Abstract
Numerous experiments in mutant and transgenic mice have implicated Hox transcription factors in development of the skeletal system, postulating a role for these proteins in cell proliferation of precursor cells and regulation of cell differentiation. Our own data from Hoxc8 and Hoxd4 transgenic mice suggest that Hoxc8 is involved in cell proliferation during cartilage development. In order to directly assess its role in cell proliferation of a specific skeletal cell type, the cartilage-producing chondrocyte, we performed morpholino-mediated knockdown experiments in normal primary chondrocytes. Through analysis of PCNA expression and staining for phosphorylated Histone 3, two cell cycle markers, we show that interference with Hoxc8 expression in chondrocytes reduces cell proliferation, but in the absence of apoptosis. Instead, cells with a knockdown in Hoxc8 expression appear to be delayed in their progression through the cell cycle. Our results provide evidence for prolonged duration of and delayed exit from M-phase, thus implicating a role for Hoxc8 in controlling cell cycle progression at this critical check point.
Keywords: Homeodomain, cell proliferation, cartilage, RNAi, knockdown, phosphorylated Histone 3, Proliferating Cell Nuclear Antigen, PCNA, S-phase, M-phase, cell cycle, transcription factor
INTRODUCTION
Hox transcription factors play important roles during development of the skeletal system, both in the patterning as well as the growth phase [1]. Targeted deletion of individual Hox genes often affects patterning of those specific skeletal elements in whose precursor cells the respective transcript is highly expressed [2]. Altered cell differentiation as well as altered migration and adhesion have been postulated to be the cause of patterning abnormalities in the skeleton [3]. For example, Yokouchi et al. showed in a competitive aggregation assay of mesenchymal cells from chick limb bud that HOXA13 expressing cells exhibited different adhesive properties compared to cells negative for the transcription factor [4]. Alternatively, altered proliferation of skeletal precursor cells has been implicated [5]. For example, Goff et al. found that overexpression of Hoxd13 was associated with decreased radioactive thymidine incorporation in tibia and increased incorporation in fibia [6]. Thus, locally differing rates of proliferation and cell survival may thus be responsible for the particular shape differences of skeletal elements [7].
A possible role in cell proliferation is supported by findings in compound mutants in which those skeletal elements that lack more than one Hox gene product may not only be mis-patterned but may also be smaller in size [8]. This reduced Hox protein expression appears to be associated with reduced cell growth or survival. Conversely, overexpression of Hox genes in cultured cells has been shown to promote cell proliferation and even tumorigenesis in vivo [9, 10]. In the developing skeleton, overexpression of Hox genes in ectopic sites may lead to defective patterning, but also to abnormalities in cartilage differentiation [11–14]. Overexpression of a Hox gene in its normal domain has been demonstrated to affect cell proliferation, typically seen as an increase in numbers of proliferating chondrocytes [13]. Collectively, these studies all show Hox transcription factor levels to be associated with the regulation of cell proliferation.
However, formation of skeletal elements is also dependent on proper commitment of precursors to the skeletogenic lineage [15], their migration to appropriate sites, and subsequent terminal differentiation to mature hypertrophic chondrocytes [16]. The temporal overlap of differentiation, migration, proliferation and survival of chondrocytes during specification and formation of skeletal elements makes it difficult to pinpoint precisely the functional role of Hox genes in chondrocytes in vivo. We therefore have employed an in vitro model of cartilage differentiation, a previously established high-density tissue culture system [17], combined with morpholino-mediated RNA interference [18] to study the role of Hox transcription factors in chondrocytes.
METHODS
Primary Chondrocyte Cultures
Preparation of primary rib chondrocytes from embryos was done by a modification of the method used by Lefebvre et al. [19]. Briefly, embryos from the FVB inbred mouse strain were isolated at 18.5 days of gestation (E 18.5); the rib cages were dissected in sterile conditions and transferred to PBS. After several washes with PBS, rib cages were digested with 0.25% collagenase type 1A (Sigma, USA) and 0.25% Trypsin for 1 hour at 37°C to remove all the soft tissues between the ribs. The rib cages were washed with PBS and the collagenase solution replenished. The cells released during continuous 90 min incubation were filtered through a 70 μm cell strainer, centrifuged at 1200 rpm for 10 min, washed with PBS and re-centrifuged at 1200 rpm for 5 min. Cells were counted with a haemocytometer, and plated at high density (105 cells/cm2) on plates coated with 0.1% gelatin in DMEM-F12 medium (Gibco, USA) containing 10% fetal bovine serum (FBS), penicillin/streptomycin (Gibco, USA), 5mM beta-glycerophosphate and 4 μM Ascorbic acid (Sigma, USA). After one day, the cells were trypsinized off and reseeded in culture plates appropriate for each experiment: Chamber slides were used for immunolocalization of Hoxc8, 6-well plates were used to extract RNA from primary chondrocyte cultures, and 12-well plates were used for the morpholino transfections to study the effect of Hoxc8 knockdown on proliferation and apoptosis.
Immunolocalization of Hoxc8
Cells plated into chamber slides were washed with PBS and fixed with 4% paraformaldehyde (PFA) for 1 hour at room temperature after various days of culture. After fixation, cells were washed thoroughly with PBS and permeabilized by incubation in 0.1% Triton X-100/1% sodium citrate/PBS for 10 min at 4°C. After washing with PBS, cells were incubated in blocking buffer (3% BSA/PBS) for 30 min at room temperature. Cells were incubated with primary monoclonal antibody against Hoxc8 [20] (Abcam, USA) diluted 1:500 in 1% BSA/PBS for 1 hour at room temperature. After mounting the cells with Vectashield mounting medium for fluorescence with DAPI (Vector Laboratories, Burlingame, CA), slides were cover slipped and examined under a Leica confocal microscope (LSM 510 META confocal scanning system).
Quantitative RT-PCR
Hoxc8 gene expression at the mRNA level was evaluated using the ABI Prism 7000 Instrument (Applied Biosystems, Foster City, CA). RNA was isolated from primary chondrocyte cultures on days 0, 3, 6, 9, 12, 15, and 18 (n=3 cultures/day) as described previously [21]. Primary chondrocytes were transferred into Trizol reagent (Invitrogen, Carlsbad, CA) and RNA was extracted according to the manufacturer’s protocol. Complementary DNA was obtained by reverse transcription (Superscript II, Invitrogen, Carlsbad, CA) of at least 5μg of RNA of each sample, following the supplier’s instructions. Purification of cDNA was done using QIAquik PCR purification columns (Qiagen, Valencia, CA). RNA as well as cDNA concentrations were measured with a NanoDrop ND-100 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Primers for amplification were designed using Primer Express software (Applied Biosystems, Foster City, CA). Hoxc8 primer sequences were 5′-CGAAGGACAAGGCCACTTAAAT-3′ (forward primer) and 5′-AGGTCTGATACCGGCTGTAAGTTT-3′ (reverse primer). Primers for two reference genes were: Gluceraldehyde-3-phosphate dehydrogenase (Gapdh) primers as provided by Applied Biosystems with the sequences 5′-CCAGAACATCATCATCCCTGCATC-3′ (forward primer) and 5′GGTAGGAACACGGAGGCC-3′ (reverse primer), and primers for Polymerase epsilon 4 (Pole4) with the sequences forward 5′-CGGGACAGGAAGCCATCTT-3′ and reverse 5′-AGCAGTAGGCATCTTTTGCGATA-3′ were used to measure Pole4 and Gapdh transcript levels in aliquots of the same cDNA samples, under the same conditions, and on the same plates as the measurements of Hoxc8 transcript levels. Quantitative real-time PCR was performed and amplified products were detected using SYBER Green PCR Master Mix (Applied Biosystems) exactly as described before [21].
RNA Interference using morpholino oligonucleotides
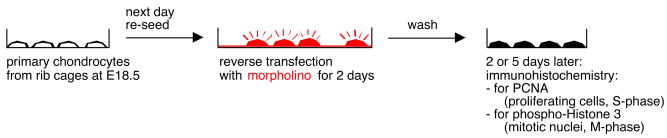
The lissamine-tagged antisense morpholino targeted to Mus musculus Hoxc8 mRNA had the sequence 5′-CATGCTGGGTACATGAAAACCCGCG-3′. The lissamine-tagged control oligonucleotide (called “standard” by the manufacturer and used interchangeably with “control” here) had the sequence 5′-CCTCTTACCTCAGTTACAATTTATA-3′, which is targeted to a splice site mutant unique to human beta-globin in thalassemia. To deliver the oligos into the cells, a weak-base amphiphilic peptide, Endo-Porter [22] (GeneTools, Philomath, OR), was added at a concentration of 4μl/ml. Freshly isolated cells that had been plated into 175 cm2 flasks the day before were washed with PBS and trypsinized using Trypsin/EDTA to be reseeded at high density into 12-well plates already containing 10μM mopholino/well (reverse transfection: transfection solution present before cells). Cells were incubated either with Hoxc8-specific mopholino or control (standard) oligo or Endo-Porter alone for 48 hours at 37°C. After 48 hours, cells were washed and the medium changed to normal chondrocyte medium (see above). Thus, the duration of transfection was for 2 days, and we refer to the time point of media change after as day 0 after transfection (see Figure 2). Immunostaining for Hoxc8 and Western blotting were done to confirm the knock down of Hoxc8 protein expression.
Figure 2.
Graphical representation of experimental approach.
Proliferation Assays
Primary chondrocyte cultures or cultures transfected with either standard or Hoxc8-specific morpholinos were fixed with 4% paraformaldehyde at −20°C for 20 min on days 2 and 5 after transfection (total days in culture: 5 and 8 days, respective, including the initial reseeding). After fixation, cells were washed thoroughly with PBS and permeabilized in 0.2% TritonX-100/PBS for 10 min. After washing with PBS, cells were incubated in blocking buffer (3%BSA/PBS) for 30 min at room temperature. Cells were incubated with mouse monoclonal antibody against proliferating nuclear antigen (PCNA) (Invitrogen) diluted 1:100 in 1% BSA/PBS, and rat monoclonal antibody against phosphorylated Histone H3 1:100 (Abcam, Cambridge, MA) overnight at 4°C. The cells were washed 3 times with PBS and then incubated with goat anti-mouse IgG secondary antibody conjugated to Alexa fluor 488 (1:500) or goat anti-rat IgG secondary antibody conjugated to Alexa fluor 555 respectively (Invitrogen) for 1 hour at room temperature. Cells mounted with mounting media containing DAPI were examined by fluorescence microscopy. Ten random pictures/well (3–4 wells/group) were taken under identical magnification and exposure conditions and images were binarized in Photoshop (Adobe, San Jose, CA). The threshold for binarization was held identical for all images of the same experiment. Using the pixel count function in Photoshop, the number of pixels above the threshold was recorded to measure the areas of positive fluorescence in each image. These areas represent the nuclei staining positive for either PCNA (green channel), anti-histone H3 (red channel) or DAPI (blue channel). The ratio between the pixel number for PCNA positive nuclei and pixel number for DAPI-stained nuclei (PCNA/DAPI) was used to determine the fraction of total signal that derived from cells in S phase of the cell cycle. The ratio of pixel number for phosphorylated-Histone H3 positive nuclei over pixel number for DAPI positive nuclei (H3/DAPI) was used to determine the fraction of signal derived from cells in mitosis (M-phase). Nuclei in mitosis (H3 positive) are larger than those of other proliferating cells (PCNA positive, compare size at the same magnification in Figure 4 Panels A and B), and therefore, a direct comparison of the pixel numbers between PCNA and H3 staining cannot be made. The major comparison in all of our assays is to be made between experimental and control samples.
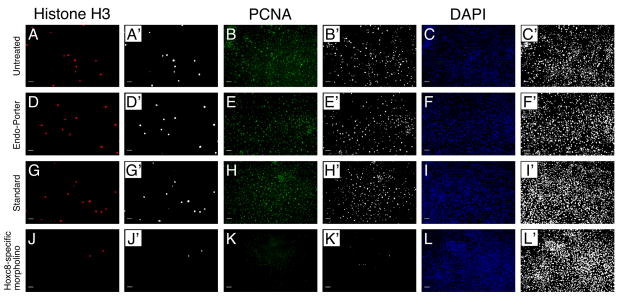
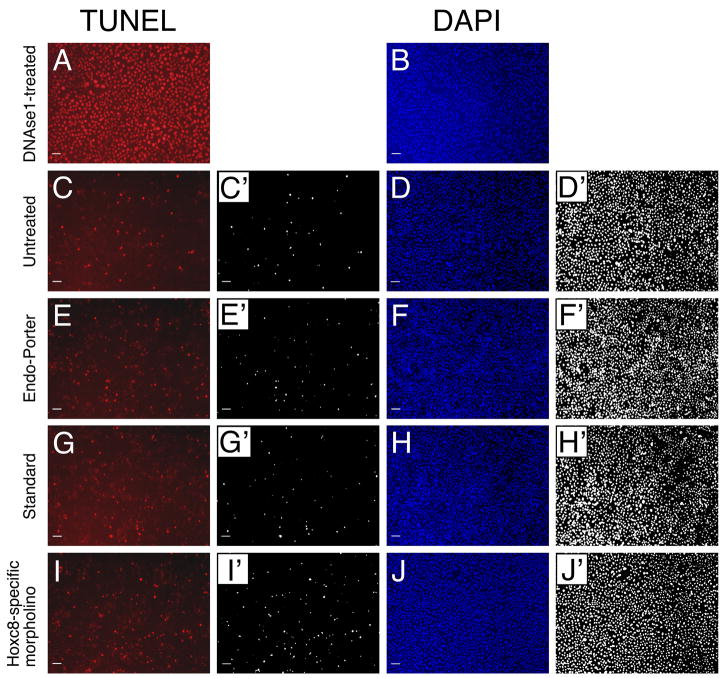
Figure 4. Morpholino-mediated knockdown of Hoxc8 affects cell proliferation.
Primary chondrocyte cultures stained with anti-phospho-histone H3 antibody (A, D, G, J), PCNA-specific antibody (B, E, H, K) and for DAPI (C, F, I, L).
(A–C): Untreated cultures
(D–F): Cultures incubated with Endo-Porter alone.
(G–I): Cultures transfected with 10μM standard oligo.
(J–L): Cultures transfected with 10μM Hoxc8-specific morpholino oligo.
The scale bar represents 100μM. All images labeled by apostrophes were converted to black and white format using the same threshold for quantification of pixel number (see Methods).
Apoptosis Assay
Apoptosis was detected using an in situ cell death detection kit, TMR red (Roche, Indianapolis, IN). Cultures were fixed on days 1 or 3 after transfection experiment. Cells were washed with PBS and fixed with cold 4% paraformaldehyde for 1 hour at room temperature. The cells were then rinsed with PBS, and permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate for 2 min on ice. The cells were incubated in TUNEL reaction mixture for 1 hour at 37°C in humidified atmosphere in the dark. Cells pre-incubated with DNase 1 (3U/ml) in 50mM Tris-HCl, pH 7.5, and 1mg/ml BSA) for 10 min (room temperature) prior to the TUNEL staining procedure served as positive controls for the apoptosis detection method. Omission of deoxynucleotidyl transferase (TdT) from the staining procedure served as the negative control for the detection method. After washing three times with PBS, cells were mounted with media containing CAPO and visualized by fluorescence microscopy. Ten random pictures/well (3 wells per experimental group) were taken and the number of apoptotic cells was determined by the pixel count method as described above.
Statistical Evaluation
Means ± SEM were calculated in Microsoft Excel by averaging of values from all images for an individual triplicate, and then between the triplicates for each sample. Results were compared between the control and experimental groups using unpaired T-tests with two-tailed P value in Prism (GraphPad Software, San Diego, CA); P<0.05 was considered significant.
RESULTS
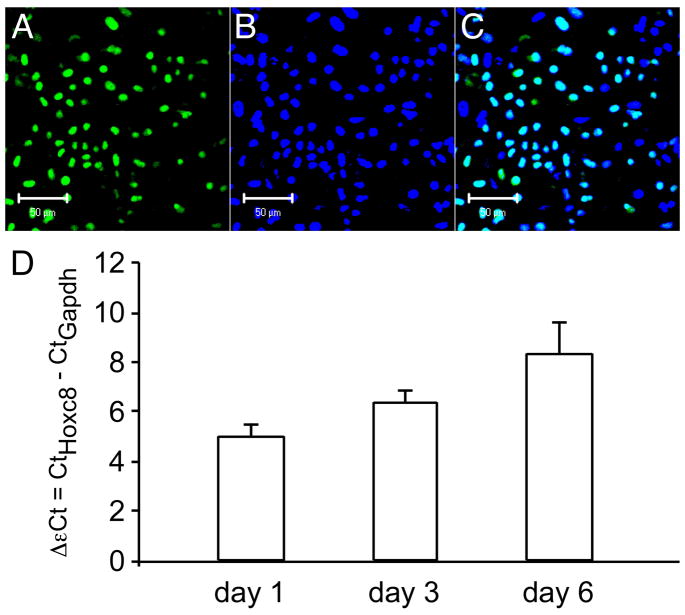
This study was designed to investigate the role of Hoxc8 in chondrocytes. We have previously shown that Hoxc8 RNA is expressed in the developing cartilage by in situ hybridization [13]. By RT-PCR, we showed transcript present in primary chondrocytes [21], as well as cultured chondrocytes [17]. We now demonstrate that Hoxc8 protein is also present in primary chondrocytes cultured at high density (Figure 1). As expected for a transcription factor, Hoxc8 protein is detected in the nuclei of cells as demonstrated by simultaneous staining with DAPI (Figure 1, B and C). We also followed the expression of Hoxc8 in normal primary chondrocytes for more than 2 weeks of differentiation in culture. While Hoxc8 expression is detected at every stage of the cultures, the strongest signal is observed in early stages up to day 6 (Figure 1), with intensity declining towards later days. Due to the development of cartilage nodules and consequent uneven distribution of differentiated cells in later stages of the culture (after 7–10 days), the decline in expression was difficult to quantify by fluorescence microscopy. Independent validation that Hoxc8 is expressed at higher levels at early stages of the cultures was obtained by quantitative RT-PCR (Figure 1, D), which detects declining levels (increasing ΔCt values) of Hoxc8 expression in chondrocytes correlated to time spent in culture.
Figure 1. Expression of Hoxc8 in primary chondrocyte cultures.
Immunolocalization of Hoxc8: Chondrocytes were fixed and immunostained for Hoxc8 on day 6 of culture. Slides were mounted with media containing DAPI and examined under a Leica confocal microscope. (A) Nuclear staining for Hoxc8; (B) DAPI staining of total cell nuclei; (C) Merged images for DAPI and Hoxc8. No signal was detected when the primary antibody was omitted. D) Quantitative RT-PCR for Hoxc8. The levels of Hoxc8 (as detected by cycle number above threshold in each sample, Ct) were normalized to the levels of the reference gene in the same sample and are expressed as ΔCt (ΔCt = CtHoxc8 − CtGapdh). Please note that lower bars (smaller ΔCt) reflect higher levels of expression.
In order to investigate the functional role of Hoxc8 in chondrocytes, we applied the morpholino antisense interference approach to primary chondrocytes isolated from mice just prior to birth. Because Hoxc8 is preferentially expressed in less differentiated chondrocytes, the early days of chondrocyte culture are therefore the appropriate subjects for functional analyses. Also, during the first week after isolation, primary chondrocyte cultures contain very few, if any, hypertrophic chondrocytes; they typically appear in later days of culture, and thus are not expected to confound our experiments. Figure 2 depicts the experimental scheme for morpholino transfections of primary cultures. Chondrocytes require acclimating to the culture for one day, after which they were re-seeded into plates that already contained morpholino oligonucleotides and/or the transfection agent (reverse transfection). We consider the two days that the cells spend in presence of morpholino oligonucleotides the period of transfection, and analyzed the effects of interference with Hoxc8 expression 2 and 5 days thereafter.
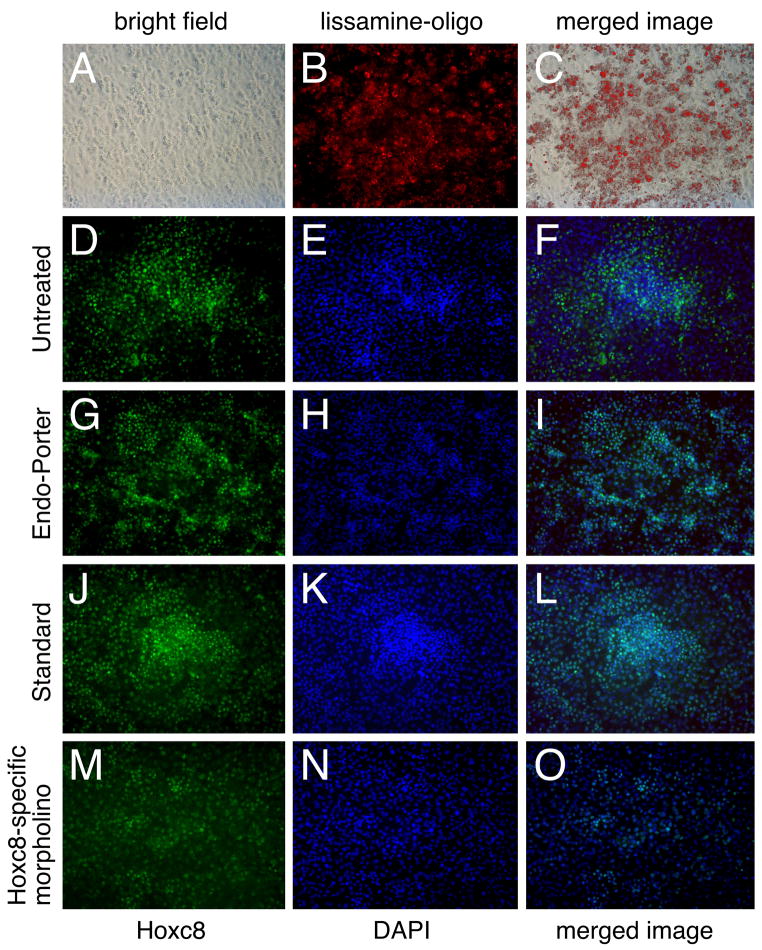
Figure 3 (Panels A–C) shows that high density cultured chondrocytes efficiently ingest lissamine-coupled morpholine oligonucleotides. The overlay of the fluorescence signal onto the bright-field image (C) as well as cell counting indicated that 76% of the cells contained morpholino after 2 days of transfection and an additional 2 days of culture. Thus, even with ongoing cell proliferation, the majority of cells in the culture are exposed to morpholino. Cultures that were incubated either with Endo-Porter alone or with the control morpholino (standard) exhibited essentially similar cell growth (compare Panels E, H, K) and Hoxc8 staining patterns (Panels D, G, J) as untreated cells, and control morpholino-incubated cells were also able to form cartilage nodules (Panels J–L). Thus, neither the transfection agent (Endo-Porter) alone, nor the standard control oligonucleotide affected chondrocyte differentiation. In contrast, the Hoxc8-specific morpholino-exposed cultures exhibited a decrease in cell number (Panel N) as well as Hoxc8 staining (Panels M and O), both in terms of fewer cells staining with the Hoxc8-specific antibody as well as reduced staining intensity. These data show that morpholino-mediated interference decreases Hoxc8 protein expression levels. The fact that lower cell numbers were present after interference with Hoxc8 expression suggested that cell proliferation or survival was affected by knockdown of Hoxc8. We therefore assessed the expression of markers for cell proliferation and also assayed the effect of Hoxc8 RNAi at different timepoints.
Figure 3. Primary chondrocytes 2 days after reverse transfection with additional 2 days of culture.
(A): Bright field picture of culture;
(B): Lissamine-tagged morpholino oligos ingested by cells;
(C): Merged image of bright field (A) and fluorescence image (B);
(D–F) Untreated cultures;
(G–I) Cultures to which only Endo-Porter was added;
(J–L) Cultures transfected with irrelevant sequence morpholino (“standard”);
(M–O) Cultures transfected with Hoxc8-specific morpholino.
(D, G, J, M): Immunofluorescence for Hoxc8.
(E, H, K, N): Nuclear staining with DAPI.
(F, I, L, O): Merged images of Hoxc8 fluorescence and DAPI staining.
Figure 4 depicts representative images from cultures left untreated (Panels A–C), exposed to Endo-Porter alone (Panels D–E) or to control oligonucleotide (Panels G–I), which all appeared indistinguishable in number of PCNA-stained or phosphorylated-Histone-3-stained cells, respectively. In contrast, cultures exposed to the Hoxc8-specific morpholino showed clearly reduced numbers of PCNA and Phospho-Histone-3-positive cells 5 days later (Panels J–L). Figure 4 also shows the graphical representation used for quantification of pixel number above threshold. These results suggest that with knockdown of Hoxc8, chondrocytes proliferate less than cells with Hoxc8 present.
One possibility is that morpholino transfection increases apoptosis; then, only cells would survive that are either untransfected or experience minimal reduction of protein levels. To account for this possibility, we directly tested for the presence of apoptotic cells 1 day after morpholino transfection. Figure 5 shows that in all experimental samples, some apoptotic cells were observed, but there were no significant differences between vehicle- or morpholino-treated cells and untreated chondrocytes. Thus, the reduction in cell numbers observed at later days in cultures exposed to Hoxc8-specific morpholino is not due to prior apoptosis of the transfected cells. This excludes morpholino toxicity as a technically confounding factor. We then conclude that the decrease in cell proliferation must be due to fewer cells entering or completing the cell cycle. This was confirmed by performing a time-course experiment. Assays for proliferation markers just 2 days after transfection (Figure 6) yielded data that are indeed consistent with the interpretation that Hoxc8 knockdown inhibits cells from entering mitosis. Even at this early time point, a reduction in PCNA-positive cells and in phospho-Histone 3 staining was detectable, although the magnitude of reduction almost doubled between days 2 and 5 after transfection. Thus, Hoxc8 knockdown reduces chondrocyte proliferation and inhibits entry into mitosis. Collectively, our data suggest that Hoxc8 is involved in regulating chondrocyte proliferation.
Figure 5. Minimal cell death in morpholino-exposed primary mouse chondrocyte cultures.
(A, B) DNAseI-treated chondrocytes as positive control for TUNEL staining
(C, D) Untreated chondrocytes
(E, F) Endo-Porter-exposed chondrocytes
(G, H) Chondrocytes transfected with standard oligo nucleotide
(I, J) Chondrocytes transfected with Hoxc8-specific morpholino
Panels labeled with apostrophe depict results used for quantification (see Methods).
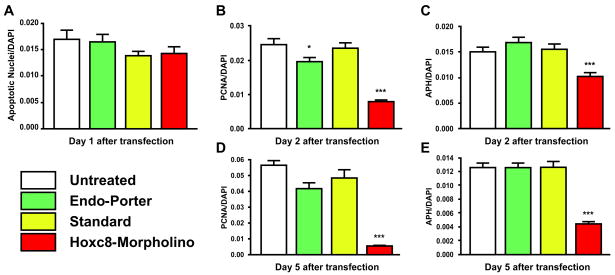
Figure 6. Knockdown of Hoxc8 in reduces cell proliferation and delays cell cycle progression.
(A): Chondrocyte cultures transfected with Hoxc8-specific morpholino show no difference in the apoptotic cell number compared to untreated and control groups on day 1 after transfection.
(B – E): Time-course of chondrocyte proliferation after morpholino transfection.
(B, D): Transfection with Hoxc8 specific antisense oligos reduced the number of cells entering S phase more than 70% on 2 days and 5 days after transfections. Although compared to the number of cells entering the S phase in untreated cultures, there is a decrease of PCNA-positive cells treated with Endo-Porter alone on day 2, there is no significant difference between untreated cells and cells transfected with the standard oligo. On day 5 there is no significant difference in the number of cells entering S phase in any of the control conditions, but PCNA-positivity is significantly reduced with Hoxc8-specific morpholino transfection.
(C, E): Transfection with Hoxc8-specific antisense oligos reduced the number of cells entering M phase to less than 60% of controls by day 2 after transfection, and to less than 30% of controls by day 5 after transfection. However, there is no significant difference between the number of cells entering the M phase in untreated culture compared to cells treated with Endo-Porter alone or with standard control (*P<0.05, ***P<0.001).
DISCUSSION
This manuscript reports on the consequences of interfering with Hoxc8 expression in mouse cartilage. In mice with a germ-line deletion of Hoxc8 [23], homeotic transformations in the thoracic skeleton are present, but cartilage appears to form normally. This was interpreted by the authors to mean that possible functional redundancy exists with other Hox transcription factors expressed in overlapping patterns to Hoxc8. Indeed, in mice in which more than one Hox gene is inactivated, more severe impairment in growth and differentiation of cartilage were found [24]. These and other studies [25–27] suggested that lack of Hox function may affect propensity, timing or progression of chondrocyte differentiation. Similarly, we and others have shown that overexpression of Hox genes affects cartilage differentiation [6, 13, 14, 28]. While suggestive and informative, these in vivo experiments also reveal an important limitation, namely that it remains unclear whether chondrocytes become impaired during commitment to the lineage or in subsequent steps of differentiation. We therefore used a high-density culture system for freshly isolated committed chondrocytes that recapitulates their differentiation to hypertrophy within a time span of 7–10 days [17]. Since Hoxc8 is expressed in primary chondrocytes at the time of isolation [21], and the purity of the chondrocyte preparation is very high [17], the cell culture system enables us to investigate in which way decreasing the initial level of Hoxc8 expression affects proliferation or differentiation of chondrocytes that were already committed to this lineage at the time of their isolation from the animal.
We used morpholinos, small modified antisense oligo-nucleotides that overlap the translation initiation site, to inhibit production of Hoxc8 protein. Binding of the morpholino to its target site is believed to be very stable [29, 30] and to inhibit de novo protein synthesis, thus producing cells negative for Hoxc8 protein as soon as they transcribe the message. In those cells that already contained the Hoxc8 protein at the time of transfection, de novo synthesis will be blocked, with the existing protein undergoing degradation, presumably at the normal rate. The half-life of Hox proteins in chondrocytes is unknown; in HeLa cells, Hoxa9 half-life was determined to be 26 hours [31], although formation of complexes with other proteins [32] or posttranslational modifications, such as phosphorylation [33], or ubiquitination may shorten half-life [31]. Assuming a half-life for Hoxc8 similar to that shown for Hoxa9, all cells successfully transfected with morpholino should be devoid of Hoxc8 protein within 2 days. Despite the finding that a fraction of cells was non-transfected, the Hoxc8-specific morpholino had distinct biological effects in the cultured chondrocytes: We found that treatment with the Hoxc8-specific morpholino reduced cell proliferation.
Conceivably, reduced cell proliferation can be associated with increased cell death or with increased differentiation to a non-proliferative status, i.e. hypertrophy. In addition, it is possible that reduced cell proliferation could be associated with de-differentiation of cells. We could not find evidence of increased apoptosis after morpholino transfection and therefore exclude this possibility from our interpretation. Similarly, we did not observe greater propensity for hypertrophy in morpholino-treated cultures within the timeframe of the incubation period until time of analysis, namely 2 and 5 days after transfection (total of 7 days in the experiment, total of 8 days after plating of the freshly isolated cells). This time frame was hosen because Hox8 expression is highest in early cultures, in which there are, if at all, very few hypertrophic cells present; we did not find indications for increased hypertrophy after morpholino treatment in those cultures that we observed for a longer time after transfection. Thus, at least at the morphological level, there is no evidence that the potential for differentiation was altered. De-differentiation of chondrocytes into a fibroblastoid state is typically observed in low-density cultures, and although reduced cell proliferation produces fewer cells, our cultures did not become so sparse as to allow de-differentiation (see Figure 3). These considerations lead us to believe that de-differentiation or hypertrophy do not have a substantial impact on our results.
Immunofluorescence analysis revealed that the measurable reduction in cell proliferation was associated with a significant reduction in PCNA-positive cells, which are in S phase of the cell cycle [34, 35]. Thus, under conditions that decrease Hoxc8 expression, chondrocytes proliferate less, and probably progress slower through the cell cycle. The low numbers of PCNA-positive cells suggests that chondrocytes either become arrested in phases prior to S-phase of the cell cycle or spend a longer time in other phases. The proportionally greater number of cells staining for phospho-Histone 3, a marker for mitosis [36], suggests that M-phase may be prolonged under conditions of decreased Hoxc8 expression. This interpretation would also account for the relative increase of phospho-Histone 3 positive cells with time in culture. Thus, fewer cells enter the cell cycle, and those that do, spend longer time in M-Phase or may even get arrested in M-phase. Only pulse-chase labeling with a DNA analog and subsequent FACS analysis [17] would be able to quantify the extent of arrest or prolongation.
In this regard, it is interesting that for HoxA10, RNAi-mediated knockdown was recently shown to arrest endometrial stromal cells in the G2/M-phase [37], suggesting that Hox proteins may play a critical role for transition through mitosis. For HoxC10 [38], cell-cycle-dependent degradation in synchronized HeLa cells prior to entry into mitosis has been reported, which indicates that levels of Hox protein at each stage of the cell cycle may be important. However, this case appears to be isolated, as other Hox proteins were detected at stable levels throughout the cell cycle [38], at least in immortalized cell lines. In contrast, we studied freshly isolated chondrocytes, cells that recapitulate chondrocyte differentiation in vitro, and continue to proliferate at rates comparable to the in vivo conditions [17]. Our results reveal that Hoxc8 is required in primary chondrocytes for timely progression through the cell cycle. This might involve the licensing protein Geminin [39], which interacts with Hox transcription factors, forming a complex with chromatin that is required for mitosis [40, 41]. It could thus be possible that the lack of Hoxc8 protein in morpholino-treated chondrocytes prevents formation of adequate levels of this complex, delaying orderly progression through the cell cycle. In addition to interactions with cell cycle proteins, Hox proteins also interact with other proteins that regulate chondrocyte proliferation, such as proteins of the Pbx family [42]. However, interactions with Pbx factors appeared to be dispensable for enhancement of the proliferative capacity by Hoxb4 overexpression in hematopoietic cells [43]; in this paradigm, the DNA-binding capacity of Hoxb4 was critical [44]. Taken together, these studies suggest that both transcriptional activity of Hox proteins, as well as interactions with cell cycle regulatory proteins, may be important for chondrocyte proliferation. Our present study provides direct evidence for the notion that Hoxc8 expression levels are important for proper regulation of chondrocyte proliferation. These results thus complement in vivo evidence [13] that implicates altered chondrocyte proliferation as the cause of cartilage defects in transgenic mice with over-expression of Hoxc8.
Acknowledgments
We thank Andrew Wall for technical support, Dr. Gabriela Pavlinkova for advice on statistics, and M. Anita Jennings for help with the immunofluorescence assays. Confocal microscopy was conducted in a facility supported in part by grant C06 RR17417 from the National Center for Research Resources, National Institutes of Health, and partly by the Nebraska EPSCoR EPS-0346476 (CFD 47.076) program. This project was supported in part by the Philip Morris External Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Capecchi MR. Function of homeobox genes in skeletal development. Ann NY Acad Sci. 1996;785:34–37. doi: 10.1111/j.1749-6632.1996.tb56241.x. [DOI] [PubMed] [Google Scholar]
- 2.Kappen C, Neubuser A, Balling R, Finnell R. Molecular basis for skeletal variation: Insights from developmental genetic studies in mice. Birth Defects Res B Dev Reprod Toxicol. 2007;80:425–50. doi: 10.1002/bdrb.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman SA. Sticky fingers: Hox genes and cell adhesion in vertebrate limb development. Bioessays. 1996;18:171–74. doi: 10.1002/bies.950180302. [DOI] [PubMed] [Google Scholar]
- 4.Yokouchi Y, Nakazato S, Yamamoto M, Goto Y, Kameda T, Iba H, Kuroiwa A. Misexpression of hoxa-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes Develop. 1995;9:2509–22. doi: 10.1101/gad.9.20.2509. [DOI] [PubMed] [Google Scholar]
- 5.Duboule D. Vertebrate hox-genes and proliferation: An alternative pathway to homeosis? Curr Opin Genet Dev. 1995;5:525–28. doi: 10.1016/0959-437x(95)90058-o. [DOI] [PubMed] [Google Scholar]
- 6.Goff DJ, Tabin CJ. Analysis of Hoxd-13 and Hoxd-11 misexprssion in chick limb buds reveals that Hox genes affect both bone condensation and growth. Development. 1997;124:627–36. doi: 10.1242/dev.124.3.627. [DOI] [PubMed] [Google Scholar]
- 7.Kappen C. Early and late functions of homeobox genes in the development of the axial skeleton. In: Buckwalter JA, Ehrlich MG, Sandell LJ, Trippel SB, editors. Skeletal growth and development: Clinical issues and basic science advances. Rosemont, IL: American Academy of Orthopedic Surgeons; 1998. pp. 147–62. [Google Scholar]
- 8.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–63. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 9.Maulbecker CC, Gruss P. The oncogenic potential of deregulated homeobox genes. Cell Growth Diff. 1993;4:431–41. [PubMed] [Google Scholar]
- 10.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of Hoxb4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–65. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 11.Pollock RA, Jay G, Bieberich CJ. Altering the boundaries of Hox3.1 expression: Evidence for antipodal gene regulation. Cell. 1992;71:911–23. doi: 10.1016/0092-8674(92)90388-s. [DOI] [PubMed] [Google Scholar]
- 12.Pollock RA, Sreenath T, Ngo L, Bieberich CJ. Gain of function mutations for paralogous hox genes: Implications for the evolution of Hox gene function. Proc Natl Acad Sci USA. 1995;92:4492–96. doi: 10.1073/pnas.92.10.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yueh YG, Gardner DP, Kappen C. Evidence for regulation of cartilage differentiation by the homeobox gene Hoxc-8. Proc Natl Acad Sci USA. 1998;95:9956–61. doi: 10.1073/pnas.95.17.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kappen C, Mello MA, Finnell RH, Salbaum JM. Folate modulates cartilage defects in Hoxd-4 transgenic mice. Genesis. 2004;39:115–66. doi: 10.1002/gene.20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors l-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–90. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 16.DeCrombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13:721–27. doi: 10.1016/s0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 17.Cormier S, Mello MA, Kappen C. Normal proliferation and differentiation of Hoxc- 8 transgenic chondrocytes in vitro. BMC Dev Biol. 2003;3:4. doi: 10.1186/1471-213X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekker SC, Larson JD. Morphant technology in model developmental systems. Genesis. 2001;30:89–93. doi: 10.1002/gene.1038. [DOI] [PubMed] [Google Scholar]
- 19.Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, DeCrombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta- galactosidase transgenic mice. Matrix Biol. 1994;14:329–35. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
- 20.Belting HG, Shashikant CS, Ruddle FH. Multiple phases of expression and regulation of mouse Hoxc-8 during early embryogenesis. J Exp Zool. 1998;282:196–22. [PubMed] [Google Scholar]
- 21.Kruger C, Talmadge C, Kappen C. Expression of folate pathway genes in the cartilage of hoxd4 and hoxc8 transgenic mice. Birth Defects Res A Clin Mol Teratol. 2006;76:216–29. doi: 10.1002/bdra.20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summerton JE. Endo-porter: A novel reagent for safe, effective delivery of substances into cells. Ann N Y Acad Sci. 2005;1058:62–75. doi: 10.1196/annals.1359.012. [DOI] [PubMed] [Google Scholar]
- 23.LeMouellic H, Lallemand Y, Brulet P. Homeosis in the mouse induced by a null mutation in the Hox-3.1 gene. Cell. 1992;69:251–64. doi: 10.1016/0092-8674(92)90406-3. [DOI] [PubMed] [Google Scholar]
- 24.van den Akker E, Fromental-Ramain C, de Graaff W, Le Mouellic H, Brulet P, Chambon P, Deschamps J. Axial skeletal patterning in mice lacking all paralogous group 8 Hox genes. Development. 2001;128:1911–21. doi: 10.1242/dev.128.10.1911. [DOI] [PubMed] [Google Scholar]
- 25.Chen F, Capecchi MR. Targeted mutations in Hoxa-9 and Hoxb-9 reveal synergistic interactions. Dev Biol. 1997;181:186–96. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- 26.Horan GS, Ramirez-Solis R, Featherstone MS, Wolgemuth DJ, Bradley A, Behringer RR. Compound mutants for the paralogous Hoxa-4, Hoxb-4, and Hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995;9:1667–77. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- 27.Horan GS, Kovacs EN, Behringer RR, Featherstone MS. Mutations in paralogous Hox genes result in overlapping homeotic transformations of the axial skeleton: Evidence for unique and redundant function. Dev Biol. 1995;169:359–72. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 28.Papenbrock T, Visconti RP, Awgulewitsch A. Loss of fibula in mice overexpressing Hoxc11. Mech Dev. 2000;92:113–23. doi: 10.1016/s0925-4773(99)00344-5. [DOI] [PubMed] [Google Scholar]
- 29.Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucl Acid Drug Dev. 1996;6:267–72. doi: 10.1089/oli.1.1996.6.267. [DOI] [PubMed] [Google Scholar]
- 30.Youngblood DS, Hatlevig SA, Hassinger JN, Iversen PL, Moulton HM. Stability of cell-penetrating peptide-morpholino oligomer conjugates in human serum and in cells. Bioconjug Chem. 2007;18:50–60. doi: 10.1021/bc060138s. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Morrone G, Zhang J, Chen X, Lu X, Ma L, Moore M, Zhou P. Cul-4a stimulates ubiquitylation and degradation of the Hoxa9 homeodomain protein. EMBO J. 2003;22:6057–67. doi: 10.1093/emboj/cdg577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen WF, Chang CP, Rozenfeld S, Sauvageau G, Humphries RK, Lu M, Lawrence HJ, Cleary ML, Largman C. Hox homeodomain proteins exhibit selective complex stabilities with Pbx and DNA. Nucleic Acids Res. 1996;24:898–906. doi: 10.1093/nar/24.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fienberg AA, Nordstedt C, Belting HG, Czernik AJ, Nairn AC, Gandy S, Greengard P, Ruddle FH. Phylogenetically conserved ck-ii phosphorylation site of the murine homeodomain protein Hoxb-6. J Exp Zool. 1999;285:76–84. [PubMed] [Google Scholar]
- 34.Celis JE, Celis A. Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: Subdivision of S phase. Proc Natl Acad Sci U S A. 1985;82:3262–66. doi: 10.1073/pnas.82.10.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bravo R. Synthesis of the nuclear protein cyclin (PCNA) and its relationship with DNA replication. Exp Cell Res. 1986;163:287–93. doi: 10.1016/0014-4827(86)90059-5. [DOI] [PubMed] [Google Scholar]
- 36.Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, Inagaki M. Identification of a novel phosphorylation site on histone h3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274:25543–49. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- 37.Lu Z, Hardt J, Kim JJ. Global analysis of genes regulated by Hoxa10 in decidualization reveals a role in cell proliferation. Mol Hum Reprod. 2008;14:357–66. doi: 10.1093/molehr/gan023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, Riva S, Peverali FA. Early mitotic degradation of the homeoprotein Hoxc10 is potentially linked to cell cycle progression. EMBO J. 2003;22:3715–24. doi: 10.1093/emboj/cdg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by Geminin binding to Cdt1. Science. 2000;290:2309–12. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 40.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator Geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–53. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 41.Kroll KL. Geminin in embryonic development: Coordinating transcription and the cell cycle during differentiation. Front Biosci. 2007;12:1395–409. doi: 10.2741/2156. [DOI] [PubMed] [Google Scholar]
- 42.Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O’Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–57. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 43.Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of Hoxb4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–65. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 44.Beslu N, Krosl J, Laurin M, Mayotte N, Humphries KR, Sauvageau G. Molecular interactions involved in Hoxb4-induced activation of HSC self-renewal. Blood. 2004;104:2307–14. doi: 10.1182/blood-2004-04-1653. [DOI] [PubMed] [Google Scholar]