Abstract
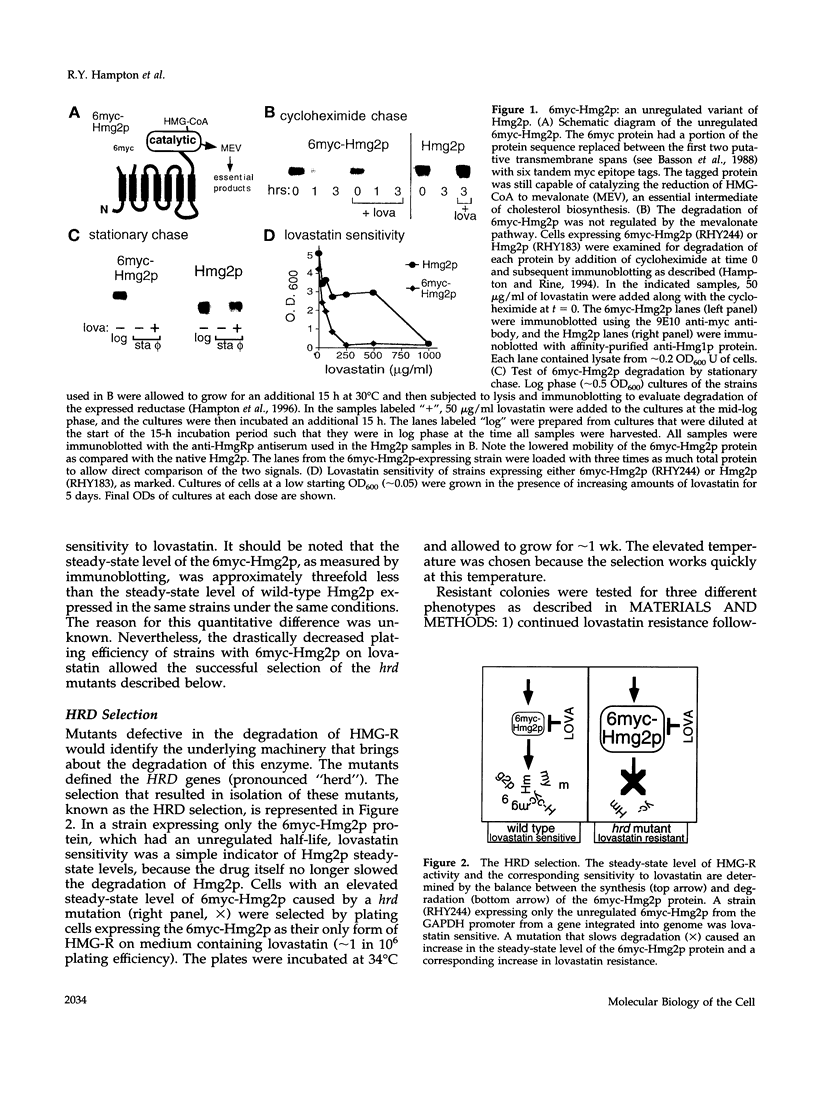
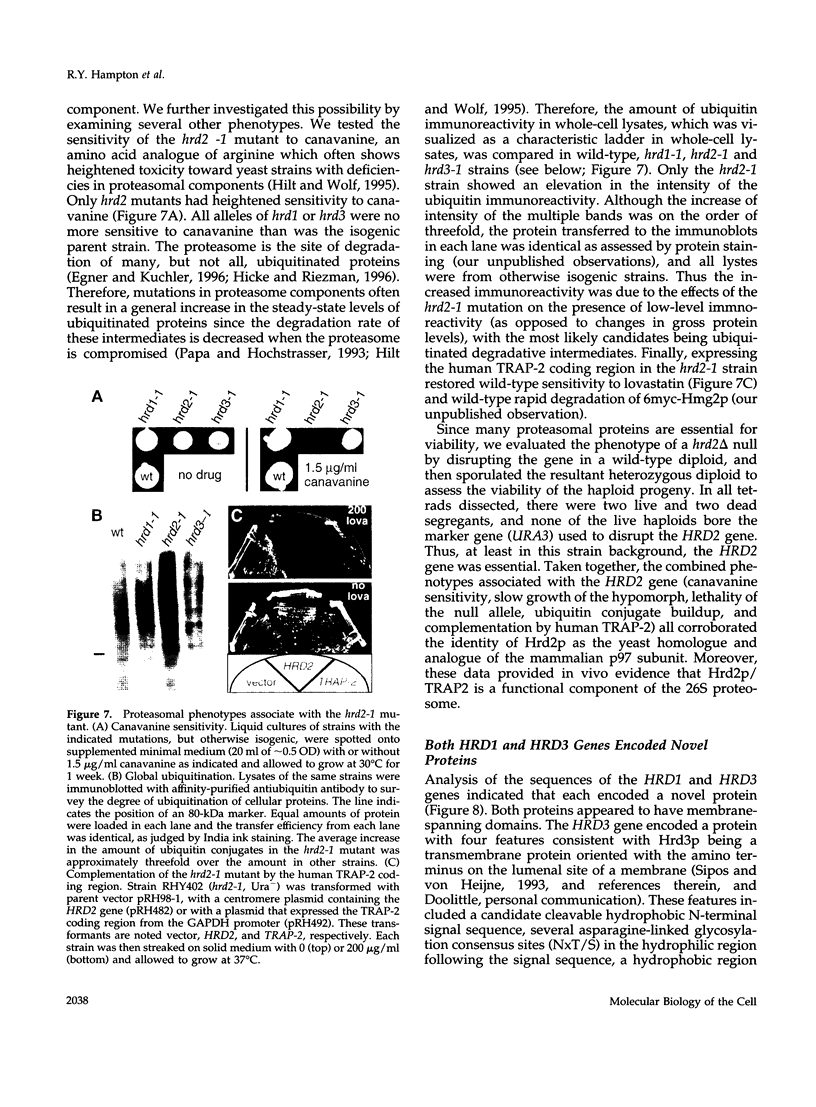
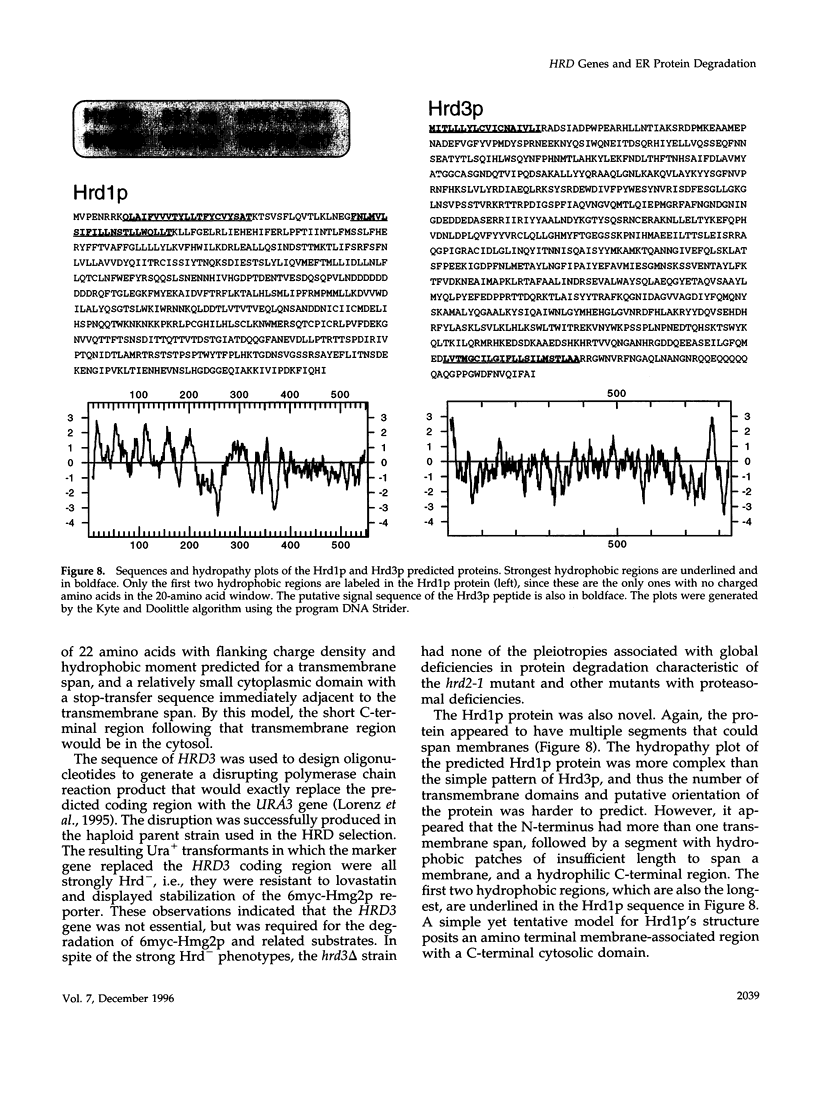
3-hydroxy-3-methylglutaryl-CoA reductase (HMG-R), a key enzyme of sterol synthesis, is an integral membrane protein of the endoplasmic reticulum (ER). In both humans and yeast, HMG-R is degraded at or in the ER. The degradation of HMG-R is regulated as part of feedback control of the mevalonate pathway. Neither the mechanism of degradation nor the nature of the signals that couple the degradation of HMG-R to the mevalonate pathway is known. We have launched a genetic analysis of the degradation of HMG-R in Saccharomyces cerevisiae using a selection for mutants that are deficient in the degradation of Hmg2p, an HMG-R isozyme. The underlying genes are called HRD (pronounced "herd"), for HMG-CoA reductase degradation. So far we have discovered mutants in three genes: HRD1, HRD2, and HRD3. The sequence of the HRD2 gene is homologous to the p97 activator of the 26S proteasome. This p97 protein, also called TRAP-2, has been proposed to be a component of the mature 26S proteasome. The hrd2-1 mutant had numerous pleiotropic phenotypes expected for cells with a compromised proteasome, and these phenotypes were complemented by the human TRAP-2/p97 coding region. In contrast, HRD1 and HRD3 genes encoded previously unknown proteins predicted to be membrane bound. The Hrd3p protein was homologous to the Caenorhabditis elegans sel-1 protein, a negative regulator of at least two different membrane proteins, and contained an HRD3 motif shared with several other proteins. Hrd1p had no full-length homologues, but contained an H2 ring finger motif. These data suggested a model of ER protein degradation in which the Hrd1p and Hrd3p proteins conspire to deliver HMG-R to the 26S proteasome. Moreover, our results lend in vivo support to the proposed role of the p97/TRAP-2/Hrd2p protein as a functionally important component of the 26S proteasome. Because the HRD genes were required for the degradation of both regulated and unregulated substrates of ER degradation, the HRD genes are the agents of HMG-R degradation but not the regulators of that degradation.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basson M. E., Thorsness M., Finer-Moore J., Stroud R. M., Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988 Sep;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C., Loayza D., Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994 Nov;5(11):1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowski R. S. Intracellular degradation of newly synthesized secretory proteins. Biochem J. 1983 Jul 15;214(1):1–10. doi: 10.1042/bj2140001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter G. A., Egan K. M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene. 1984 Dec;32(3):263–274. doi: 10.1016/0378-1119(84)90002-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bour S., Schubert U., Strebel K. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J Virol. 1995 Mar;69(3):1510–1520. doi: 10.1128/jvi.69.3.1510-1520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang H. L., Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991 Mar 28;350(6316):313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Papa F. R., Swaminathan S., Ursic D., Rasmussen T. P., Culbertson M. R., Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol. 1995 Nov;15(11):6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., Moomaw C. R., Zagnitko O. P., Proske R. J., Chu-Ping M., Afendis S. J., Swaffield J. C., Slaughter C. A. PA700, an ATP-dependent activator of the 20 S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J Biol Chem. 1994 Aug 19;269(33):20878–20884. [PubMed] [Google Scholar]
- Dice J. F., Chiang H. L. Peptide signals for protein degradation within lysosomes. Biochem Soc Symp. 1989;55:45–55. [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J Biol Chem. 1983 Jun 25;258(12):7272–7275. [PubMed] [Google Scholar]
- Egner R., Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996 Jan 8;378(2):177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- Eriksson P., André L., Ansell R., Blomberg A., Adler L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol Microbiol. 1995 Jul;17(1):95–107. doi: 10.1111/j.1365-2958.1995.mmi_17010095.x. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Freemont P. S. The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993 Jun 11;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Grant B., Greenwald I. The Caenorhabditis elegans sel-1 gene, a negative regulator of lin-12 and glp-1, encodes a predicted extracellular protein. Genetics. 1996 May;143(1):237–247. doi: 10.1093/genetics/143.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant E. P., Michalek M. T., Goldberg A. L., Rock K. L. Rate of antigen degradation by the ubiquitin-proteasome pathway influences MHC class I presentation. J Immunol. 1995 Oct 15;155(8):3750–3758. [PubMed] [Google Scholar]
- Hampton R. Y., Koning A., Wright R., Rine J. In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc Natl Acad Sci U S A. 1996 Jan 23;93(2):828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R. Y., Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994 Apr;125(2):299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996 Jan 26;84(2):277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hilt W., Wolf D. H. Proteasomes of the yeast S. cerevisiae: genes, structure and functions. Mol Biol Rep. 1995;21(1):3–10. doi: 10.1007/BF00990964. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin and intracellular protein degradation. Curr Opin Cell Biol. 1992 Dec;4(6):1024–1031. doi: 10.1016/0955-0674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995 Apr;7(2):215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Ubiquitin-phytochrome conjugates. Pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989 Mar 25;264(9):4998–5005. [PubMed] [Google Scholar]
- Jensen T. J., Loo M. A., Pind S., Williams D. B., Goldberg A. L., Riordan J. R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995 Oct 6;83(1):129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kornitzer D., Raboy B., Kulka R. G., Fink G. R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994 Dec 15;13(24):6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecureux L. W., Wattenberg B. W. The regulated degradation of a 3-hydroxy-3-methylglutaryl-coenzyme A reductase reporter construct occurs in the endoplasmic reticulum. J Cell Sci. 1994 Sep;107(Pt 9):2635–2642. doi: 10.1242/jcs.107.9.2635. [DOI] [PubMed] [Google Scholar]
- Lorenz M. C., Muir R. S., Lim E., McElver J., Weber S. C., Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995 May 26;158(1):113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996 Feb;132(3):291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C., Lu Y., Campbell A., Fang H., Green N. A mutation affecting signal peptidase inhibits degradation of an abnormal membrane protein in Saccharomyces cerevisiae. J Biol Chem. 1995 Jul 21;270(29):17139–17147. doi: 10.1074/jbc.270.29.17139. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Oda M. N., Scott S. V., Hefner-Gravink A., Caffarelli A. D., Klionsky D. J. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996 Mar;132(6):999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F. R., Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993 Nov 25;366(6453):313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Papavassiliou A. G., Treier M., Chavrier C., Bohmann D. Targeted degradation of c-Fos, but not v-Fos, by a phosphorylation-dependent signal on c-Jun. Science. 1992 Dec 18;258(5090):1941–1944. doi: 10.1126/science.1470918. [DOI] [PubMed] [Google Scholar]
- Pearce D. A., Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem. 1995 Sep 8;270(36):20879–20882. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- Roitelman J., Simoni R. D. Distinct sterol and nonsterol signals for the regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1992 Dec 15;267(35):25264–25273. [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993 Nov 5;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Schena M., Picard D., Yamamoto K. R. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 1991;194:389–398. doi: 10.1016/0076-6879(91)94029-c. [DOI] [PubMed] [Google Scholar]
- Schork S. M., Thumm M., Wolf D. H. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995 Nov 3;270(44):26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Förtsch B., Rautenstrauss B., Wolf D. H., Schweizer E. Differential proteolytic sensitivity of yeast fatty acid synthetase subunits alpha and beta contributing to a balanced ratio of both fatty acid synthetase components. Eur J Biochem. 1992 Feb 1;203(3):607–614. doi: 10.1111/j.1432-1033.1992.tb16590.x. [DOI] [PubMed] [Google Scholar]
- Scott S. V., Klionsky D. J. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J Cell Biol. 1995 Dec;131(6 Pt 2):1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L., von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993 May 1;213(3):1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Song H. Y., Dunbar J. D., Zhang Y. X., Guo D., Donner D. B. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J Biol Chem. 1995 Feb 24;270(8):3574–3581. [PubMed] [Google Scholar]
- Tierney D. J., Haas A. L., Koop D. R. Degradation of cytochrome P450 2E1: selective loss after labilization of the enzyme. Arch Biochem Biophys. 1992 Feb 14;293(1):9–16. doi: 10.1016/0003-9861(92)90358-4. [DOI] [PubMed] [Google Scholar]
- Tsuji E., Misumi Y., Fujiwara T., Takami N., Ogata S., Ikehara Y. An active-site mutation (Gly633-->Arg) of dipeptidyl peptidase IV causes its retention and rapid degradation in the endoplasmic reticulum. Biochemistry. 1992 Dec 1;31(47):11921–11927. doi: 10.1021/bi00162a035. [DOI] [PubMed] [Google Scholar]
- Ward C. L., Omura S., Kopito R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995 Oct 6;83(1):121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996 Mar 8;84(5):769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Wilcox C. A., Redding K., Wright R., Fuller R. S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992 Dec;3(12):1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]