2958. doi: 10.1128/JB.188.8.2945-2958.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramamurthy T, Yoshino K-I, Huang X, et al. The novel heat-stable enterotoxin subtype gene (ystB) of Yersinia enterocolitica: nucleotide sequence and distribution of the yst genes. Microbial Pathogenesis. 1997;23(4):189–200. doi: 10.1006/mpat.1997.0146. [DOI] [PubMed] [Google Scholar]
- 30.Bhagat N, Virdi JS. Distribution of virulence-associated genes in Yersinia enterocolitica biovar 1A correlates with clonal groups and not the source of isolation. FEMS Microbiology Letters. 2007;266:177–183. doi: 10.1111/j.1574-6968.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- 31.Howard SL, Gaunt MW, Hinds J, Witney AA, Stabler R, Wren BW. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. Journal of Bacteriology. 2006;188(10):3645–3653. doi: 10.1128/JB.188.10.3645-3653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]