Abstract
Purpose of review
This report summarizes emerging clinical and preclinical data pertaining to the use of CCR5 monoclonal antibodies (mAbs) as therapies for HIV-1 infection. The epitope specificity of CCR5 mAbs is discussed in relation to its critical impact on antiviral activity and CCR5 antagonism. We compare and contrast mAbs and small-molecule CCR5 antagonists in terms of their binding and antiviral properties. Two CCR5 mAbs have entered clinical testing and have successfully completed proof-of-concept studies in HIV-infected individuals, providing initial information on the potential therapeutic utility of these agents.
Recent findings
New studies support the view that the most potently antiviral CCR5 mAbs recognize the second extracellular loop of CCR5 either exclusively or in combination with the amino terminus. Studies have revealed fundamental differences in how mAbs and small molecules bind CCR5 and inhibit HIV-1. CCR5 mAbs and small-molecule CCR5 antagonists have demonstrated consistent antiviral synergy and limited or no viral cross-resistance in independent studies. Single intravenous infusions of CCR5 mAbs significantly reduced HIV-1 RNA levels in infected individuals for 2–3 weeks without appreciable toxicity.
Summary
CCR5 mAbs have demonstrated broad and potent antiviral activity in vitro. Clinical studies have established CCR5 mAbs as potent antiretroviral agents with prolonged activity following a single dose. CCR5 mAbs represent both a distinct class of CCR5 inhibitor and a novel approach to HIV-1 therapy.
Keywords: CCR5, monoclonal antibody, PRO 140, HGS004
Introduction
CCR5 monoclonal antibodies (mAbs) recently entered proof-of-concept trials in HIV-infected individuals with only CCR5-tropic (R5) virus detectable, and results from the first two studies are available. Single, well-tolerated infusions of CCR5 mAbs resulted in potent, rapid and prolonged reductions in viral load, and the single-dose antiviral effects compared favorably with those obtained after 10 to 14 days of treatment with small-molecule CCR5 antagonists. The emerging clinical and laboratory data support the view that CCR5 mAbs offer several potential advantages over existing antiretroviral therapies in terms of potency, tolerability, dosing frequency and other factors.
CCR5 Structure and Roles in Immune Function and HIV-1 Infection
CCR5 biology was reviewed recently [1]. The following paragraphs summarize critical aspects and recent developments. HIV-1 entry into host cells proceeds through a coordinated series of events mediated by the viral envelope glycoproteins gp120/gp41 and host cell receptors. HIV-1 gp120 first binds its primary receptor, CD4 [2], and then undergoes a conformational change that exposes a binding site for a chemokine receptor [3;4], principally CCR5 [5–7] or CXCR4 [8], that acts as a fusion co-receptor. These events trigger gp41-mediated fusion of the viral and cellular membranes. CCR5 is the predominant co-receptor used by HIV-1 for transmission and during the early stages of infection. In contrast, CXCR4-using variants are rarely transmitted but can become more apparent later in disease [9–14].
CCR5 belongs to a family of G-protein-coupled receptors (GPCRs) that respond to stimuli ranging from photons to proteins. CCR5 is expressed on a number of cell types, including activated/memory CD4+ and CD8+ T cells, macrophages, NK cells, NKT cells, microglia and astrocytes. On CD4+ T cells, CCR5 is a marker for a T helper 1 (Th1) phenotype. CCR5 regulates cell migration, activation and polarization through multiple kinase pathways. Its natural agonists are the chemokines CCL3 (MIP-1α), CCL3L1 (MIP-1αP), CCL4 (MIP-1β) and CCL5 (RANTES), which are soluble ~8 kDa cytokines. HIV-1 infection does not require CCR5 signaling.
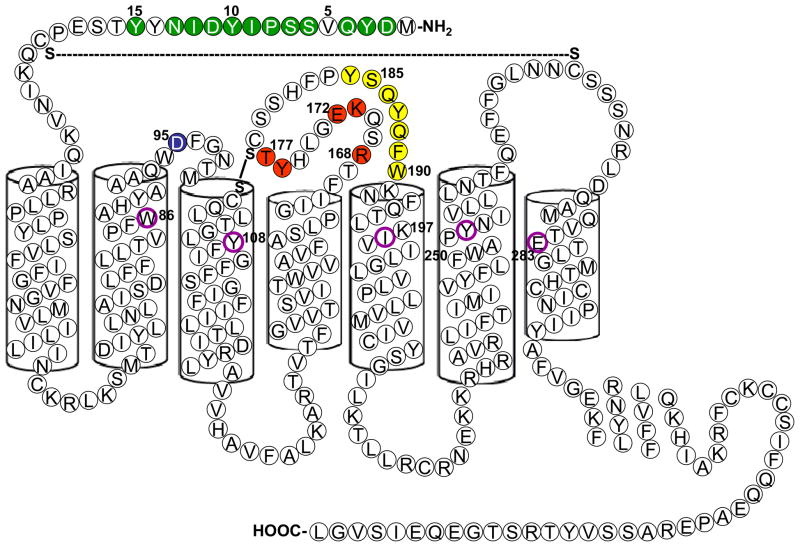
CCR5 spans the plasma membrane seven times in a serpentine manner (Figure 1) [15]. The extracellular portions represent potential targets for HIV-inhibitory mAbs and comprise an amino-terminal domain (Nt) and three extracellular loops (ECL1, ECL2 and ECL3). Sulfation of Nt tyrosines promotes gp120 binding and facilitates HIV-1 entry [16;17].
Figure 1. Schematic diagram of CCR5 illustrating the binding sites for mAbs and small molecules.
Amino acids are indicated in the single-letter code. Residues implicated in mAb binding are color-coded by filled symbols in the extracellular domain or sub-domain. Green = Nt; blue = ECL1; red = amino-terminal portion of ECL2; yellow = carboxy-terminal portion of ECL2. The specificities of individual mAbs are listed in Table 1. A putative binding site for maraviroc and vicriviroc in the transmembrane helices (residues W86, Y108, I198, Y251 and E283) is illustrated with magenta circles. The assignment of the seven transmembrane helices [15] is indicated with cylinders.
Numerous coding region, regulatory region and copy-number polymorphisms exist in the genes for CCR5 and its ligands. The geographic distributions of these polymorphisms suggest that these genes have evolved in response to differing environmental pathogens. Polymorphisms in CCR5 and its ligands critically affect HIV-1 transmission and disease progression [9–14;18–20]. A number of polymorphisms have a greater disease-modifying effect than heterozygosity for the well-studied 32 basepair deletion in the CCR5 coding region (Δ32). Δ32 is an inactivating mutation that affords nearly complete protection from HIV-1 infection when present in two copies. Δ32 homozygotes lack a functional CCR5 receptor. This genetic absence of CCR5 has no obvious effect on human health, but it has been associated with increased risk for symptomatic disease from flavivirus infections [21;22]. The findings support development of agents that block HIV-1 without abrogating CCR5’s role in normal immune function.
Generation of CCR5 mAbs
Numerous CCR5 mAbs have been described [23–33]. Most mAbs were generated by immunizing wild-type mice with rodent cell lines engineered to express human CCR5. Typically, hybridomas were generated and then screened for production of mAbs that bind CCR5. In other cases, hybridomas were initially screened for inhibition of HIV-1 envelope-mediated cell fusion. This latter approached yielded mAbs with potent antiviral activity [25;32].
Alternatively, CCR5 mAbs were generated in mice transgenic for the heavy and light chain genes for human antibodies [31] or were isolated from a phage antibody library constructed from non-immunized human donors [28]. Another mAb was isolated from a phage antibody library derived from an immunized rabbit [30]. Additional mAbs were raised against an Nt peptide [29].
As a class, GPCRs are not highly immunogenic in their native states embedded within the cell membrane. The extracellular portion of CCR5 comprises just 90 amino acids distributed over four domains. The largest of these domains are the Nt and ECL2 at approximately 30 amino acids each. It perhaps is not surprising that significant efforts were expended to generate CCR5 mAbs: Wu et al. identified CCR5 mAbs in one of eight hybridoma fusions [23], while other groups reported screening between 10,000 and 25,000 hybridoma supernatants to identify six to seven novel CCR5 mAbs [25;32].
Epitope specificity
The most potent HIV-inhibitory mAbs described to date recognize conformational epitopes. For such mAbs, specificity has been mapped using CCR5 point mutants [25;27;33;34], CCR5 deletants, and/or CCR5 chimeras that contain extracellular regions from homologous chemokine receptors [26;27;33;34]. These approaches have yielded results that are broadly consistent, with CCR5 point mutants providing the greatest precision. For example, independent groups have mapped the epitope for mAb 2D7 to ECL2 using CCR5/CCR2b chimeras [24;27;33;34]. The 2D7 epitope was mapped to ECL2 residues K171 and E172 using CCR5 alanine point mutants [25;27;33;34].
Table 1 lists the epitopes recognized by mAbs that have been mapped using CCR5 point mutants, and the amino acids involved in mAb binding are illustrated in Figure 1. For these mAbs, the dominant epitopes lie within the Nt and ECL2, which are the largest extracellular regions and show significant divergence from mouse CCR5. As illustrated in Figure 1, ECL2 can be divided into amino-terminal and carboxy-terminal regions based on patterns of mAb reactivity [27;34].
Table 1. Amino acids implicated in mAb binding to CCR5 as determined using CCR5 point mutants.
Mutation of the indicated amino acids was reported to reduce mAb binding to CCR5 as determined by flow cytometry.
| Antibody | Epitope | Reference(s) |
|---|---|---|
| 2D7 | Q170, K171, E172, W190 | [25;27;33;34] |
| 502 | D2, Y3 | [27] |
| 519 | D2, Y3, Q4 | [27] |
| 45501 | K171, E172 | [27] |
| 45517 | K171, E172 | [27] |
| 45523 | K171, E172, D95 | [27] |
| 45529 | Y184, S185, Q186, Y187, Q188, F189 | [27] |
| 45531 | Y184, S185, Q186, Y187, Q188, F189 | [27] |
| 45533 | K171, E172 | [27] |
| 45549 | K171, E172 | [27] |
| CTC2 | S6, S7, Y10, D11 | [27] |
| CTC5 | D2 | [27] |
| CTC8 | Y10, D11, I12, N13 | [27] |
| CTC9 | D2, Y3 | [27] |
| CTC12 | S7, I9, Y10, D11 | [27] |
| MC-6 | K171, E172 | [33] |
| MC-7 | S7, P8, Y10, D11 | [33] |
| MCR35.4 | Y184, S185, Q186, Y187, Q188, F189 | [27] |
| MCR40.3 | Y184, S185, Q186, Y187, Q188, F189 | [27] |
| PA8 | N13, Y15 | [25] |
| PA9 | D2, Y3, Q4, S7, P8, N13, Y176, T177 | [25] |
| PA10 | D2, Y3, Q4, P8, N13, Y176, T177 | [25] |
| PA11 | Q4 | [25] |
| PA12 | Q4 | [25] |
| PA14 | D2, R168, Y176 | [25] |
| RoAb12 | K171, E172, W190 | [34] |
| RoAb14 | K171, E172, W190 | [34] |
| RoAb18 | K171, E172, W190 | [34] |
Most potently antiviral mAbs bind residues in ECL2 alone or in combination with Nt residues [25;27;34]. Compared to ECL2 mAbs, mAbs that bind exclusively to the Nt typically have less potent antiviral activity [24;25;27]. In contrast, Nt mAbs are more potent than ECL2 mAbs in blocking binding of soluble gp120/CD4 complexes to CCR5 [25;27;34]. This finding presumably reflects the multiple roles of CCR5 in binding gp120 and triggering membrane fusion.
These findings are consistent with a two-site model for gp120-CCR5 interactions [35;36]. In the model for subtype B viruses, the bridging sheet and V3 stem on gp120 bind to tyrosine-sulfated forms of the CCR5 Nt, whereas the V3 crown interacts with ECL2 [16;37–40]. The model suggests that optimal inhibition of HIV-1 may be obtained with a mAb that occludes HIV-1’s access to both ECL2 and Nt, either by directly binding a multidomain epitope or by steric hindrance.
The binding sites for CCR5 mAbs are distinct from those for small-molecule CCR5 antagonists. These differences in CCR5 binding translate into important differences in antiviral properties, as described below. The available small-molecule CCR5 inhibitors bind the hydrophobic cavity formed by the transmembrane helices. Notably, E283 in the seventh transmembrane region is a principal site of interaction for small molecules. A recent study mapped the binding sites for maraviroc and vicriviroc to an identical set of amino acids [41], as indicated in Figure 1.
Antiviral activity in vitro
Although numerous CCR5 mAbs have been described, few broadly and potently inhibit HIV-1. As discussed above, epitope specificity critically influences antiviral activity. Antiviral activity did not correlate with CCR5 binding affinity for mAbs to unrelated epitopes [31]; however, antiviral activity tracked CCR5 binding affinity for mAbs to similar epitopes [34;42].
Given the multitude of CCR5 mAbs developed by independent groups, no systematic comparison of antiviral activities has been performed. However, a number of mAbs were tested using PhenoSense™ HIV-1 Entry (Monogram Biosciences, South San Francisco, CA), a single-cycle assay that utilizes HIV-1 envelope-complemented reporter viruses [43–46]. The validated, reproducible nature of this assay enables some limited cross-study generalizations. Based on the published information, the most potent CCR5 mAbs demonstrated 50% inhibitory concentrations (IC50s) in the range of 0.1–1.0 μg/mL (0.67–6.7 nM), with an approximately 1 log10 variation across diverse viral isolates. The mAbs afford essentially complete inhibition at higher concentrations [43–46].
CCR5 mAbs have demonstrated similar potencies for viruses derived from different genetic subtypes [32;47;48], stages of disease [49], and adult and pediatric infections [50]. CCR5 expression levels show considerable person-to-person variation [23;51;52] and have been reported to affect both HIV-1 infectivity [23] and the potency of CCR5 inhibitors in vitro [52]. CCR5 mAbs efficiently inhibited CCR5-mediated entry of dual/mixed (R5X4) viruses in cell lines that express CCR5 but not CXCR4 [25;32;50]; however, limited inhibition of R5X4 viruses was observed in cultures of peripheral blood mononuclear cells [47–49].
CCR5 antagonism
In contrast to the two-site model for gp120, chemokines principally bind CCR5 via ECL2 [24;53;54]. The binding sites for gp120 and chemokines on CCR5 therefore are overlapping but distinct, and the antiviral and antagonist activities of CCR5 mAbs are dissociable. ECL2 mAbs often inhibit HIV-1 and chemokine signaling with similar efficiencies [24;25;32]; Nt mAbs typically display minimal CCR5 antagonism but less potent antiviral activity [25;32;33;55]. Rarely, CCR5 mAbs have been reported to possess agonist or partial agonist activity [33]; most mAbs do not activate CCR5 at any concentration.
Amongst the mAbs described in the published literature, the mAb PA14 binds a unique epitope spanning ECL2 and Nt [25]. PA14 preferentially inhibited HIV-1 at concentrations that did not block the natural activity of CCR5 in vitro, although CCR5 antagonism was observed at higher concentrations [25]. When compared with the ECL2 mAb 2D7 in parallel testing, PA14 was a more potent HIV-1 inhibitor and a less potent CCR5 antagonist. The findings indicate that PA14 can distinguish fine differences in the binding sites for HIV-1 and chemokines on CCR5.
Synergy with other antiretroviral agents
Three in vitro studies examined the antiviral activity of CCR5 mAbs in combination with small-molecule CCR5 antagonists [56–58]. The antibodies examined were PA14, PRO 140 (humanized PA14), 2D7, RoAb13, RoAb14, 2D7 and 45523. The small-molecule CCR5 antagonists included maraviroc, vicriviroc, aplaviroc, SCH-C and TAK-779. Antiviral synergy was reported by each group for most studied combinations of CCR5 mAbs and small-molecule antagonists, and the synergy was attributed to co-binding of CCR5 [56;57]. One notable exception was mAb 45523 used in combination with either maraviroc or aplaviroc, where synergy was not observed due to competition for CCR5 binding [57]. In parallel studies, additive rather than synergistic effects were observed for combinations of small-molecule CCR5 inhibitors [56;57]. The findings provide a rationale to combine CCR5 mAbs and small-molecule antagonists in the clinic and further underscore the mechanistic differences between these classes of CCR5 inhibitors.
Synergy also was reported for combinations of CCR5 mAbs that bind distinct epitopes, with the highest synergy observed between Nt and ECL2 mAbs [25;57]. Additive to synergistic effects were reported between CCR5 mAbs and enfuvirtide, a peptide inhibitor of gp41 membrane fusion [56;57]. Additivity was observed between CCR5 mAbs that bind similar or overlapping epitopes.
Cross-resistance between CCR5 mAbs and small-molecule CCR5 antagonists
Viruses resistant to small-molecule CCR5 antagonists were generated by serial passage of virus in the presence of increasing concentrations of inhibitor in vitro. These viruses typically retained an R5 phenotype and acquired the ability to utilize inhibitor-bound receptor [59–64]. In vivo resistance has reflected the emergence of resistant R5 viruses as well as the outgrowth of pre-existing R5X4 viruses [65;66]. In single-cycle antiviral assays, viral resistance to small-molecule antagonists was manifest as a reduction in the maximum percent inhibition at high inhibitor concentrations rather than a change in IC50 [59–63;66], consistent with the view that small-molecule CCR5 antagonists act as allosteric inhibitors [67–69].
Several small-molecule resistant viruses were tested for susceptibility to CCR5 mAbs. Despite demonstrating high-level resistance to the small-molecule CCR5 antagonists, the viruses remained susceptible or even hyper-susceptible to inhibition by CCR5 mAbs [59–61;66;70]. The lack of cross-resistance between mAbs and small-molecule CCR5 antagonists likely reflects differences in their modes of CCR5 binding (Figure 1) and mechanisms of HIV-1 inhibition (competitive v. allosteric).
There is limited information at present regarding forced viral resistance to CCR5 mAbs. Additional studies are needed to determine whether such viruses retain an R5 phenotype and are susceptible to inhibition by small-molecule CCR5 antagonists.
Human clinical studies
Two CCR5 mAbs have been tested in HIV-infected individuals [42;51]. Both mAbs are of the human IgG4 isotype, and the studies shared several design similarities. In each case, the mAbs were studied as single, escalating intravenous infusions to HIV-infected subjects with HIV-1 RNA >5,000 copies/mL, CD4 > 250 cells/μL, only CCR5-tropic (R5) virus detectable, and no concurrent antiretroviral therapy. Co-receptor tropism was assessed using the first-generation Trofile™ assay (Monogram Biosciences) [43]. In both studies, subjects were followed for 56–58 days post-treatment to assess tolerability, pharmacokinetics (PK) and antiviral effects.
Unlike the development programs for small-molecule CCR5 antagonists [71–73], the phase 1 programs for the CCR5 mAbs did not examine drug-drug or food interactions. Such studies were not necessary given that mAbs are injected and are catabolized by proteolysis within cells of the reticuloendothelial system. This process is distinct from and does not interfere with the typical metabolic pathways for small-molecule drugs.
HGS004 (CCR5mAb004, Human Genome Sciences, Rockville, MD) [42]
HGS004 is a human mAb that binds ECL2 and inhibits R5 HIV-1 entry and chemokine signaling with similar efficiencies [31]. In a phase 1 clinical trial, 63 subjects were randomized to receive placebo or HGS004 at doses of 0.4, 2, 8, 20 and 40 mg/kg. Subjects were mostly male (86%) with a mean age of 41 years and mean HIV-1 RNA levels of 25,100 copies/mL. All subjects completed the study. Two 2mg/kg subjects experienced infusion-related uticarial rash that responded to diphenhydramine, and all subsequent subjects were pre-treated with diphenhydramine prior to infusion.
Significant reductions in HIV-1 RNA were observed at doses of 8 mg/kg and higher. Plasma HIV-1 RNA reductions of >1 log10 were observed in 14 of 26 subjects (54%) treated with 8, 20 or 40 mg/kg HGS004. Mean viral load reductions of 1 log10 were observed at day 14 for the 8 and 20 mg/kg groups. At 40 mg/kg, the mean viral load reduction was approximately 0.8 log10 at days 14 and 21. Three of ten 40 mg/kg subjects experienced a change in co-receptor tropism to dual/mixed virus on study. One of these individuals experienced a transient 1 log10 reduction in HIV-1 RNA; the others had no significant antiviral response. Co-receptor tropism changes also were observed in one subject each in the 0.4 and 20 mg/kg groups but not in other groups.
PK data were non-linear. Although the maximum serum concentrations were dose proportional, overall exposure (area under the concentration-time curve, AUC) increased more than proportionally with dose. The mean terminal serum half-life ranged from 4.7 to 7.9 days across the different dose levels. The mean CCR5 receptor occupancy was approximately 80% at day 28 for each of the three highest dose groups. Significant increases in CD4 and CD8 cell counts were observed in all HGS004 groups, and this finding was hypothesized to reflect redistribution of CCR5-expressing cells from peripheral tissues into the blood.
PRO 140 (Progenics Pharmaceuticals, Inc., Tarrytown, NY) [51]
PRO 140 is a humanized form of the mouse mAb PA14, which binds an epitope spanning ECL2 and Nt. PA14 and PRO 140 have been characterized for breadth and potency of antiviral activity in several preclinical studies [25;47;49;50;56;74].
The first HIV trial of PRO 140 was a randomized, double-blind, placebo-controlled study in 39 individuals with early-stage disease. Cohorts were randomized 3:10 to receive a single infusion of placebo or PRO 140 at doses of 0.5, 2 or 5 mg/kg. PRO 140 was generally well tolerated, and no dose-limiting toxicity or pattern of toxicity was observed. There was no requirement to pre-medicate with antihistamines. There were no remarkable laboratory or electrocardiogram findings.
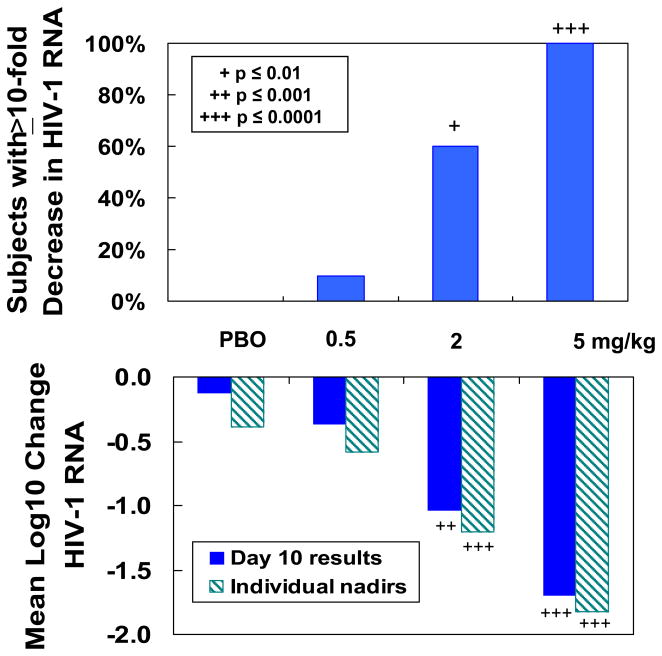
Rapid, dose-dependent and highly significant reductions in HIV-1 RNA were observed (Figure 2). Mean maximum (nadir) viral load reductions of 0.39±0.20, 0.58±0.30, 1.20±0.63 (p=0.0002) and 1.83±0.41 log10 (p<0.0001) were observed for the placebo, 0.5 mg/kg, 2 mg/kg and 5 mg/kg groups, respectively. At day 10, the mean log10 declines in viral load were 0.13±0.24, 0.37±0.54, 1.04±0.45 (p=0.0001) and 1.70±0.49 (p<0.0001) for the placebo and ascending dose groups. Mean viral load reductions of >1 log10 persisted for 2–3 weeks post-treatment in the 5 mg/kg group. All 5 mg/kg subjects had an antiviral response of ≥1.0 log10 reduction in HIV-1 RNA (Figure 2). These single-dose antiviral effects are the largest reported for any HIV-1 drug and compare favorably with those observed following 10 to 14 days of treatment with small-molecule CCR5 antagonists [75–77].
Figure 2. Antiviral activity of single-dose PRO 140 in HIV-infected adults.
Subjects received single intravenous infusions of placebo (PBO) or PRO 140 at doses of 0.5 mg/kg, 2 mg/kg or 5 mg/kg. Plasma HIV-1 RNA levels were monitored for 58 days. Top panel: Percentage of subjects in each treatment group who experienced a ≥ 10-fold reduction in HIV-1 RNA at any timepoint post-treatment. Bottom panel: Mean log10 changes in HIV-1 RNA for each treatment group. Study Day 10 represents nine days post-treatment. An individual nadir represents the maximum reduction experienced by a subject at any time-point post-treatment.
The area under the PRO 140 concentration-time curve from time zero to infinity (AUC∞) values increased more than proportionally with dose, averaging 11.1, 74.3 and 278 mg × day/L for the ascending dose groups. Mean terminal serum half-lives were 3.9 and 3.5 days for the 2 and 5 mg/kg dose groups, respectively. All tests for anti-PRO 140 antibodies were negative with the exception of a single low-titer result at Day 59 for a 5 mg/kg subject. The antibodies had no obvious effect on PK metrics or antiviral response. At 5 mg/kg PRO 140, there was a trend (p=0.055) towards increased CD4+ cells over baseline. There was no depletion of CCR5+ cells following treatment; however, significant receptor occupancy (p<0.05) was observed for 2–4 weeks in all PRO 140 groups. This study established PRO 140 as a potent antiretroviral agent with prolonged activity.
Conclusions
CCR5 mAbs broadly and potently inhibit R5 HIV-1 in vitro, and potent antiviral activity has been demonstrated in HIV-infected individuals. CCR5 mAbs represent a novel approach to HIV-1 therapy and offer several potential advantages over existing therapies in terms of infrequent (e.g., weekly to monthly) dosing, favorable tolerability, and limited drug-drug or food interactions. CCR5 mAbs are distinct from small-molecule CCR5 antagonists in terms of their binding sites on CCR5 and mechanisms of HIV-1 inhibition. CCR5 mAbs and small-molecule antagonists can be considered distinct classes of CCR5 inhibitors based on their potent antiviral synergy and lack of viral cross-resistance. Clinical proof of concept has been obtained using intravenously administered CCR5 mAbs. Additional studies of intravenously and subcutaneously administered CCR5 mAbs have been initiated.
Acknowledgments
We thank Patricia Schneider for expert graphical assistance.
Funding: Public Health Service award AI066329 from the National Institute of Allergy and Infectious Diseases
References
- 1.Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 2.Maddon PJ, Dalgleish AG, McDougal JS, et al. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 3.Trkola A, Dragic T, Arthos J, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Gerard NP, Wyatt R, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 5.Dragic T, Litwin V, Allaway GP, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC- CKR-5. Nature. 1996;381:667–73. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 6.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 9.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 10.Smith MW, Dean M, Carrington M, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 11.O’Brien WA, Hartigan PM, Martin D, et al. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 12.Ioannidis JP, Rosenberg PS, Goedert JJ, et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 13.van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moyle GJ, Petropoulos C, Goodrich J, et al. Prevalence and predictive factors for CCR5 and CXCR4 co-receptor usage in a large cohort of HIV-1 positive individuals. XV International AIDS Conference, Abstract WePeB5725; Bangkok, Thailand. July 11–16, 2004; 2004. [Google Scholar]
- 15.Seibert C, Ying W, Gavrilov S, et al. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology. 2006;349:41–54. doi: 10.1016/j.virol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Cormier EG, Persuh M, Thompson DA, et al. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Nat Acad Sci USA. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farzan M, Mirzabekov T, Kolchinsky P, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez E, Dhanda R, Bamshad M, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 20.Dolan MJ, Kulkarni H, Camargo JF, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 21.Lim JK, Louie CY, Glaser C, et al. Genetic deficiency of chemokine receptor CCR5 is a strong risk factor for symptomatic West Nile virus infection: a meta-analysis of 4 cohorts in the US epidemic. J Infect Dis. 2008;197:262–265. doi: 10.1086/524691. These studies indicate that a genetic absence of CCR5 was associated with increased risk for symptomatic and severe disease association with infection by West Nile virus and tick-borne encephalitis virus. The findings illustrate a role for CCR5 in normal immune function in humans. The findings support the development of agents that preferentially block R5 HIV-1 while preserving normal CCR5 function in whole or in part. [DOI] [PubMed] [Google Scholar]
- 22.Kindberg E, Mickiene A, Ax C, et al. A deletion in the chemokine receptor 5 (CCR5) gene is associated with tickborne encephalitis. J Infect Dis. 2008;197:266–269. doi: 10.1086/524709. These studies indicate that a genetic absence of CCR5 was associated with increased risk for symptomatic and severe disease association with infection by West Nile virus and tick-borne encephalitis virus. The findings illustrate a role for CCR5 in normal immune function in humans. The findings support the development of agents that preferentially block R5 HIV-1 while preserving normal CCR5 function in whole or in part. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Paxton W, Kassam N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, LaRosa G, Kassam N, et al. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson WC, Rabut GE, Nagashima KA, et al. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–55. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill CM, Kwon D, Jones M, et al. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. Virology. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 27.Lee B, Sharron M, Blanpain C, et al. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 28.Osbourn JK, Earnshaw JC, Johnson KS, et al. Directed selection of MIP-1 alpha neutralizing CCR5 antibodies from a phage display human antibody library. Nat Biotechnol. 1998;16:778–781. doi: 10.1038/nbt0898-778. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Frade JM, Vila-Coro AJ, Martin A, et al. Similarities and differences in RANTES-and (AOP)-RANTES-triggered signals: implications for chemotaxis. J Cell Biol. 1999;144:755–765. doi: 10.1083/jcb.144.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberger P, Sutton JK, Rader C, et al. Generation and characterization of a recombinant human CCR5-specific antibody. A phage display approach for rabbit antibody humanization. J Biol Chem. 2000;275:36073–36078. doi: 10.1074/jbc.M002765200. [DOI] [PubMed] [Google Scholar]
- 31.Roschke V, Clark S, Branco L, et al. Characterization of a panel of novel human monoclonal antibodies that specifically antagonize CCR5 and block HIV-1 entry. 44th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Abstract 2871; Washington, D C. October 30 - November 2, 2004; 2004. [Google Scholar]
- 32.Ji C, Brandt M, Dioszegi M, et al. Novel CCR5 monoclonal antibodies with potent and broad-spectrum anti-HIV activities. Antiviral Res. 2007;74:125–137. doi: 10.1016/j.antiviral.2006.11.003. The report describes the generation, antiviral properties and antagonist activities of a novel panel of CCR5 mAbs with potent antiviral activity. The mAbs were broadly active against HIV-1 isolates from diverse genotype and against viruses that were resistant to enfuvirtide or maraviroc. [DOI] [PubMed] [Google Scholar]
- 33.Blanpain C, Vanderwinden JM, Cihak J, et al. Multiple active states and oligomerization of CCR5 revealed by functional properties of monoclonal antibodies. Mol Biol Cell. 2002;13:723–737. doi: 10.1091/mbc.01-03-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Rao E, Dioszegi M, et al. The second extracellular loop of CCR5 contains the dominant epitopes for highly potent anti-human immunodeficiency virus monoclonal antibodies. Antimicrob Agents Chemother. 2007;51:1386–1397. doi: 10.1128/AAC.01302-06. Epitope specificity, affinity and binding kinetics were determined for a panel of CCR5 mAbs with potent antiviral activity. These mAbs mapped to either the amino or carboxy portions of ECL2. The authors conclude that these regions contain the dominant epitopes for mAbs with potent anti-HIV-1 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doranz BJ, Lu ZH, Rucker J, et al. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cormier EG, Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76:8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cormier EG, Tran DN, Yukhayeva L, et al. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol. 2001;75:5541–9. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizzuto CD, Wyatt R, Hernandez-Ramos N, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 39.Rizzuto CD, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 40.Huang CC, Lam SN, Acharya P, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondru R, Zhang J, Ji C, et al. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol Pharmacol. 2008;73:789–800. doi: 10.1124/mol.107.042101. The authors utilize site-directed mutagenesis and docking models to compare the CCR5 binding sites for five small-molecule CCR5 antagonists of clinical interest, including maraviroc and vicriviroc. The structurally diverse antagonists were proposed to fit within the same pocket formed by the transmembrane helices of CCR5. The transmembrane binding pocket for small-molecule CCR5 antagonists can be compared with the extracellular epitopes recognized by CCR5 mAbs. [DOI] [PubMed] [Google Scholar]
- 42.Lalezari J, Yadavalli GK, Para M, et al. Safety, pharmacokinetics, and antiviral activity of HGS004, a novel fully human IgG4 monoclonal antibody against CCR5, in HIV-1-infected patients. J Infect Dis. 2008;197:721–727. doi: 10.1086/527327. The authors describe the results of a phase 1 study of the CCR5 mAb HGS004 in 63 subjects with HIV-1 infection. The single-blind, randomized, placebo-controlled study evaluated the tolerability, pharmacokinetics, receptor occupancy and antiviral effects of single escalating doses ranging to 40 mg/kg HGS004. The report concluded that this CCR5 mAb was generally well tolerated and demonstrated meaningful antiviral activity when administered to subjects with R5 HIV-1. [DOI] [PubMed] [Google Scholar]
- 43.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binley JM, Wrin T, Korber B, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson WC, Maddon PJ. Resistance to HIV-1 entry inhibitors. Current Drug Targets --Infectious Disorders. 2003;3:255–262. doi: 10.2174/1568005033481015. [DOI] [PubMed] [Google Scholar]
- 47.Trkola A, Ketas TJ, Nagashima KA, et al. Potent, broad-spectrum inhibition of human immunodeficiency virus type 1 by the CCR5 monoclonal antibody PRO 140. J Virol. 2001;75:579–588. doi: 10.1128/JVI.75.2.579-588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cilliers T, Nhlapo J, Coetzer M, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003;77:4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rusert P, Kuster H, Joos B, et al. Virus Isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. The Journal of Virology. 2005;79:8454–8469. doi: 10.1128/JVI.79.13.8454-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shearer WT, Deville JG, Samson PM, et al. Susceptibility of pediatric HIV-1 isolates to recombinant CD4-IgG(2) (PRO 542) and humanized mAb to the chemokine receptor CCR5 (PRO 140) J Allergy Clin Immunol. 2006;118:518–521. doi: 10.1016/j.jaci.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 51.Jacobson JM, Saag MS, Thompson MA, et al. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. J Infect Dis. 2008 doi: 10.1086/592169. in press. The authors describe the results of a phase 1 study of the CCR5 mAb PRO 140 in 39 subjects with HIV-1 infection. The randomized, double-blind, placebo-controlled study evaluated the tolerability, pharmacokinetics, receptor occupancy and antiviral effects of single escalating doses ranging to 5 mg/kg PRO 140. The trial established clear proof of concept for PRO 140 as a potent antiretroviral agent with extended activity after a single dose. [DOI] [PubMed] [Google Scholar]
- 52.Ketas TJ, Kuhmann SE, Palmer A, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samson M, LaRosa G, Libert F, et al. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 54.Dragic T, Trkola A, Lin SW, et al. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–85. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vila-Coro AJ, Mellado M, Martin dA, et al. HIV-1 infection through the CCR5 receptor is blocked by receptor dimerization. Proc Natl Acad Sci U S A. 2000;97:3388–3393. doi: 10.1073/pnas.050457797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murga J, Franti M, Pevear DC, et al. Potent antiviral synergy between monoclonal antibody and small-molecule CCR5 inhibitors of human immunodeficiency virus type 1. Antimicrobial Agents and Chemotherapy. 2006;50:3289–3296. doi: 10.1128/AAC.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji C, Zhang J, Dioszegi M, et al. CCR5 small-molecule antagonists and monoclonal antibodies exert potent synergistic antiviral effects by cobinding to the receptor. Mol Pharmacol. 2007;72:18–28. doi: 10.1124/mol.107.035055. The study confirms and significantly extends prior reports that CCR5 mAbs synergize with small-molecule CCR5 antagonists and with other CCR5 mAbs to unrelated epitopes. Synergy between CCR5 inhibitors was attributed to cobinding of the receptor. [DOI] [PubMed] [Google Scholar]
- 58.Safarian D, Carnec X, Tsamis F, et al. An anti-CCR5 monoclonal antibody and small molecule CCR5 antagonists synergize by inhibiting different stages of human immunodeficiency virus type 1 entry. Virology. 2006;352:477–484. doi: 10.1016/j.virol.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 59.Trkola A, Kuhmann SE, Strizki JM, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci USA. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marozsan AJ, Kuhmann SE, Morgan T, et al. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 61.Kuhmann SE, Pugach P, Kunstman KJ, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westby M, Smith-Burchnell C, Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogert RA, Wojcik L, Buontempo C, et al. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology. 2008;373:387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Pugach P, Marozsan AJ, Ketas TJ, et al. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. This study describes the phenotypes of viruses selected in vitro for resistance to vicriviroc and AD101, a chemically related compound. Each escape mutant virus was observed to be cross-resistant to maraviroc, aplaviroc and other small-molecule CCR5 antagonists when assayed in cultures of primary CD4+ T cells. However, the escape mutant viruses remained susceptible to inhibition by mAb PRO 140. The findings extend prior studies that reported different resistance profiles for mAb and small-molecule inhibitors of CCR5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westby M, Lewis M, Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsibris AM, Sagar M, Gulick RM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. This study characterized the genotypes and phenotypes of viruses isolated from a subject following failure of a vicriviroc-containing regimen. The isolates exhibited high-level resistance to vicriviroc, cross-resistance to another small-molecule CCR5 antagonist, and increased susceptibility to inhibition of the CCR5 mAb HGS004. The findings for these clinical isolates complement similar findings reported previously for viruses selected in vitro for resistance to small-molecule antagonists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watson C, Jenkinson S, Kazmierski W, Kenakin T. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol Pharmacol. 2005;67:1268–1282. doi: 10.1124/mol.104.008565. [DOI] [PubMed] [Google Scholar]
- 68.Kenakin T. How pharmacological receptor theory can guide new drug discovery CCR5 HIV inhibitors in AIDS as a case study. Proc West Pharmacol Soc. 2007;50:1–7. [PubMed] [Google Scholar]
- 69.Haworth B, Lin H, Fidock M, et al. Allosteric effects of antagonists on signalling by the chemokine receptor CCR5. Biochem Pharmacol. 2007;74:891–897. doi: 10.1016/j.bcp.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 70.Pugach P, Ketas TJ, Michael E, Moore JP. Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology. 2008;377:401–407. doi: 10.1016/j.virol.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan PL, Weatherley B, McFadyen L. A population pharmacokinetic meta-analysis of maraviroc in healthy volunteers and asymptomatic HIV-infected subjects. Br J Clin Pharmacol. 2008;65 (Suppl 1):76–85. doi: 10.1111/j.1365-2125.2008.03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adkison KK, Shachoy-Clark A, Fang L, et al. Pharmacokinetics and short-term safety of 873140, a novel CCR5 antagonist, in healthy adult subjects. Antimicrob Agents Chemother. 2005;49:2802–2806. doi: 10.1128/AAC.49.7.2802-2806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson BM, Song IH, Adkison KK, et al. Evaluation of the drug interaction potential of aplaviroc, a novel human immunodeficiency virus entry inhibitor, using a modified cooperstown 5 + 1 cocktail. J Clin Pharmacol. 2006;46:577–587. doi: 10.1177/0091270006287291. [DOI] [PubMed] [Google Scholar]
- 74.Ketas TJ, Frank I, Klasse PJ, et al. Human immunodeficiency virus type 1 attachment, coreceptor and fusion inhibitors are active against both direct and trans infection of primary cells. J Virol. 2003;77:2762–2767. doi: 10.1128/JVI.77.4.2762-2767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fatkenheuer G, Pozniak AL, Johnson MA, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 76.Lalezari J, Thompson M, Kumar P, et al. 873140, a novel CCR5 antagonist: Antiviral activity and safety during short-term monotherapy in HIV-infected adults. 44th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Abstract 2871; Washington, D C. October 30 - November 2, 2004; 2004. [Google Scholar]
- 77.Schurmann D, Fatkenheuer G, Reynes J, et al. Antiviral activity, pharmacokinetics and safety of vicriviroc, an oral CCR5 antagonist, during 14-day monotherapy in HIV-infected adults. AIDS. 2007;21:1293–1299. doi: 10.1097/QAD.0b013e3280f00f9f. [DOI] [PubMed] [Google Scholar]