Abstract
Trimethylamine-N-oxide (TMAO) is a naturally occurring osmolyte that stabilizes proteins against denaturation. Although the impact of TMAO on the folding thermodynamics of many proteins has been well characterized, far fewer studies have investigated its effects on protein folding kinetics. In particular, no previous studies have used Φ-value analysis to determine whether TMAO may alter the structure of the folding transition state. Here we have measured the effects on folding kinetics of 16 different amino acid substitutions distributed across the structure of the Fyn SH3 domain both in the presence and absence of TMAO. The folding and unfolding rates in TMAO, on average, improved to equivalent degrees, with a twofold increase in the protein folding rate accompanied by a twofold decrease in the unfolding rate. Importantly, TMAO caused little alteration to the Φ-values of the mutants tested, implying that this compound minimally perturbs the folding transition state structure. Furthermore, the solvent accessibility of the transition state was not altered as reflected in an absence of a TMAO-induced change in the denaturant  factors. Through TMAO-induced folding studies, a
factors. Through TMAO-induced folding studies, a  factor of 0.5 was calculated for this compound, suggesting that the protein backbone, which is the target of action of TMAO, is 50% exposed in the transition state as compared to the native state. This finding is consistent with the equivalent effects of TMAO on the folding and unfolding rates. Through thermodynamic analysis of mutants, we also discovered that the stabilizing effect of TMAO is lessened with increasing temperature.
factor of 0.5 was calculated for this compound, suggesting that the protein backbone, which is the target of action of TMAO, is 50% exposed in the transition state as compared to the native state. This finding is consistent with the equivalent effects of TMAO on the folding and unfolding rates. Through thermodynamic analysis of mutants, we also discovered that the stabilizing effect of TMAO is lessened with increasing temperature.
Keywords: osmolytes, trimethylamine-N-oxide, folding transition state, folding kinetics, protein stability, osmolyte-induced stability, SH3 domains, Φ-value analysis
Introduction
Protecting osmolytes are small organic compounds found in a wide variety of living organisms that can protect cellular proteins against denaturation in harsh environmental conditions, such as high temperature, high salt, or high urea.1,2 Osmolytes are chemically diverse and can be grouped into three major classes: methylamine compounds, polyols, and certain amino acids.2 These compounds have been shown to offer general protection of proteins against denaturation both in vivo and in vitro. A large body of work has demonstrated that osmolytes are preferentially excluded from the vicinity of the peptide backbone.3–13 The resulting energetically unfavorable interaction between these compounds and the peptide backbone causes a destabilization of the unfolded state of proteins relative to the native state such that the energy gap between the two states increases.14–16 Thus, destabilization of the denatured state is the primary mechanism by which osmolytes increase the overall stability of proteins (reviewed in Ref.17).
While much has been learned in recent years about the mechanism by which osmolytes increase the thermodynamic stability of proteins at equilibrium, few studies have addressed the effects of these compounds on protein folding kinetics. In one study, the effects of several protecting osmolytes on the folding and unfolding rates of a single variant of the FKBP12 protein were measured.18 An increase in the folding rate and a decrease in the unfolding rate of this protein in approximately equal magnitudes were observed in the presence of these compounds. Since the peptide backbone plays a dominant role in the energetics of osmolyte-induced stability, the effect of osmolytes on folding kinetics likely reflects the degree of backbone exposure in the folding transition state structure. Thus, the osmolyte-induced deceleration of the unfolding rate seen in this study, which implies destabilization of the transition state with respect to the native state, was interpreted to mean that the folding transition state structure possesses more exposed backbone atoms than the native state. Similar observations were made in a different study investigating the effects of protecting osmolyte trimethylamine-N-oxide (TMAO) on the folding kinetics of RNase HII.19 Another study showed that osmolytes accelerated the folding rate of Barstar by stabilizing an early intermediate in the folding pathway of this protein.20 Since all of the above studies addressed the folding properties of only a single protein, and not multiple site-directed mutants, Φ-value analysis21 could not be used to assess potential alterations in the structure of the folding transition state.
To gain further insight into the mechanism of action of osmolytes, in the present study we have determined the folding and unfolding rates of a series of site-directed mutants of the Fyn SH3 domain in the presence and absence of the protecting osmolyte TMAO. TMAO was used for these studies because its properties have been extensively characterized, and it was shown to have the largest effect on the folding kinetics of FKBP12.18 The folding properties and folding transition state structure of the Fyn SH3 domain have been thoroughly described by studies in our laboratory and others.22–27 This domain, the structure of which is composed of two orthogonally packed β-sheets (see Fig. 1), displays reversible two-state folding kinetics. Its folding transition state is highly polarized with strands b through d being highly structured and strands a and e, which come together to the front β-sheet (see Fig. 1), being mostly unstructured. This transition state structure is conserved among all SH3 domains tested,28 and has been verified by a variety of experimental techniques and simulations.29–32 Thus, the Fyn SH3 domain provides an ideal model system for determining the effects of TMAO on a folding transition state structure.
Figure 1.
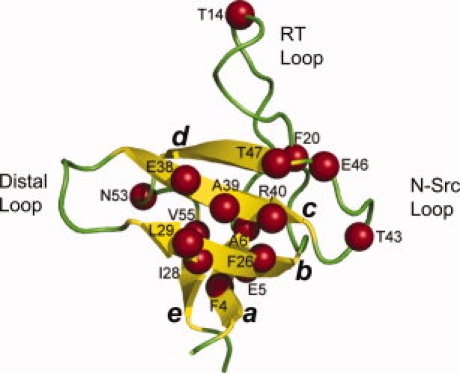
The structure of the Fyn SH3 domain highlighting the positions of mutations used in this work (designated with spheres). This figure was generated by the program PyMol (DeLano Scientific, Palo Alto, CA) using the coordinates of the crystal structure of the Fyn SH3 domain (PDB code 1SHF).
Since the effect of TMAO on the folding behavior of a series of single amino acid substitutions within a single protein has never been assessed, the impact that TMAO might have on folding kinetic Φ-values and by extension the detailed structure of the folding transition state is unknown. For this reason, the present study was undertaken with the primary goal of ascertaining whether TMAO induces any significant changes in the structure of the transition state of the Fyn SH3 domain. To achieve this aim, we characterized the folding properties of site-directed mutants at positions spread across the sequence of this protein in the presence and absence of TMAO. Further insight into the effects of TMAO on the folding transition state was gained by establishing conditions under which TMAO was required to induce the folding of the Fyn SH3 domain. This approach allowed us to establish the dependency of the folding rate on the concentration of TMAO, and assess the backbone exposure to TMAO of the folding transition state. These studies provide the most comprehensive analysis to date of the effects of an osmolyte on protein folding kinetics, and thus provide new insight into the mechanism of action of these compounds.
Results
To address the mechanism of TMAO-mediated stabilization of proteins, we investigated the effect of this osmolyte on the folding kinetics and the thermodynamic stability of the WT Fyn SH3 domain and 16 site-directed mutants distributed across the structure of this protein. The positions of these amino acid substitutions lie in both structured and unstructured regions of the folding transition state structure (see Fig. 1). Positions 4, 6, 20, 26, 28, 39, and 55 lie in the hydrophobic core of the native state structure of this domain, while the other positions substituted lie in exposed positions. The equilibrium and folding kinetic properties of the hydrophobic core mutants have been characterized previously.24,33 The other mutants used in this study were selected from a collection of site directed variants that have recently been characterized27,34 or are currently under investigation in our laboratory, but have not yet been published. Rather than only characterizing substitutions with Ala as is done in most studies, we have used a variety of substitutions here to ensure that the observed overall effects of TMAO on these mutants is not unduly influenced by the type of substitution being used. We have previously shown that substitutions with a variety of amino acids at any given position cause little alteration of the folding transition state structure24,25,27,35 and that similar Φ-values are obtained independent of the substitution examined.
Thermal denaturation of Fyn SH3 domain in the presence of TMAO
The temperature-induced unfolding curves of the WT Fyn SH3 domain and its mutants both in the presence and the absence of TMAO are sigmoidal and have characteristic features of a cooperative two-state transition [Fig. 2(A)]. Consistent with previous observations that osmolytes protect proteins against thermal denaturation, TMAO at a concentration of 1M significantly increased the melting temperature of the WT Fyn SH3 domain and its mutants (Table I). Interestingly, the extent of the modulation of the stability varied among the mutants. A strong negative correlation (r = −0.85) was detected between the change in presence of osmolyte of the Tm values  and the melting temperature of the mutants (Tm), indicating that proteins with lower thermal stability exhibit a larger degree of stabilization by TMAO [Fig. 3(A)]. For example, the F20A mutant (Tm = 50°C) exhibited an increase in Tm of more than 10°C in TMAO, while the Tm of WT (Tm = 75.6°C) increased by only 4°C.
and the melting temperature of the mutants (Tm), indicating that proteins with lower thermal stability exhibit a larger degree of stabilization by TMAO [Fig. 3(A)]. For example, the F20A mutant (Tm = 50°C) exhibited an increase in Tm of more than 10°C in TMAO, while the Tm of WT (Tm = 75.6°C) increased by only 4°C.
Figure 2.
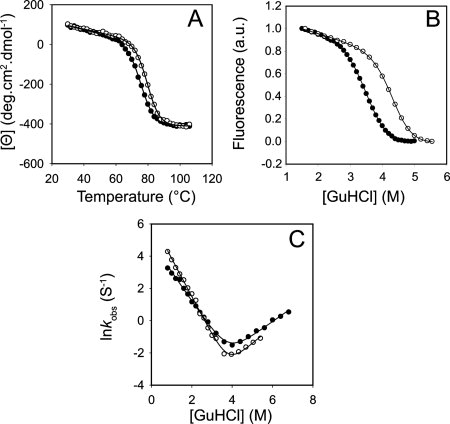
The effect of TMAO on the equilibrium stability and the folding kinetics of the WT Fyn SH3 domain. The temperature-induced unfolding curve (A), the equilibrium GuHCl-induced melt profile (B), and the kinetic chevron plot of the WT Fyn SH3 domain (C) in the absence (closed circles) and in the presence (open circles) of 1M TMAO.
Table I.
The Folding Kinetics and the Thermodynamic Stability Parameters of the Fyn SH3 Domain Mutants in the Presence and in the Absence of TMAO
| kf (S−1) | mkf | ku (S−1) | mku | Tm (°C) | ΔHf→u (Tm)a (kcal mol−1) | 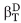 |
Φb | |
|---|---|---|---|---|---|---|---|---|
| 0M TMAO | ||||||||
| WT | 56 ± 2 | 2.10 ± 0.03 | 0.42 ± 0.01 | 0.87 ± 0.02 | 75.6 ± 0.2 | 60.1 | 0.70 ± 0.01 | N/A |
| F4V | 45 ± 3 | 1.90 ± 0.07 | 0.90 ± 0.08 | 0.62 ± 0.07 | 72.4 ± 0.3 | 54.7 | 0.75 ± 0.01 | 0.20 ± 0.07 |
| E5V | 80 ± 10 | 2.04 ± 0.06 | 0.24 ± 0.01 | 0.99 ± 0.04 | 80.9 ± 0.4 | 53.8 | 0.67 ± 0.01 | 0.33 ± 0.07 |
| A6S | 51 ± 4 | 2.24 ± 0.07 | 3.9 ± 0.1 | 0.76 ± 0.03 | 61.2 ± 0.3 | 46.1 | 0.75 ± 0.01 | 0.04 ± 0.03 |
| T14R | 54 ± 3 | 2.08 ± 0.04 | 0.36 ± 0.02 | 0.92 ± 0.04 | 77.4 ± 0.2 | 53.5 | 0.69 ± 0.01 | — |
| F20A | 24 ± 2 | 1.5 ± 0.1 | 9.2 ± 0.4 | 0.77 ± 0.04 | 50 ± 2 | 32.5 | 0.66 ± 0.01 | 0.21 ± 0.02 |
| F26V | 30 ± 2 | 2.1 ± 0.1 | 10.6 ± 0.4 | 0.80 ± 0.03 | 58.6 ± 0.6 | 42.3 | 0.72 ± 0.01 | 0.16 ± 0.01 |
| I28A | 3.0 ± 0.3 | 2.3 ± 0.2 | 1.6 ± 0.1 | 0.90 ± 0.07 | 54 ± 1 | 40.8 | 0.75 ± 0.02 | 0.69 ± 0.03 |
| L29Y | 23 ± 1 | 2.19 ± 0.04 | 0.79 ± 0.02 | 0.80 ± 0.02 | 61.3 ± 0.6 | 49.0 | 0.73 ± 0.01 | 0.58 ± 0.03 |
| E38A | 25 ± 1 | 1.93 ± 0.04 | 0.75 ± 0.02 | 0.77 ± 0.03 | 68.1 ± 0.1 | 45.2 | 0.72 ± 0.02 | 0.58 ± 0.04 |
| A39G | 6.1 ± 0.3 | 1.95 ± 0.06 | 0.62 ± 0.02 | 0.89 ± 0.04 | 62.3 ± 0.3 | 42.1 | 0.69 ± 0.01 | 0.85 ± 0.03 |
| R40N | 3.3 ± 0.2 | 2.10 ± 0.06 | 0.75 ± 0.02 | 0.79 ± 0.02 | 57.8 ± 0.4 | 41.0 | 0.73 ± 0.01 | 0.83 ± 0.02 |
| T43N | 22.5 ± 0.7 | 2.09 ± 0.03 | 0.54 ± 0.01 | 0.81 ± 0.02 | 72.1 ± 0.1 | 52.9 | 0.72 ± 0.01 | 0.78 ± 0.04 |
| E46A | 41 ± 2 | 2.22 ± 0.05 | 0.58 ± 0.03 | 0.91 ± 0.06 | 73.1 ± 0.1 | 53.7 | 0.71 ± 0.01 | 0.49 ± 0.09 |
| T47A | 18 ± 1 | 2.25 ± 0.05 | 0.58 ± 0.02 | 0.81 ± 0.04 | 68.9 ± 0.2 | 54.1 | 0.73 ± 0.01 | 0.77 ± 0.05 |
| N53V | 76 ± 2 | 2.10 ± 0.02 | 1.75 ± 0.05 | 0.97 ± 0.02 | 70.1 ± 0.2 | 57.4 | 0.68 ± 0.01 | −0.27 ± 0.04 |
| V55L | 79 ± 2 | 2.03 ± 0.01 | 1.11 ± 0.01 | 0.98 ± 0.02 | 71.8 ± 0.3 | 56.3 | 0.67 ± 0.01 | −0.58 ± 0.07 |
| Average | 2.07 ± 0.06 | 0.84 ± 0.03 | 49.1 | 0.71 ± 0.01 | ||||
| 1M TMAO | ||||||||
| WT | 150 ± 10 | 2.33 ± 0.06 | 0.24 ± 0.02 | 1.1 ± 0.1 | 79.7 ± 0.4 | 62.2 | 0.68 ± 0.01 | N/A |
| F4V | 84 ± 6 | 2.08 ± 0.05 | 0.43 ± 0.03 | 0.7 ± 0.1 | 79.1 ± 0.5 | 55.1 | 0.76 ± 0.01 | 0.48 ± 0.08 |
| E5V | 160 ± 20 | 2.06 ± 0.07 | 0.08 ± 0.02 | 1.3 ± 0.3 | 85.6 ± 0.3 | 60.6 | 0.59 ± 0.03 | 0.4 ± 0.1 |
| A6S | 120 ± 10 | 2.3 ± 0.1 | 1.6 ± 0.1 | 0.84 ± 0.07 | 69.5 ± 0.5 | 45.7 | 0.74 ± 0.01 | 0.11 ± 0.05 |
| T14R | 130 ± 10 | 2.21 ± 0.06 | 0.12 ± 0.01 | 1.4 ± 0.2 | 82.7 ± 0.3 | 63.9 | 0.61 ± 0.02 | −0.3 ± 0.1 |
| F20A | 46 ± 2 | 1.92 ± 0.08 | 3.2 ± 0.2 | 0.52 ± 0.06 | 62 ± 1 | 39.3 | 0.77 ± 0.02 | 0.31 ± 0.02 |
| F26V | 59 ± 3 | 2.08 ± 0.05 | 5.7 ± 0.3 | 1.08 ± 0.07 | 67.4 ± 0.6 | 56.5 | 0.66 ± 0.01 | 0.22 ± 0.01 |
| I28A | 6.8 ± 0.6 | 2.6 ± 0.2 | 1.5 ± 0.4 | 1.0 ± 0.2 | 65.0 ± 0.5 | 52.0 | 0.63 ± 0.01 | 0.58 ± 0.02 |
| L29Y | 56 ± 4 | 2.30 ± 0.06 | 0.36 ± 0.03 | 1.0 ± 0.1 | 72.1 ± 0.1 | 53.7 | 0.71 ± 0.01 | 0.70 ± 0.08 |
| E38A | 62 ± 5 | 2.29 ± 0.07 | 0.62 ± 0.05 | 1.2 ± 0.1 | 76.6 ± 0.4 | 57.3 | 0.67 ± 0.01 | 0.47 ± 0.05 |
| A39G | 15.7 ± 0.8 | 2.06 ± 0.05 | 0.35 ± 0.02 | 0.9 ± 0.1 | 71.5 ± 0.3 | 56.3 | 0.69 ± 0.01 | 0.86 ± 0.04 |
| R40N | 8.4 ± 0.6 | 2.32 ± 0.07 | 0.38 ± 0.02 | 0.91 ± 0.05 | 66.7 ± 0.2 | 51.7 | 0.72 ± 0.01 | 0.86 ± 0.03 |
| T43N | 68 ± 6 | 2.32 ± 0.07 | 0.22 ± 0.01 | 1.0 ± 0.1 | 76.4 ± 0.2 | 54.1 | 0.70 ± 0.01 | 1.1 ± 0.2 |
| E46A | 81 ± 6 | 2.23 ± 0.05 | 0.18 ± 0.01 | 0.8 ± 0.1 | 81.5 ± 0.3 | 61.4 | 0.73 ± 0.01 | — |
| T47A | 39 ± 3 | 2.11 ± 0.06 | 0.25 ± 0.02 | 1.0 ± 0.1 | 74.1 ± 0.2 | 52.0 | 0.68 ± 0.01 | 1.0 ± 0.1 |
| N53V | 133 ± 8 | 2.13 ± 0.04 | 0.98 ± 0.09 | 1.1 ± 0.1 | 76.6 ± 0.2 | 56.2 | 0.67 ± 0.01 | 0.07 ± 0.05 |
| V55L | 160 ± 10 | 1.97 ± 0.05 | 0.44 ± 0.04 | 1.3 ± 0.2 | 79.1 ± 0.3 | 58.5 | 0.61 ± 0.01 | −0.2 ± 0.1 |
| Average | 2.20 ± 0.08 | 1.0 ± 0.1 | 55.1 | 0.68 ± 0.02 | ||||
The ΔHf→u (Tm) values are accompanied by a 5% error (based on repetitions).
Because of the small magnitude of their ΔΔGf→u, no Φ-values were calculated for T14R in the absence and for E46A in the presence of TMAO.
Figure 3.
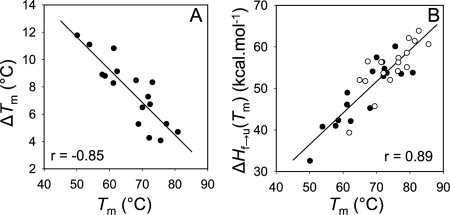
Analysis of the temperature-induced unfolding data of the Fyn SH3 domain mutants. The correlation between melting temperature of the Fyn SH3 mutants (Tm) and the extent of increase in the melting temperature in 1M TMAO 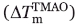 (A). The correlation between Tm and ΔHf→u (Tm) in the presence (open circles) or in the absence (closed circles) of TMAO (B). The ΔHf→u (Tm) values were calculated from manual fitting of the temperature melts, as described in Materials and methods. The ΔHf→u (Tm) versus Tm data in the absence and in the presence of TMAO were also individually fit, and slopes identical to what reported in panel B were obtained.
(A). The correlation between Tm and ΔHf→u (Tm) in the presence (open circles) or in the absence (closed circles) of TMAO (B). The ΔHf→u (Tm) values were calculated from manual fitting of the temperature melts, as described in Materials and methods. The ΔHf→u (Tm) versus Tm data in the absence and in the presence of TMAO were also individually fit, and slopes identical to what reported in panel B were obtained.
Analysis of the temperature-induced unfolding curves also yielded a value for the ΔHf→u(Tm) for each mutant. The plots of ΔHf→u(Tm) versus Tm for all of the mutants tested with or without TMAO could be fit by straight lines. The slopes of the lines fitting the two sets of data were almost identical, and hence a single line could fit all of the data just as well [Fig. 3(B)]. These results indicate that the ΔCp, which is equivalent to the slope of the ΔHf→u(Tm) versus Tm plot, is constant for all of mutants tested, and TMAO does not significantly alter the ΔCp value of the Fyn SH3 domain. It should be noted that the ΔCp value obtained from the plot in Figure 3(B) (0.73 kcal mol−1 degree−1) is close to the value of 0.68 kcal mol−1 degree−1 that we previously obtained for this domain through the combined analysis of temperature melts and denaturant melts performed at a variety of temperatures.33
The effect of TMAO on the folding kinetics of Fyn SH3 domain mutants
To further investigate the mechanism of the TMAO-mediated increase in the stability of the Fyn SH3 domain, we measured the folding (kf) and the unfolding (ku) rates of this protein and its mutants in the presence and absence of TMAO. The plots of the logarithm of the observed rate constants versus guanidine hydrochloride (GuHCl) concentration (“chevron” plot) for the WT protein under both conditions display straight folding and unfolding arms, which indicates a two-state transition [Fig. 2(C)]. The chevron plots of all the mutants also exhibited straight folding and unfolding arms (data not shown). To minimize the errors associated with linear extrapolation of the observed rates to zero denaturant concentration, we have reported the folding and the unfolding rates of the WT Fyn SH3 domain and its mutants at 0.5 and 5M concentrations of GuHCl, respectively. It can be seen that the folding rates of the mutants in the absence of TMAO vary over a 25-fold range, and the unfolding rates vary over 40-fold range (Table I). To verify the kinetics data and ensure that two-state folding was maintained for all mutants, we plotted the Tm values at equilibrium for each mutant against the change in the overall stability (ΔΔGf→u) of the mutant as calculated from the kinetic data (Table I). It can be seen that the kinetics data agree well with the equilibrium data both in the absence and the presence of TMAO (see Fig. 4).
Figure 4.
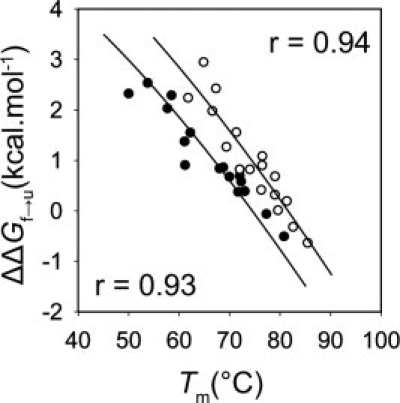
The correlation between the melting temperature and the change in the equilibrium stability of the Fyn SH3 domain mutants in the absence (closed circles, r = 0.93) and in the presence (open circles, r = 0.94) of 1M TMAO. Fitting was performed as described previously24 using a constant ΔCp value of 0.73 kcal mol−1 degree−1, with the stability of the WT protein as the only free parameter.
By comparing the folding rates of the WT and each mutant in the presence and absence of TMAO, the change in the energy gap between the unfolded state and the transition state occurring as the result of TMAO addition (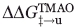 ) can be calculated. In the same manner, comparison of the unfolding rates allows calculation of the change in energy gap between the folded state and the transition state occurring as a result of TMAO addition (
) can be calculated. In the same manner, comparison of the unfolding rates allows calculation of the change in energy gap between the folded state and the transition state occurring as a result of TMAO addition (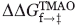 ). By dividing the
). By dividing the 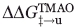 value of each mutant by the total stabilization engendered by TMAO, a ΦTMAO can be calculated, which quantitates the relative effect of TMAO on the folding and unfolding rates. For most positions, ΦTMAO is between 0.5 and 0.6, indicating that TMAO increased the folding rate and decreased the unfolding rate in almost equal proportion (Table II). The same conclusion can be reached by analysis of the folding and unfolding rates themselves (Table I), which shows that on average the folding rates of the mutants increase by 2.3-fold and the unfolding rates decrease by twofold upon addition of TMAO. Only the I28A stands out as a substantial outlier from this trend because the small unfolding arm of the chevron plot of this mutant precludes an accurate determination of its unfolding rate in the presence of TMAO. The average overall increase in thermodynamic stability resulting from TMAO addition was 0.88 kcal mol−1 (Table II). In contrast to the Tm measurements, the magnitude of stabilization resulting from TMAO was similar for all the mutants, and did not increase for less stable mutants.
value of each mutant by the total stabilization engendered by TMAO, a ΦTMAO can be calculated, which quantitates the relative effect of TMAO on the folding and unfolding rates. For most positions, ΦTMAO is between 0.5 and 0.6, indicating that TMAO increased the folding rate and decreased the unfolding rate in almost equal proportion (Table II). The same conclusion can be reached by analysis of the folding and unfolding rates themselves (Table I), which shows that on average the folding rates of the mutants increase by 2.3-fold and the unfolding rates decrease by twofold upon addition of TMAO. Only the I28A stands out as a substantial outlier from this trend because the small unfolding arm of the chevron plot of this mutant precludes an accurate determination of its unfolding rate in the presence of TMAO. The average overall increase in thermodynamic stability resulting from TMAO addition was 0.88 kcal mol−1 (Table II). In contrast to the Tm measurements, the magnitude of stabilization resulting from TMAO was similar for all the mutants, and did not increase for less stable mutants.
Table II.
Changes in the Folding Energetics of the Fyn SH3 Domain as a Result of TMAO Addition
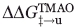 (kcal mol−1) (kcal mol−1) |
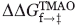 (kcal mol−1) (kcal mol−1) |
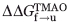 (kcal mol−1) (kcal mol−1) |
ΦTMAO | |
|---|---|---|---|---|
| WT | −0.57 ± 0.05 | −0.33 ± 0.05 | −0.90 ± 0.07 | 0.63 ± 0.06 |
| F4V | −0.37 ± 0.06 | −0.43 ± 0.07 | −0.80 ± 0.09 | 0.45 ± 0.08 |
| E5V | −0.4 ± 0.2 | −0.58 ± 0.07 | −1.02 ± 0.17 | 0.4 ± 0.2 |
| A6S | −0.48 ± 0.07 | −0.53 ± 0.04 | −1.01 ± 0.08 | 0.47 ± 0.07 |
| T14R | −0.51 ± 0.06 | −0.65 ± 0.06 | −1.16 ± 0.08 | 0.44 ± 0.06 |
| F20A | −0.37 ± 0.05 | −0.62 ± 0.04 | −0.99 ± 0.06 | 0.38 ± 0.06 |
| F26V | −0.39 ± 0.06 | −0.37 ± 0.03 | −0.76 ± 0.07 | 0.51 ± 0.07 |
| I28A | −0.48 ± 0.09 | 0.0 ± 0.2 | −0.5 ± 0.2 | 1.0 ± 0.4 |
| L29Y | −0.52 ± 0.05 | −0.46 ± 0.05 | −0.98 ± 0.07 | 0.53 ± 0.06 |
| E38A | −0.55 ± 0.06 | −0.11 ± 0.05 | −0.66 ± 0.08 | 0.8 ± 0.1 |
| A39G | −0.56 ± 0.04 | −0.34 ± 0.04 | −0.90 ± 0.05 | 0.62 ± 0.06 |
| R40N | −0.55 ± 0.05 | −0.40 ± 0.03 | −0.95 ± 0.06 | 0.57 ± 0.05 |
| T43N | −0.65 ± 0.06 | −0.53 ± 0.04 | −1.18 ± 0.07 | 0.55 ± 0.05 |
| E46A | −0.41 ± 0.05 | −0.69 ± 0.04 | −1.10 ± 0.07 | 0.37 ± 0.05 |
| T47A | −0.45 ± 0.05 | −0.49 ± 0.05 | −0.94 ± 0.08 | 0.47 ± 0.06 |
| N53V | −0.33 ± 0.03 | −0.34 ± 0.06 | −0.67 ± 0.07 | 0.49 ± 0.06 |
| V55L | −0.41 ± 0.04 | −0.55 ± 0.05 | −0.96 ± 0.07 | 0.43 ± 0.06 |
| Average | 0.51 ± 0.07 | |||
The effect of TMAO on the folding transition state structure of the Fyn SH3 domain
The mkf and mku values (the dependence of lnkf and lnku on [GuHCl], respectively) derived from the kinetic experiments provide information on the relative solvent exposure of the folding transition state.36 While there is variation in the mkf and mku values between the mutants tested (generally ± 10%), a small but significant increase in these values was observed in the presence of TMAO. In TMAO, the average mkf value increased by 7.5%, and the average mku value increased by 20%. The degree of m value changes were not correlated to the stability (Table I). No significant correlations were observed between any m values and ΔΔG values, which could indicate Hammond-like behavior.37,38 βT values, which are derived for the mkf and mku values, reflect the degree of solvent exposure of the folding transition state relative to the native state. As shown in Table I, the average denaturant βT factor of WT and mutants in the presence of TMAO (0.68 ± 0.02) is identical to the average βT factor of 0.70 ± 0.01 obtained in the absence of this compound. Deviations from the average βT factor by the mutants in the presence, and in the absence of TMAO, were found to be less than 10% in all cases.
To ascertain whether the presence of TMAO causes any alteration to the structure of the folding transition state, we performed Φ-value analysis on the mutants examined here. Φ-values are calculated from free energies derived from the folding and unfolding rates of each mutant, whereby a Φ-value of 1 indicates that the substitution perturbs the free energy of the transition state as much as the native state, while a value of zero indicates the free energy of the transition state is unperturbed. Φ-values are generally interpreted as providing a quantitative estimate of the degree of native structure formation around a given residue in the folding transition state with a high Φ-value being taken to indicate that the residue possesses a high degree of native structure in the transition state.21,39 In Figure 5, we plotted the Φ-value of each mutant obtained in the presence of 1M TMAO versus those obtained in the absence of this compound (Φ-values are reported in Table I). The strong linear correlation (r = 0.91) between the Φ-values in the presence and the absence of TMAO suggests that TMAO does not dramatically alter the structure of the folding transition state of the Fyn SH3 domain.
Figure 5.
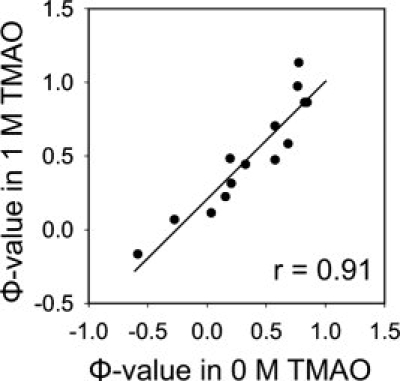
The Φ-values of the Fyn SH3 domain mutants obtained in the presence and absence of 1M TMAO. Φ-values were determined from folding and unfolding rates reported in Table I as described in Materials and methods.
TMAO-induced folding of the Fyn SH3 domain
To directly characterize the kinetics of TMAO-induced folding, we carried out an experiment in which a partially denatured sample of the WT Fyn SH3 domain (in 3M GuHCl) was rapidly mixed with increasing amounts of TMAO, keeping the final GuHCl concentration constant at 3M. The mixing resulted in folding of the domain such that the evolution of the folded state proved to be a single exponential function of the mixing time [Fig. 6(A)]. As illustrated in Figure 6(B), a strong linear dependence (r = 0.99) was detected between ln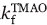 and [TMAO]. Similarly, the equilibrium stability (ΔGf→u) of the domain (from equilibrium GuHCl-induced unfolding of the WT protein) increased linearly (r = 0.99) with increasing the concentration of TMAO [Fig. 6(C)], as previously shown for other proteins.40 Consistent with the experiments described earlier, the meq value of the domain with respect to GuHCl increased with increasing TMAO. This increase occurred in a linear fashion over the range of TMAO concentrations tested [Fig. 6(D)]. It is notable that the GuHCl meq value of 2.0 kcal mol−1 M−1 for the WT domain in 1M TMAO that can be derived from our kinetic experiments (Table I, meq = RT(mkf + mku) is identical to that determined form the equilibrium denaturant melts (Table III)).
and [TMAO]. Similarly, the equilibrium stability (ΔGf→u) of the domain (from equilibrium GuHCl-induced unfolding of the WT protein) increased linearly (r = 0.99) with increasing the concentration of TMAO [Fig. 6(C)], as previously shown for other proteins.40 Consistent with the experiments described earlier, the meq value of the domain with respect to GuHCl increased with increasing TMAO. This increase occurred in a linear fashion over the range of TMAO concentrations tested [Fig. 6(D)]. It is notable that the GuHCl meq value of 2.0 kcal mol−1 M−1 for the WT domain in 1M TMAO that can be derived from our kinetic experiments (Table I, meq = RT(mkf + mku) is identical to that determined form the equilibrium denaturant melts (Table III)).
Figure 6.
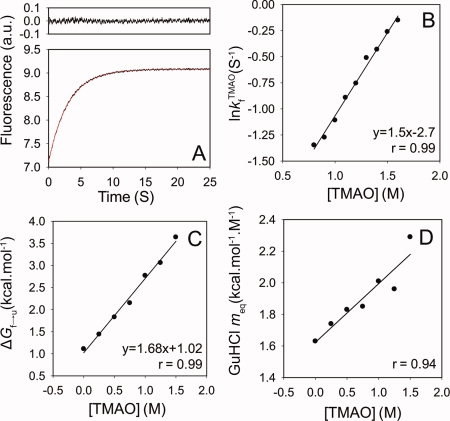
TMAO-induced folding of the denatured (in 3M GuHCl) Fyn SH3 domain. The evolution of the folded state as a function of the mixing time with 1M TMAO (A). The lower panel represents the actual kinetic trace overlaid with the best-fit single exponential function and the upper panel represents the residuals of the fit. The dependence on [TMAO] of the logarithm of the folding rate (B), the overall stability (C), and the GuHCl meq value of the WT Fyn SH3 domain (D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table III.
TMAO-Induced Folding of GuHCl-Denatured WT Fyn SH3 Domain
| [TMAO] (M) | GuHCl meq (kcal mol−1 M−1) | ΔGf→u (kcal mol−1) |
|---|---|---|
| 0 | 1.6 ± 0.1 | 1.1 ± 0.1 |
| 0.25 | 1.8 ± 0.1 | 1.4 ± 0.1 |
| 0.5 | 1.8 ± 0.2 | 1.8 ± 0.2 |
| 0.75 | 1.9 ± 0.2 | 2.2 ± 0.1 |
| 1 | 2.0 ± 0.4 | 2.8 ± 0.3 |
| 1.25 | 2.0 ± 0.3 | 3.1 ± 0.2 |
| 1.5 | 2.3 ± 0.5 | 3.6 ± 0.3 |
To assess the solvent exposure of the folding transition state in 3M GuHCl as probed by folding in TMAO, we calculated a βT value for the Fyn SH3 domain from these experiments. This value was calculated from an mkf value for TMAO (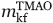 ) determined from the slope of the ln
) determined from the slope of the ln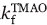 versus [TMAO] plot [Fig. 6(B)] and an meq (
versus [TMAO] plot [Fig. 6(B)] and an meq (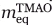 ) value determined from the ΔGf→u versus [TMAO] plot [Fig. 6(C)] using the formula:
) value determined from the ΔGf→u versus [TMAO] plot [Fig. 6(C)] using the formula:
The calculated 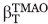 factor for this osmolyte is equal to 0.52.
factor for this osmolyte is equal to 0.52.
Discussion
The goal of this study was to investigate the effect of osmolytes on the folding transition state structure of proteins. To this end, our work presents the first analysis of the effects of TMAO on a large collection of mutants of a single domain. Through this effort, we have found that the Φ-values of 15 individual residues distributed across the Fyn SH3 domain structure were changed very little when folding kinetics experiments were performed in 1M TMAO. These data clearly indicate that the structure of the folding transition state is not altered by this compound. It is particularly notable that positions with low degrees of structure formation in the transition state (i.e., exhibiting low Φ in the absence of osmolyte) retained their low Φ-values in the presence of TMAO. Thus, this osmolyte does not induce structure formation in the unstructured regions of the folding transition state of the Fyn SH3 domain. This situation contrasts with the effect of the trifluoroethanol (TFE), which has been shown to promote formation of secondary structure,41 alter the position of the folding transition state along the reaction coordinate42 and exert opposite effects on protein stability as a function of concentration.43,44 Further supporting the conclusion that TMAO does not alter the folding transition state structure, the βT value of the WT and mutant proteins was similar in the presence or absence of TMAO, implying that the degree of exposure of the transition state to solvent is not perturbed by TMAO. The consistency of the effects of TMAO on the folding kinetics across all the mutants tested suggests that the effect of TMAO on folding energetics is nonspecific with respect to a wide variety of side chain chemistries, solvent exposures, and secondary structure motifs at the site of mutation. This finding supports previous work implicating the peptide backbone most strongly in the mechanism of TMAO-induced stabilization of proteins (reviewed in Ref.17).
A surprising finding was the significant increase in the m value of GuHCl caused by the presence of TMAO, which was detected in both kinetic and equilibrium folding experiments. The increase in meq in TMAO partially accounts for the large increases in ΔGf→u value of this domain when extrapolated to 0M GuHCl (i.e., the increase in stability is not as large at 3M GuHCl, Table III). Increases in m values are often interpreted as being due to a change in the solvent accessibility of the unfolded state with respect to the folded state; however, here we do not see a concomitant increase in ΔCp as would be expected with a change in unfolded state structure.45 There is evidence that TMAO does not affect the strength of hydrophobic interactions.46 Since the hydrophobic effect is the major determinant of the positive ΔCp value of protein unfolding, the lack of change in the ΔCp value of the Fyn SH3 domain in the presence of TMAO may be explained by this observation. We hypothesize that the increase in m value may arise from solvent effects. There is evidence that TMAO alters the structure of water in the vicinity of peptides,47 and such alterations of solvent structure might in turn modify the chemical potential of GuHCl leading to apparent alterations in the GuHCl m values of the protein. In contrast, urea in mixtures with osmolytes has been shown to exert its denaturing effects on proteins independent of the protecting osmolytes, and no alteration in the urea unfolding m values of proteins have been observed.48,49 The increase in the GuHCl m value in the presence of TMAO may stem from its ionic character.
It is well established that osmolytes, such as TMAO, increase protein stability by interacting unfavorably with the peptide backbone. Since more backbone is exposed in the unfolded state, osmolytes preferentially destabilize the unfolded state relative to the native state resulting in a larger energy gap between the two states. With respect to folding kinetics, if TMAO only destabilized the unfolded state and not the folding transition state, then the presence of TMAO would result in only an acceleration of the folding rate with no change in the unfolding rate. However, TMAO induced both an increase in the folding rate of the WT and mutant Fyn SH3 domains, and an almost equivalent decrease in the unfolding rate (i.e., ΦTMAO ∼0.5), implying that the folding transition state is substantially destabilized by TMAO relative to the native state. Strikingly, our investigation of TMAO-induced folding of a partially denatured sample of the Fyn SH3 domain revealed a βT value of 0.5 for TMAO, indicating that the folding transition state exposes approximately 50% more surface area than the native state. This level of solvent exposure of the transition state relative to the native provides a satisfying explanation for why TMAO has equal effects on both the folding and unfolding rates of this domain. It is interesting to note that the GuHCl βT value (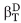 ) of 0.725 of the Fyn SH3 domain is somewhat higher than the βT of TMAO. This difference may be the result of measuring the
) of 0.725 of the Fyn SH3 domain is somewhat higher than the βT of TMAO. This difference may be the result of measuring the 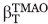 in the presence of 3M GuHCl. However, it is also possible that the transition state “solvent exposure” probed by TMAO is different than that probed by GuHCl. While strong evidence exists showing that the effect of TMAO is manifested almost exclusively through interactions with the peptide backbone,14 denaturants such as GuHCl and urea may operate through more complex mechanisms involving side chain interactions. Thus, the level of transition state exposure revealed through denaturant studies may reflect, to some degree, burial of both backbone and side chains.
in the presence of 3M GuHCl. However, it is also possible that the transition state “solvent exposure” probed by TMAO is different than that probed by GuHCl. While strong evidence exists showing that the effect of TMAO is manifested almost exclusively through interactions with the peptide backbone,14 denaturants such as GuHCl and urea may operate through more complex mechanisms involving side chain interactions. Thus, the level of transition state exposure revealed through denaturant studies may reflect, to some degree, burial of both backbone and side chains.
In support of the generality of our observations on the effects of TMAO on the Fyn SH3 domain, we found that the effects of TMAO on the equilibrium folding properties of this domain were similar to those reported for other systems. Previous studies have shown that osmolytes increase protein stability and that the two-state folding behavior of proteins is maintained in the presence of these compounds.40,50–52 Consistent with these observations, analyses of the temperature- (or GuHCl-) induced unfolding curves as well as the GuHCl-mediated folding kinetics data of the Fyn SH3 domain mutants suggest that the TMAO increases the thermodynamic stability of the Fyn SH3 domain and that two-state folding behavior is preserved in the presence of TMAO (See Fig. 2 for WT data). The lack of change in the ΔCp [Fig. 3(B)] and ΔHf→u(Tm) values (Table I) of the Fyn SH3 domain in the presence of TMAO suggests that the mechanism of TMAO-mediated increase in the thermodynamic stability of this protein is largely entropic in nature, as was recently reported for other proteins.53 These observations collectively suggest that the thermodynamic signature of the effects of TMAO on the stability of the Fyn SH3 domain is similar to what has been previously demonstrated for other proteins. Thus, we conclude that the effects of TMAO on the folding transition state of the Fyn SH3 domain are likely to apply to other protein model systems as well.
Materials and Methods
Mutagenesis and protein purification
Mutations were generated using a PCR-based strategy, and recombinantly expressed as hexahistidine fusions. Protein purification was carried out as described previously.22 Recombinant proteins were used without cleaving the hexahistidine tag. Sample purity was verified through SDS-PAGE and protein concentrations were determined by UV absorbance at 280 nm as described previously.54 All experiments were performed in 50 mM sodium phosphate buffer (pH = 7.0) supplemented with 100 mM NaCl.
Folding kinetics studies
The kinetics of folding and unfolding were monitored by Trp fluorescence using a Bio-Logic SFM-4 stopped flow device (BioLogic Instruments, Claix, France), as described in detail.27 All of the kinetics measurements were performed at 25°C. Where appropriate, all the buffer and denaturant solutions were prepared in 1M TMAO (Sigma-Aldrich Canada, Oakville, ON). For each mutant, Φ-values in the presence and absence of TMAO were determined from folding kinetics rates using the standard equations.55 The errors reported for kf,ku, mkf, mku, and Tm values are the uncertainties associated with fitting the kinetic guanidine- (or equilibrium temperature-) induced unfolding data to appropriate equations. The errors reported for Φ and 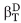 values are by propagating the fitting errors according to the equation:
values are by propagating the fitting errors according to the equation:
where u = f(xi) and dxi is the residual errors of xi.
To quantify the effect of TMAO on the stability of the folding transition state (‡) of the Fyn SH3 domain, a ΦTMAO parameter was defined as 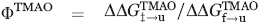
The 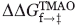 and
and 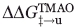 were calculated from the folding (kf) and unfolding (ku) rates obtained in the absence and the presence (denoted with a +) of TMAO according to
were calculated from the folding (kf) and unfolding (ku) rates obtained in the absence and the presence (denoted with a +) of TMAO according to 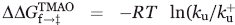 ),
), 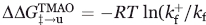 ), and
), and 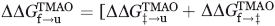 .
.
TMAO jump experiment
The TMAO jump experiment was performed using a GuHCl denatured sample of WT Fyn SH3 domain in 3M GuHCl where the domain is approximately half unfolded. In this experiment, a sample of WT Fyn SH3 domain in 3M GuHCl was rapidly mixed with increasing concentrations of osmolyte TMAO (prepared in 3M GuHCl) and the evolution of the folded state was recorded as a function of TMAO mixing time. The kinetics traces were subsequently fit to appropriate single exponential functions to extract the time constant of the TMAO-induced folding using BioKine software (BioLogic Instruments, Claix, France).
Equilibrium GuHCl-induced unfolding studies of the Fyn SH3 domain
Equilibrium GuHCl melts were monitored using a protein concentration of 1 μM on an Aviv Spectrofluorometer ATF 105 (Aviv Instruments, Lakewood, NJ) monitoring Trp fluorescence emission at 340 nm with excitation at 295 nm. The melt profiles were then fit to appropriate equations assuming two-state transition in IGOR Pro (WaveMetrics, Portland, OR) using a nonlinear least squares method as described previously.22
Temperature-induced unfolding studies of the Fyn SH3 domain
For each mutant, the change in the ellipticity (mdeg) at 220 nm in the absence or in the presence of 1M TMAO was monitored on an Aviv Circular Dichroism spectrometer model 62A DS (Aviv Instruments, Lakewood, NJ). The melting temperature (Tm), and change in enthalpy upon unfolding at Tm (ΔHf→u(Tm)) values for each mutant were determined with manual fitting of the melting curves.56 This method does not require an assumption of the ΔCp value of a mutant protein.
Concluding Remarks
In this work, we demonstrate that the osmolyte TMAO does not alter the structure of the folding transition state of the Fyn SH3 domain. Based on our results, we conclude that TMAO can reliably be used for the purposes of Φ-value analysis on very unstable mutants that would otherwise be unamenable for folding kinetic analysis. In this regard, of special interest is the observation that the effects of TMAO on the folding energetics of the Fyn SH3 domain is nonspecific with respect to a wide range of side chain chemistries and does not exhibit dependence on the secondary and tertiary structure contexts surrounding amino acid side chains. Thus, the use of TMAO could provide an important extension to our knowledge about the folding properties of a variety of highly destabilized mutants.
Acknowledgments
The authors thank Dr. Julie Forman-Kay for the use of Stopped-flow device. S.L is a recipient of a CIHR Training Grant in Protein Folding fellowship. A.Z-A is supported by a doctoral Canada Graduate Scholarship (CGS-D3) from the Natural Sciences and Engineering Research Council of Canada. This work was supported by an operating grant to A.R.D. from the Canadian Institutes of Health Research (MOP-13609).
Glossary
Abbreviations:
- CD
circular dichroism
- GuHCl
guanidine hydrochloride
- SH3
src homology 3
- TFE
trifluoroethanol
- TMAO
trimethylamine-N-oxide.
References
- 1.Yancey PH, Somero GN. Counteraction of urea destabilization of protein structure by methylamine osmoregulatory compounds of elasmobranch fishes. Biochem J. 1979;183:317–323. doi: 10.1042/bj1830317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa T, Timasheff SN. Mechanism of stabilization of proteins by glycerol and sucrose. Seikagaku. 1982;54:1255–1259. [PubMed] [Google Scholar]
- 4.Arakawa T, Timasheff SN. Preferential interactions of proteins with solvent components in aqueous amino acid solutions. Arch Biochem Biophys. 1983;224:169–177. doi: 10.1016/0003-9861(83)90201-1. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa T, Timasheff SN. The mechanism of action of Na glutamate, lysine HCl, and piperazine-N,N′-bis(2-ethanesulfonic acid) in the stabilization of tubulin and microtubule formation. J Biol Chem. 1984;259:4979–4986. [PubMed] [Google Scholar]
- 6.Arakawa T, Timasheff SN. Mechanism of poly(ethylene glycol) interaction with proteins. Biochemistry. 1985;24:6756–6762. doi: 10.1021/bi00345a005. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa T, Timasheff SN. The stabilization of proteins by osmolytes. Biophys J. 1985;47:411–414. doi: 10.1016/S0006-3495(85)83932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arakawa T, Bhat R, Timasheff SN. Preferential interactions determine protein solubility in three-component solutions: the MgCl2 system. Biochemistry. 1990;29:1914–1923. doi: 10.1021/bi00459a036. [DOI] [PubMed] [Google Scholar]
- 9.Timasheff SN. The control of protein stability and association by weak interactions with water: how do solvents affect these processes? Annu Rev Biophys Biomol Struct. 1993;22:67–97. doi: 10.1146/annurev.bb.22.060193.000435. [DOI] [PubMed] [Google Scholar]
- 10.Lin TY, Timasheff SN. Why do some organisms use a urea-methylamine mixture as osmolyte? Thermodynamic compensation of urea and trimethylamine N-oxide interactions with protein. Biochemistry. 1994;33:12695–12701. doi: 10.1021/bi00208a021. [DOI] [PubMed] [Google Scholar]
- 11.Saunders AJ, Davis-Searles PR, Allen DL, Pielak GJ, Erie DA. Osmolyte-induced changes in protein conformational equilibria. Biopolymers. 2000;53:293–307. doi: 10.1002/(SICI)1097-0282(20000405)53:4<293::AID-BIP2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Weatherly GT, Pielak GJ. Second virial coefficients as a measure of protein–osmolyte interactions. Protein Sci. 2001;10:12–16. doi: 10.1110/ps.29301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schellman JA. Protein stability in mixed solvents: a balance of contact interaction and excluded volume. Biophys J. 2003;85:108–125. doi: 10.1016/S0006-3495(03)74459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Bolen DW. The peptide backbone plays a dominant role in protein stabilization by naturally occurring osmolytes. Biochemistry. 1995;34:12884–12891. doi: 10.1021/bi00039a051. [DOI] [PubMed] [Google Scholar]
- 15.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 16.Qu Y, Bolen CL, Bolen DW. Osmolyte-driven contraction of a random coil protein. Proc Natl Acad Sci USA. 1998;95:9268–9273. doi: 10.1073/pnas.95.16.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolen DW, Baskakov IV. The osmophobic effect: natural selection of a thermodynamic force in protein folding. J Mol Biol. 2001;310:955–963. doi: 10.1006/jmbi.2001.4819. [DOI] [PubMed] [Google Scholar]
- 18.Russo AT, Rosgen J, Bolen DW. Osmolyte effects on kinetics of FKBP12 C22A folding coupled with prolyl isomerization. J Mol Biol. 2003;330:851–866. doi: 10.1016/s0022-2836(03)00626-0. [DOI] [PubMed] [Google Scholar]
- 19.Mukaiyama A, Koga Y, Takano K, Kanaya S. Osmolyte effect on the stability and folding of a hyperthermophilic protein. Proteins. 2008;71:110–118. doi: 10.1002/prot.21660. [DOI] [PubMed] [Google Scholar]
- 20.Pradeep L, Udgaonkar JB. Osmolytes induce structure in an early intermediate on the folding pathway of barstar. J Biol Chem. 2004;279:40303–40313. doi: 10.1074/jbc.M406323200. [DOI] [PubMed] [Google Scholar]
- 21.Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell KL, Davidson AR. Mutagenesis of a buried polar interaction in an SH3 domasequence conservation provides the best prediction of stability effects. Biochemistry. 1998;37:16172–16182. doi: 10.1021/bi981788p. [DOI] [PubMed] [Google Scholar]
- 23.Plaxco KW, Guijarro JI, Morton CJ, Pitkeathly M, Campbell ID, Dobson CM. The folding kinetics and thermodynamics of the Fyn-SH3 domain. Biochemistry. 1998;37:2529–2537. doi: 10.1021/bi972075u. [DOI] [PubMed] [Google Scholar]
- 24.Northey JG, Di Nardo AA, Davidson AR. Hydrophobic core packing in the SH3 domain folding transition state. Nat Struct Biol. 2002;9:126–130. doi: 10.1038/nsb748. [DOI] [PubMed] [Google Scholar]
- 25.Northey JG, Maxwell KL, Davidson AR. Protein folding kinetics beyond the phi value: using multiple amino acid substitutions to investigate the structure of the SH3 domain folding transition state. J Mol Biol. 2002;320:389–402. doi: 10.1016/S0022-2836(02)00445-X. [DOI] [PubMed] [Google Scholar]
- 26.Di Nardo AA, Korzhnev DM, Stogios PJ, Zarrine-Afsar A, Kay LE, Davidson AR. Dramatic acceleration of protein folding by stabilization of a nonnative backbone conformation. Proc Natl Acad Sci USA. 2004;101:7954–7959. doi: 10.1073/pnas.0400550101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarrine-Afsar A, Dahesh S, Davidson AR. Protein folding kinetics provides a context-independent assessment of beta-strand propensity in the Fyn SH3 domain. J Mol Biol. 2007;373:764–774. doi: 10.1016/j.jmb.2007.07.059. [DOI] [PubMed] [Google Scholar]
- 28.Zarrine-Afsar A, Larson SM, Davidson AR. The family feud: do proteins with similar structures fold via the same pathway? Curr Opin Struct Biol. 2005;15:42–49. doi: 10.1016/j.sbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Grantcharova VP, Baker D. Folding dynamics of the src SH3 domain. Biochemistry. 1997;36:15685–15692. doi: 10.1021/bi971786p. [DOI] [PubMed] [Google Scholar]
- 30.Martinez JC, Serrano L. The folding transition state between SH3 domains is conformationally restricted and evolutionarily conserved. Nat Struct Biol. 1999;6:1010–1016. doi: 10.1038/14896. [DOI] [PubMed] [Google Scholar]
- 31.Riddle DS, Grantcharova VP, Santiago JV, Alm E, Ruczinski I, Baker D. Experiment and theory highlight role of native state topology in SH3 folding. Nat Struct Biol. 1999;6:1016–1024. doi: 10.1038/14901. [DOI] [PubMed] [Google Scholar]
- 32.Grantcharova VP, Riddle DS, Baker D. Long-range order in the src SH3 folding transition state. Proc Natl Acad Sci USA. 2000;97:7084–7089. doi: 10.1073/pnas.97.13.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Nardo AA, Larson SM, Davidson AR. The relationship between conservation, thermodynamic stability, and function in the SH3 domain hydrophobic core. J Mol Biol. 2003;333:641–655. doi: 10.1016/j.jmb.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 34.Zarrine-Afsar A, Wallin S, Neculai AM, Neudecker P, Howell PL, Davidson AR, Chan HS. Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding. Proc Natl Acad Sci USA. 2008;105:9999–10004. doi: 10.1073/pnas.0801874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mok YK, Elisseeva EL, Davidson AR, Forman-Kay JD. Dramatic stabilization of an SH3 domain by a single substitution: roles of the folded and unfolded states. J Mol Biol. 2001;307:913–928. doi: 10.1006/jmbi.2001.4521. [DOI] [PubMed] [Google Scholar]
- 36.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 37.Matouschek A, Fersht AR. Application of physical organic chemistry to engineered mutants of proteins: Hammond postulate behavior in the transition state of protein folding. Proc Natl Acad Sci USA. 1993;90:7814–7818. doi: 10.1073/pnas.90.16.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez IE, Kiefhaber T. Hammond behavior versus ground state effects in protein folding: evidence for narrow free energy barriers and residual structure in unfolded states. J Mol Biol. 2003;327:867–884. doi: 10.1016/s0022-2836(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 39.Matouschek A, Fersht AR. Protein engineering in analysis of protein folding pathways and stability. Methods Enzymol. 1991;202:82–112. doi: 10.1016/0076-6879(91)02008-w. [DOI] [PubMed] [Google Scholar]
- 40.Mello CC, Barrick D. Measuring the stability of partly folded proteins using TMAO. Protein Sci. 2003;12:1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kentsis A, Sosnick TR. Trifluoroethanol promotes helix formation by destabilizing backbone exposure: desolvation rather than native hydrogen bonding defines the kinetic pathway of dimeric coiled coil folding. Biochemistry. 1998;37:14613–14622. doi: 10.1021/bi981641y. [DOI] [PubMed] [Google Scholar]
- 42.Yiu CP, Mateu MG, Fersht AR. Protein folding transition states: elicitation of Hammond effects by 2,2,2-trifluoroethanol. Chembiochem. 2000;1:49–55. doi: 10.1002/1439-7633(20000703)1:1<49::AID-CBIC49>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 43.Main ER, Jackson SE. Does trifluoroethanol affect folding pathways and can it be used as a probe of structure in transition states? Nat Struct Biol. 1999;6:831–835. doi: 10.1038/12287. [DOI] [PubMed] [Google Scholar]
- 44.Hamada D, Chiti F, Guijarro JI, Kataoka M, Taddei N, Dobson CM. Evidence concerning rate-limiting steps in protein folding from the effects of trifluoroethanol. Nat Struct Biol. 2000;7:58–61. doi: 10.1038/71259. [DOI] [PubMed] [Google Scholar]
- 45.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Athawale MV, Dordick JS, Garde S. Osmolyte trimethylamine-N-oxide does not affect the strength of hydrophobic interactions: origin of osmolyte compatibility. Biophys J. 2005;89:858–866. doi: 10.1529/biophysj.104.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hovagimyan KG, Gerig JT. Interactions of trimethylamine N-oxide and water with cyclo-alanylglycine. J Phys Chem B. 2005;109:24142–24151. doi: 10.1021/jp055075+. [DOI] [PubMed] [Google Scholar]
- 48.Holthauzen LM, Bolen DW. Mixed osmolytes: the degree to which one osmolyte affects the protein stabilizing ability of another. Protein Sci. 2007;16:293–298. doi: 10.1110/ps.062610407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul S, Patey GN. The influence of urea and trimethylamine-N-oxide on hydrophobic interactions. J Phys Chem B. 2007;111:7932–7933. doi: 10.1021/jp0733668. [DOI] [PubMed] [Google Scholar]
- 50.Auton M, Bolen DW. Additive transfer free energies of the peptide backbone unit that are independent of the model compound and the choice of concentration scale. Biochemistry. 2004;43:1329–1342. doi: 10.1021/bi035908r. [DOI] [PubMed] [Google Scholar]
- 51.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu P, Bolen DW. Osmolyte-induced protein folding free energy changes. Proteins. 2006;63:290–296. doi: 10.1002/prot.20868. [DOI] [PubMed] [Google Scholar]
- 53.Singh R, Haque I, Ahmad F. Counteracting osmolyte trimethylamine N-oxide destabilizes proteins at pH below its pKa. Measurements of thermodynamic parameters of proteins in the presence and absence of trimethylamine N-oxide. J Biol Chem. 2005;280:11035–11042. doi: 10.1074/jbc.M410716200. [DOI] [PubMed] [Google Scholar]
- 54.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarrine-Afsar A, Davidson AR. The analysis of protein folding kinetic data produced in protein engineering experiments. Methods. 2004;34:41–50. doi: 10.1016/j.ymeth.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Allen DL, Pielak GJ. Baseline length and automated fitting of denaturation data. Protein Sci. 1998;7:1262–1263. doi: 10.1002/pro.5560070524. [DOI] [PMC free article] [PubMed] [Google Scholar]


