Abstract
Stochastic ‘noise’ arises from random thermal fluctuations in the concentration of protein, RNA or other molecules within the cell and is an unavoidable aspect of life at the single-cell level. Evidence is accumulating that this biochemical noise critically influences cellular auto-regulatory circuits and can ‘flip’ genetic switches to drive probabilistic fate decisions in bacteria, viruses, cancer, and stem cells. Here, we review how stochastic gene expression in key auto-regulatory proteins can control fate determination between latency and productive replication in both phage-λ and HIV-1. We highlight important new results that synthetically manipulate auto-regulatory circuitry and noise, to bias HIV-1’s ability to enter proviral latency. We argue that an appreciation of noise in gene expression may shed light on the mystery of animal virus latency and that strategies to manipulate noise may have impact on anti-viral therapeutics.
Introduction
Consider a clonal cell population (i.e. genetically identical or isogenic cells) where all cells are divided from a single parent cell. The cells in the population will exhibit considerable cell-to-cell variation in the level of any specific gene product and this variation is referred to as the stochastic noise [1–3]. The origin of this noise is biochemical: intracellular processes are driven by reactant molecules randomly diffusing and colliding within the cell and are thus inherently stochastic. Specifically, noise in gene expression can arise from the random timing in individual reactions associated with promoter remodeling, transcription and translation [4–6]. Moreover, intercellular differences in the amount of cellular components (for example, RNA polymerase, transcription factors and ribosomes) also cause variations in expression levels. Measurements in live, single cells have shown that gene expression noise can lead to large statistical fluctuations in protein and mRNA levels in both prokaryotes and eukaryotes [7–10]. These fluctuations (i.e. noise) can have significant effects on biological function and phenotype.
On the one hand, noise can be problematic for essential proteins whose levels have to be tightly maintained within certain bounds for optimal performance [11] (e.g. noise can ‘corrupt’ information processing by cells [12]). However, on the other hand, noise could conceivably confer a benefit to the organism. Recent studies show that increasing noise in stress-related yeast proteins can confer a selective advantage by allowing a larger fraction of the population to have stress-response protein levels above a threshold [2,13]. When confronted with two different pathways, cells may also exploit noise in key regulatory proteins for a probabilistic pathway selection and cell-fate determination [3,14]. Such stochastic fate decisions can create phenotypic heterogeneity across isogenic (i.e. clonal) cell populations, and heterogeneity is beneficial in fluctuating extracellular environments [15,16]. For example, during growth of a bacterial colony, a fraction of E. coli cells stochastically enter a persister state, a slow growing state that can elude the action of antibiotics [17]. While the persister strategy lowers the average colony growth rate under normal conditions, it pays off if E. coli randomly encounters antibiotics in its environment since the persister cells survive with high probability and resume normal growth after antibiotic treatment.
Like prokaryotes (and many other organisms), viruses also live in environments that change unpredictably. For viruses, fluctuating environments can be due to large variations in the number of susceptible host cells at any given time. In these fluctuating host populations, noise-driven probabilistic decisions (i.e. entry into latency or reactivation from latency) may provide a virus with a fitness advantage [18,19] similar to the fitness advantage that noise can confer in prokaryotes [20–22]. In fact, a body of evidence now shows that noise can influence a fate decision between productive replication and dormancy (i.e. latency or lysogeny) in two viral systems: Human Immunodeficiency Virus type 1 (HIV-1) and bacteriophage-λ (phage-λ) [23,24].
Here, we compare and contrast the transcriptional regulatory mechanisms that phage-λ and HIV-1 use to exploit noise and control fate determination. In particular, we review recent evidence demonstrating that HIV-1 proviral latency, like phage-λ lysogeny, is driven in part by stochastic gene expression, and that HIV-1 proviral latency is thus an inherent property of the virus [24,25]. Importantly, we highlight recent results showing that altering HIV-1 auto-regulatory circuitry alters noise in HIV-1 gene expression and biases the HIV-1 fate decision for productive replication versus latency [26].
Stochastic fate determination in phage-λ
Since the 1950’s it has been known that genetically identical phage-infected bacteria, grown in the same environment, can undergo a ‘developmental bifurcation’ where one fraction of infected cells enter a lytic replication state and other infected cells enter a lysogenic state where phage-λ remains dormant [27]. In the late 1990’s Arkin and colleagues developed computational models for the phage-λ gene network which argued that stochastic noise in gene expression together with an underlying ‘bistability’ in phage-λ’s gene network, could account for the lysis-lysogeny developmental bifurcation [23]. The phage-λ viral gene circuit consists of a double negative feedback loop, where proteins CI and Cro mutually repress each other’s expression. This feedback creates a bistable epigenetic switch where protein levels can lock into two different states: high Cro and low CI (which corresponds to lysis) or low Cro and high CI (which corresponds to lysogeny) [28]. Random timing in individual biochemical reactions during the early phase of infection can create small intercellular differences in the concentration of these repressor proteins. These differences are significantly amplified by the Cro-CI mutual repression causing an initial homogenous cell population to bifurcate into two sub-populations that are locked into different states and have different developmental outcomes [23]. Similar stochastic models have also been proposed to explain why only a fraction of lysogens become lytic when induced with ultraviolet (UV) irradiation [29].
Importantly, phage-λ encodes a negative feedback mechanism where CI represses its own transcription and this negative feedback may stabilize the lysogenic state against stochastic fluctuations. Such negative auto-regulatory loops are common gene network motifs within cells that suppress noise in protein levels [30]. Tight regulation of CI ensures that CI levels don’t become small by random chance, which may cause a spontaneous transition to the lytic state. Negative feedback also prevents CI levels from becoming large, and high CI levels may desensitize the lysogenic state to induction signals [31].
Deterministic factors such as cell volume and MOI can bias probabilistic fate decisions
In the early 1970s, Phillippe Kourilsky performed a set of experiments that demonstrated how the frequency of lysogeny was dependent on the number of input phage-λ particles per cell (i.e. the multiplicity-of-infection or MOI) and that increased MOI correlated with increased lysogeny frequency [32]. Later experiments by Ira Herskowitz [33] showed that nutritional state of the cell also heavily biased lysogeny. These experimental correlations between MOI (or nutritional state) and lysogeny were in fact partly the inspiration for Arkin’s stochastic lysis-lysogeny model and this stochastic model generates variable fractions of lytic and lysogenic cells at different MOIs, nutritional states, and even different cell volumes [23]. In fact, high MOI and small cell volume have a striking equivalence: if many phage infect a large cell, or if a single phage infects a much smaller cell, in both cases the initial concentration of lysogeny factors can be very high. A fascinating recent study [34] demonstrates that cell size (like MOI) can indeed be a strong predictor of phage-λ infection outcome and that infections of smaller cells increase the probability of lysogeny, while infections of larger cells are biased towards lysis. Most strikingly, this study shows that even for the very largest and very smallest cells a fraction of cells consistently choose an outcome which is opposite to what would be predicted by cell volume alone. For example, some percentage of large cells become lysogens when the bacterial size dictates that they should become lytic. The fact that a percentage of cells enter the “wrong” fate, strongly implies that phage-λ’s fate decision is probabilistic. Dramatically, the authors capture this probabilistic decision-making by direct single-cell observation of two daughter cells (of the same size) that take opposite developmental outcomes: one daughter cell becomes lytic, the other daughter develops into a lysogen. A deterministic model where cell size dictates the fate outcome cannot explain this striking observation. In fact, the lysis-lysogeny decision is apparently probabilistic even in the face of a strong deterministic pressure such as cell size. The probabilistic/stochastic model where the fate decision is ‘biased’ or nudged in one direction by deterministic pressures such as bacterial cell volume appears to be the most parsimonious with the available data.
Noise induced variability in lysis times: implications for non-latent viruses
Apart from its role in decision-making, another important consequence of noise is that it can create significant variability in lysis times. This variability was recently quantified in phage-λ by monitoring the gene expression at different stages of the lytic cascade after induction with UV irradiation [35]. The time taken to complete different lytic stages was found to be uncorrelated across cells. This result suggests that variation in lysis times arises due to random fluctuations in cascade components and not because the lytic cascade proceeds at different rates in different cells [35]. Randomness in the time taken to complete the lytic phase can cause variability in the viral burst size (the number of progeny virus released per host cell), which has been well documented in phage-λ [36]. Presumably, randomness in lysis times and burst size due to stochasticity in viral molecular components is a general feature across all viruses.
Stochastic fate determination in HIV-1
Like phage-λ, the Human Immunodeficiency Virus type 1 (HIV-1) can also enter one of two developmental fates: upon infecting a CD4+ T lymphocyte HIV-1 can either enter an active replication state (productive infection) or enter a post-integration/proviral latent state (an analog of phage lysogeny). A substantial body of evidence has confirmed that HIV-1 proviral latent cells are quiescent for viral production and that viral gene expression is shut off during viral latency [37,38]. These proviral latent cells are considered the most significant obstacle thwarting HIV-1 eradication from a patient [39,40] since latent cells can ‘reactivate’ during interruption of highly active anti-retroviral therapy (HAART) to generate rapid viral rebounds that re-establish pre-treatment HIV-1 levels [41].
Stochastic Noise in the HIV-1 Tat auto-regulatory loop is sufficient to drive latency
While many host factors have been implicated in controlling HIV-1 replication and latency [42–44], there has been no conclusive identification of host cellular factors that direct an infected T lymphocyte to become latent. Importantly, the HIV-1 Tat protein (Trans-Activator of Transcription) is absolutely essential for active replication and latent reactivation [42,45,46]. Tat transactivation drives active replication by mediating hyper-phosphorylation of RNA polymerase II to enhance transcriptional elongation from HIV’s Long-Terminal Repeat (LTR) promoter [39,42,47]. Tat transactivation thus comprises an essential positive-feedback loop that drives HIV lytic replication by auto-stimulating its own gene expression 50–100 fold above basal levels and simultaneously up-regulating the expression of HIV Rev (the essential viral mRNA export factor) [48]. Our group and others have shown that stochastic fluctuations in Tat gene expression can act as a molecular switch and allow HIV-1 to enter a transcriptionally dormant state [24,49].
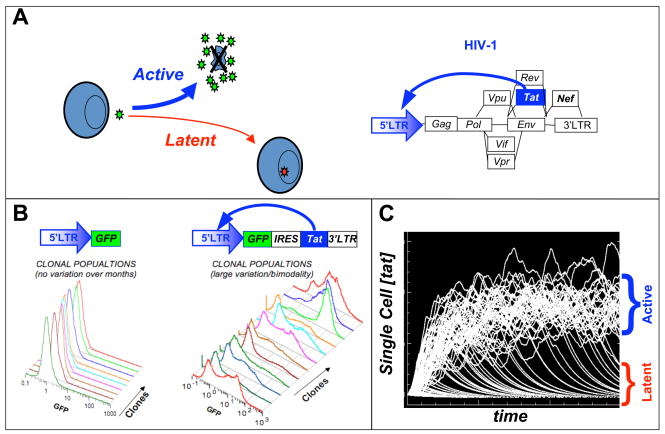
To study noise in Tat expression, sub-genomic HIV-1 based lentiviral vectors encoding only Tat and the green fluorescent protein (GFP) as a reporter were used to infect Jurkat T cells and construct isogenic (clonal) populations [24]. Strikingly, single-cell level analysis of these isogenic populations by flow cytometry (Fig 1b), showed significant phenotypic bifurcation in isogenic Tat-positive populations despite all cells being genetically identical and grown from a single infected parent cell. The phenotypic bifurcation in isogenic Tat positive-feedback population was evidenced as a fraction of the infected cells exhibiting very high gene expression levels (on) and fraction of infected cells exhibited no viral gene expression (off). Importantly, isogenic populations infected with control lentiviral vectors lacking functional Tat positive feedback did not display this phenotypic bifurcation. Subsequent analysis showed that stochastic fluctuations in Tat gene expression were of primary importance in influencing this developmental bifurcation (or decision) in the HIV Tat positive-feedback circuit.
Figure 1. A minimal HIV Tat positive-feedback circuit is sufficient to generate a ‘latency’ decision in the presence of noise.
(A) The HIV-1 ‘proviral’ genome with Tat positive feedback displayed and the HIV-1 developmental bifurcation between active replication and proviral latency after infection of a CD4+ T lymphocyte. (B) Jurkat T cells infected with either a control LTR-GFP or feedback LTR-GFP-IRES-Tat lentiviral vector, were single-cell sorted by FACS, grown into clonal populations and subsequently analyzed by flow cytometry. (C) Single-cell Tat trajectories of an HIV-1 gene-circuit model. Each trajectory represents the level of Tat within a single-cell over time. Significant fluctuations in Tat levels exist and can drive a phenotypic bifurcation between two states.
The result that stochastic Tat gene expression could drive latency, coupled with important work by Eric Verdin’s group [42] to develop state-of-the-art and accepted cell-culture models of HIV-1 latency, highlighted the fact that proviral latency was not a consequence of HAART or due to complex T cell maturation phenomena that occur only in vivo. The result that minimal Tat circuits can reproduce latency has the important implication that proviral latency is an inherent property of the HIV-1 lifecycle and is due fundamental physical and chemical processes underlying the virus’s regulatory network. Since proviral latency appears to be an inherent property of HIV-1 gene expression that has been conserved, it is likely that latency plays a role in the natural history of HIV-1 replication and likely provides the virus with a fitness advantage in the natural setting.
Unlike phage-λ, the HIV-1 Tat auto-regulatory loop lacks bistability
Based on the established bistability model in phage-λ [23], it seemed that phenotypic bifurcation in minimal HIV Tat circuits might be a result of biochemical noise driving Tat levels stochastically above or below a bistable threshold in HIV Tat. Molecularly, bistability requires some from of self-cooperative threshold, such as protein homo-multimerization or cooperative binding. The presence of such as threshold is typically measured by calculating a ‘Hill’ coefficient from a dose-response curve, as is done in the study enzyme kinetics. A Hill coefficient ≥ 2 necessarily implies the presence of a self-cooperative threshold, whereas a Hill coefficient ≤ 1 rules out the presence of a self-cooperative threshold and necessarily rules out bistability. The Hill coefficient for the HIV Tat circuit was measured using quantitative single-cell imaging and found to equal one [25], thereby ruling out bistability in the HIV Tat circuit. Instead, it appears that HIV utilizes a different mechanism, excitable circuitry, to decide between alternate fates. In the excitable circuit model, cell fate is determined by the duration of transient Tat expression pulses (Fig 1c) and Tat mediated positive feedback acts to extend the lifetime of the these pulses. More specifically, short-lived Tat pulses allow for latency and long-lived Tat pulses drive active viral replication and reactivation from latency.
Weakening feedback strength alters noise and reduces entry into HIV-1 latency
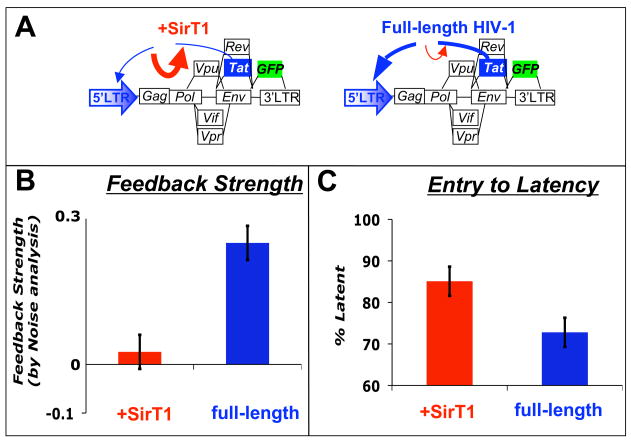
The excitable circuit model presented a clear avenue for altering the HIV fate decision in single cells. In particular, if the Tat positive feedback strength could be diminished, then the duration of the Tat pulses would be shortened and infected cells would be more likely to enter latency. To test this excitable circuit model, a concept was borrowed from electrical circuit analysis: gene expression noise was exploited as an experimental probe. Specifically, by measuring the duration of stochastic fluctuations in Tat, feedback strength can be directly quantified through noise autocorrelation analysis [50]. This gene-expression noise autocorrelation method was used to determine Tat positive-feedback strength in minimal and full-length HIV-1 virus at the single-cell level [26]. Most importantly, in the presence of a known down regulator of Tat function, the histone deacetylase SirT1 (Fig 2a) [51], feedback strength was significantly diminished (Fig 2a–b). Strikingly, cells with reduced feedback strength showed a significant shortening in the duration of Tat expression transients and an increase in the proportion of cells entering latency in this system (Fig 2c) [26]. Thus, by manipulating feedback strength and the accompanying noise frequency profile, the HIV-1 latency decision can be altered, potentially for therapeutic benefit, as previously proposed [52].
Figure 2. Weakening HIV-1 Tat positive feedback biases HIV-1 towards latency and limits reactivation.
(A) Schematics of full-length HIV-1 with nef replaced by a gfp reporter (blue) and the SirT1 over expression effect (red). (B) SirT1 over expression deacetylase Tat and reduces positive-feedback strength (red) as evidenced from gene expression noise analysis. (C) SirT1 over-expressing cells have a higher probability of entering latency and show decreased latent reactivation [26].
Conclusion
Deterministic models are clearly insufficient to explain the probabilistic nature of phage-λ lysogeny and HIV-1 proviral latency. Conclusively stating that a biological process is stochastic is a strong statement and requires significantly more evidence than simply observing a distribution. However, we believe that extensive single-cell evidence now exists for the stochastic nature of a variety of biological fate-determination processes, for comprehensive reviews see [3,14], and specifically for viral latency decisions. One set of experiments that lends strong support to the stochastic model are noise perturbation experiments that eliminate or diminish the stochastic noise in a specific process and test whether the process retains a probabilistic phenotype. Recent studies in a number of systems [20–22] have succeeded in specifically manipulating the noise in gene expression, without manipulating the deterministic mean in expression levels, and demonstrated that fate outcome is significantly altered. For HIV-1, these noise manipulation experiments provide strong evidence that proviral latency is significantly influenced by underlying stochastic noise in gene expression [26]. Similar noise manipulation experiments in phage-λ would lend strong support to the stochastic model of lysis-lysogeny in phage-λ.
From the available evidence, viruses appear to exploit noise in gene expression by encoding auto-regulatory positive feedback gene circuits (or double negative feedback gene circuits) that may amplify noise to generate probabilistic ‘molecular switches’ for latency. If this proves correct, stochastic noise in gene expression noise may represent a novel therapy target to manipulate viral fate and this has been demonstrated in principle by manipulating HIV-1’s Tat positive feedback circuit to bias the virus’s ability to enter latency and reactivate [26]. An understanding of noise in gene expression may also shed light upon latency in other viruses such as herpesviruses where latency is an integral of part of the viral life cycle. Herpesvirus reactivation from latency exhibits a probabilistic phenotype, which is evocative of a role for noise in gene expression. Stochastic gene expression from latent herpersvirus genomes has been reported in a beta herpesvirus [53] and recently hypothesized as a mechanism for reactivation from latency in an alpha herpesvirus [54]. A systems-level understanding of viral gene-network architecture, and its associated molecular noise, will likely be crucial to unraveling the mystery of animal viral latency.
Acknowledgments
We thank Mike Simpson, Cynthia Bolovan-Fritts, Winnie Wen, Melissa Wong, Brandon Razooky, and members of the Weinberger laboratory for helpful discussions. LSW acknowledges support from the Pew Charitable Trust Young Scholars in the Biomedical Sciences program and by NIH award K25GM083395.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 2.Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. The authors provide a comprehensive review of stochastic gene expression in the context of different biological systems ranging from microbes to metazoans. The benefits and negative effects of noise are discussed in detail. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 6.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci U S A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 8.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 10.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Libby E, Perkins TJ, Swain PS. Noisy information processing through transcriptional regulation. Proc Natl Acad Sci U S A. 2007;104:7151–7156. doi: 10.1073/pnas.0608963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop AL, Rab FA, Sumner ER, Avery SV. Phenotypic heterogeneity can enhance rare-cell survival in ‘stress-sensitive’ yeast populations. Mol Microbiol. 2007;63:507–520. doi: 10.1111/j.1365-2958.2006.05504.x. [DOI] [PubMed] [Google Scholar]
- 14.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. Reviews stochastic fate determination in various systems including entry into competency in B. subtilis and generation of alternative color vision photoreceptors in D. Melanogaster. Mechanisms by which cells choose fate stochastically and the benefits of doing so are discussed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–475. doi: 10.1038/ng.110. Using the galactose utilization network of Saccharomyces cerevisiae, the authors show that noise-driven transitions between multiple phenotypes can provide a fitness advantage in the presence of unforeseen environmental fluctuations. [DOI] [PubMed] [Google Scholar]
- 16.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 17.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 18.Mittler JE. Evolution of the genetic switch in temperate bacteriophage. I. Basic theory. J Theor Biol. 1996;179:161–172. doi: 10.1006/jtbi.1996.0056. [DOI] [PubMed] [Google Scholar]
- 19.Stumpf MP, Laidlaw Z, Jansen VA. Herpes viruses hedge their bets. Proc Natl Acad Sci U S A. 2002;99:15234–15237. doi: 10.1073/pnas.232546899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. The authors show that noise in the regulatory protein ComK causes the transition to competent state in Bacillus subtilis. Experimentally reducing this Comk noise decreased the transition probability causing a lower number of competent cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 22.Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS ONE. 2008;3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Weinberger LS, Shenk T. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 2007;5:e9. doi: 10.1371/journal.pbio.0050009. This paper shows that the HIV-1 Tat auto-regulatory positive feedback loop lacks bistability and functions as an excitable circuit. The Tat feedback circuit provides a pulse of gene expression activity upon transactivation, which always decays to zero after some period of time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinberger LS, Dar RD, Simpson ML. Nat Genet. Vol. 40. 2008. Transient-mediated fate determination in a transcriptional circuit of HIV; pp. 466–470. The paper shows that tat auto-regulatory positive feedback loop functions to extend the duration of Tat stochastic gene expression transients. The lifetime of these transients may play a critical role in determining HIV-1 infected cell-fate. In particular, experimentally manipulating the feedback strength changed the duration of these gene expression transients and biased HIV-1 fate decision between productive replication and latency. [DOI] [PubMed] [Google Scholar]
- 27.Lieb M. The establishment of lysogenicity in Escherichia coli. J Bacteriol. 1953;65:642–651. doi: 10.1128/jb.65.6.642-651.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ptashne M. A genetic switch: phage lambda revisited. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 29.Tian T, Burrage K. Bistability and switching in the lysis/lysogeny genetic regulatory network of bacteriophage lambda. J Theor Biol. 2004;227:229–237. doi: 10.1016/j.jtbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 31.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kourilsky P. Lysogenization by bacteriophage lambda. I. Multiple infection and the lysogenic response. Mol Gen Genet. 1973;122:183–195. doi: 10.1007/BF00435190. [DOI] [PubMed] [Google Scholar]
- 33.Herskowitz I, Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- 34.St-Pierre F, Endy D. Determination of cell fate selection during phage lambda infection. Proc Natl Acad Sci U S A. 2008;105:20705–20710. doi: 10.1073/pnas.0808831105. The authors argue that pre-existing heterogeneity at the time of infection can bias the lysis-lysogeny decision towards either fate. In particular, cell volume at the start of infection was shown to play a key role in determining the outcome of phage-λ infection, with a 2-fold increase in cell volume resulting in a 4-fold decrease in the probability of lysogeny. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amir A, Kobiler O, Rokney A, Oppenheim AB, Stavans J. Noise in timing and precision of gene activities in a genetic cascade. Mol Syst Biol. 2007;3:71. doi: 10.1038/msb4100113. The paper measures the randomness in timing of events as phage-λ proceeds though different stages of the lytic cascade. The authors report that the time taken to complete different stages of the cascade are uncorrelated and the noise in the timing of successive stages of the cascade decreases as the lytic cascade progresses. This is consistent with a model where the lytic cascade is made up of a number of relatively independent stages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delbruck M. The Burst Size Distribution in the Growth of Bacterial Viruses (Bacteriophages) J Bacteriol. 1945;50:131–135. doi: 10.1128/JB.50.2.131-135.1945. [DOI] [PubMed] [Google Scholar]
- 37.Seth N, Kaufmann D, Lahey T, Rosenberg ES, Wucherpfennig KW. Expansion and contraction of HIV-specific CD4 T cells with short bursts of viremia, but physical loss of the majority of these cells with sustained viral replication. J Immunol. 2005;175:6948–6958. doi: 10.4049/jimmunol.175.10.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 39.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 40.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. The review focuses on the clinical relevance of HIV latency and argues that the latent reservoir remains the major obstacle in finding a cure. [DOI] [PubMed] [Google Scholar]
- 41.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 42.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. Embo J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. Embo J. 2007 doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. Embo J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TS, Peterlin BM. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol. 2003;77:8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. The multifactorial nature of HIV-1 latency. Trends Mol Med. 2004;10:525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 49.Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog. 2009;5:e1000260. doi: 10.1371/journal.ppat.1000260. The paper investigates how different host transcription factor binding sites at the HIV-1 LTR promoter contribute to controlling viral stochastic gene expression in the Tat feedback circuit. They report that certain cis-acting elements at the promoter function to reduce transcriptional noise and may play an important role in stabilizing HIV latency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 51.Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinberger LS, Schaffer DV, Arkin AP. Theoretical design of a gene therapy to prevent AIDS but not human immunodeficiency virus type 1 infection. J Virol. 2003;77:10028–10036. doi: 10.1128/JVI.77.18.10028-10036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grzimek NK, Dreis D, Schmalz S, Reddehase MJ. Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs. J Virol. 2001;75:2692–2705. doi: 10.1128/JVI.75.6.2692-2705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson RL, Preston CM, Sawtell NM. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 2009;5:e1000352. doi: 10.1371/journal.ppat.1000352. The authors hypothesize that in response to stress, stochastic gene expression of a key transactivator protein VP16 from latent viral genome initiates lytic gene expression and reactivation from latency. [DOI] [PMC free article] [PubMed] [Google Scholar]