Abstract
Among eukaryotic organisms a vast majority of Box H/ACA ribonucleoproteins (RNPs) are responsible for the post-transcriptional introduction of pseudouridine (Ψ) into ribosomal RNAs (rRNA) and spliceosomal small nuclear RNAs (snRNA), thus influencing protein translation and pre-mRNA splicing, respectively. Additionally, a few distinct Box H/ACA RNPs are involved in the processing of rRNA, and the stabilization of vertebrate telomerase RNA. Thus, whether directly or indirectly, Box H/ACA RNPs impact major steps of gene expression, as well as play a role in maintaining genome integrity. Box H/ACA RNPs each consist of a unique Box H/ACA RNA and a set of four common core proteins. While the RNA component is responsible for dictating site-specificity, the four core proteins impact numerous aspects of RNP function including both stability and catalytic potential. Interestingly, mutations have been identified in the core proteins of the Box H/ACA RNP, resulting in a rare inherited bone marrow failure syndrome referred to as dyskeratosis congenita. This review discusses our current understanding of the roles of the protein components of the Box H/ACA RNP, and provides a framework to understand how mutations in the Box H/ACA RNP contribute to disease pathology.
Keywords: Box H/ACA RNA, Box H/ACA RNP, pseudouridine, dyskeratosis congenita, pre-mRNA splicing, ribosome biogenesis, telomerase
Introduction
Recent years have seen an increase in the number of noncoding RNAs identified, as well as an expansion of the known processes noncoding RNAs participate in (Mattick and Makunin 2006). Box H/ACA RNAs are one of the largest classes of noncoding RNA, and are present in all eukaryotes and archaea (Dennis and Omer 2005; Yu et al. 2005; Terns and Terns 2006). Throughout evolution, Box H/ACA RNAs have acquired the ability to engage in a range of cellular processes, including the formation of pseudouridine residues, ribosomal RNA (rRNA) processing, as well as the proper maintenance of telomeric DNA (Kiss 2002; Terns and Terns 2002; Meier 2005; Yu et al. 2005). Box H/ACA RNAs reside within ribonucleoprotein complexes (RNPs) each consisting of, in addition to one unique Box H/ACA RNA, several core proteins. For the vast majority of Box H/ACA RNPs, i.e., those responsible for pseudouridylation, a set of four common core proteins suffices to form a functionally competent RNP (Baker et al. 2005; Charpentier et al 2005; Meier et al 2005; Yu et al. 2005; Terns and Terns 2006).
Structural and Functional Variation Among the Box H/ACA RNPs
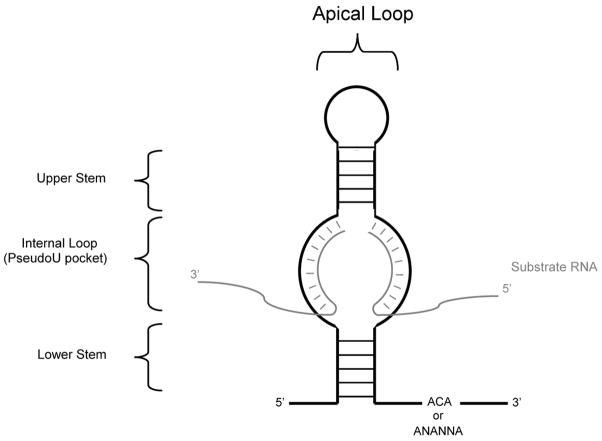
The fundamental building block of a Box H/ACA RNA consists of an imperfect hairpin structure followed immediately by a single stranded region containing either the variant sequence 5′-ANANNA-3′ (Box H, where N is any nucleotide) or the trinucleotide 5′-ACA-3′ (Box ACA) (Balakin et al. 1996; Ganot et al. 1997; Ni et al. 1997; Terns and Terns 2006). Residing within each hairpin is a large internal loop (Figure 1). Structural disparities exist among Box H/ACA RNAs of different functional classes allowing for unique interactions with substrates and proteins, ultimately impacting both localization and function (Eliceiri 2005; Meier 2005; Terns and Terns 2006).
Figure 1.
Fundamental building block of a Box H/ACA RNA. The minimal components of Box H/ACA RNAs are a lower stem, internal loop, upper stem, apical loop, and Box H or Box ACA. The Box H or Box ACA is typically located 3 nucleotides upstream of the 3′ end. The internal loop is capable of base-pairing with complementary sequences within the substrate RNA. The uridine residue targeted for pseudouridylation (Ψ), as well as the adjacent downstream nucleotide (N), are positioned at the base of the upper stem approximately 14–16 nucleotides upstream of either Box H or Box ACA and are left unpaired so as to remain accessible for isomerization.
Box H/ACA sno/scaRNAs direct site-specific pseudouridylation
Most Box H/ACA RNPs are involved in the posttranscriptional conversion of uridine to pseudouridine (Figure 2) (Yu et al. 2005; Meier 2005; Terns and Terns 2006). The structure of archaeal Box H/ACA RNAs which guide pseudouridylation varies, consisting of one to three hairpins (Dennis and Omer 2005) (Figure 1); whereas eukaryotic Box H/ACA RNAs contain two hairpins that are uniquely arranged to form a two-domain “hairpin-hinge-hairpin-tail” structure (Tang et al. 2002; Yu et al. 2005; Terns and Terns 2006; Reichow et al. 2007). Box H/ACA RNPs involved in directing site-specific pseudouridylation recognize their substrates through base-pairing interactions between the internal loop sequence (the guide sequence) of the Box H/ACA RNA and two short stretches of sequences within the substrate that flank the target uridine (Figure 1). The target uridine is positioned at the base of the upper-stem, which is 14–16 nucleotides upstream of either Box H or Box ACA and is left unpaired, remaining accessible for isomerization (Yu et al. 2005). Box H/ACA RNPs that are involved in the pseudouridylation of rRNA are localized to the nucleolus (snoRNPs; small nucleolar RNP) whereas the RNPs that guide pseudouridylation of spliceosomal snRNAs are primarily localized within Cajal bodies (scaRNPs; small cajal-body specific RNP) (Kiss 2002). While nucleolar targeting requires an intact Box H and Box ACA, the retention of Box H/ACA RNPs within the Cajal body requires an additional sequence element referred to as the CAB box (5′-UGAG-3′) located in the apical loop of either hairpin (Lange et al. 1999; Narayanan et al. 1999; Ruhl et al 2000, Richard et al. 2003). In 2006, Fu and Collins demonstrated that the Sm proteins, SmB and SmD3, are necessary for Cajal body retention and their interaction with scaRNAs depends on the CAB box (Fu and Collins, 2006).
Figure 2.
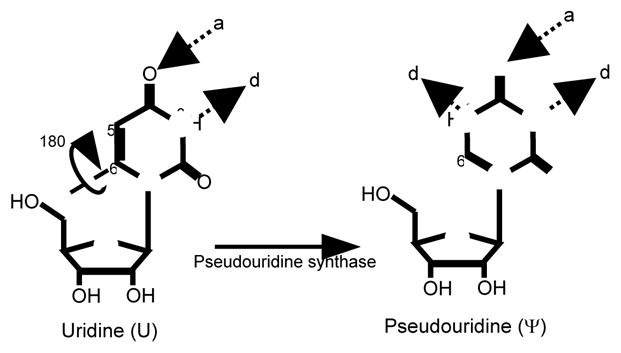
Pseudouridine is the 5-ribosyl isomer of uridine. Pseudouridine is formed from uridine by breakage of the glycosidic bond between N1 and C1′, 180° rotation of the base along the C6-N3 axis, and reformation of the glycosidic bond between C5 and C1′. Pseudouridine has one more hydrogen bond donor (d) than uridine, while hydrogen bond acceptors (a) are the same.
Telomere maintenance requires a Box H/ACA RNA
Vertebrate telomerase is a specialized Box H/ACA RNP responsible for the addition of telomeric DNA repeats onto the ends of eukaryotic chromosomes (Mitchell et al. 1999; Collins 2000; Meier 2005; Terns and Terns 2006). While the 5′ half of telomerase RNA (TR) contains a large pseudoknot domain as well as the template region, the 3′ half adopts the hairpin-hinge-hairpin-tail structure of eukaryotic Box H/ACA RNAs. Interestingly, the Box H/ACA domain of TR is essential for TR accumulation, 3′ end processing and nuclear trafficking (Mitchell et al. 1999; Lukowiak et al. 2001; Fu and Collins 2003; Jady et al. 2004). Furthermore, TR has been shown to localize to cajal bodies, and this localization is similarly dependent on SmB and SmD3, as well as a CAB box within TR (Jady et al. 2004; Zhu et al. 2004, Fu and Collins 2007). It should be noted that while TR contains a Box H/ACA domain, in vitro data indicates that TR is not capable of pseudouridylating a synthetic substrate complementary to the internal loops of the TR hairpins (Meier 2005).
Box H/ACA RNAs direct rRNA cleavage
In eukaryotic organisms, a few Box H/ACA RNAs are also involved in the processing of rRNA (Meier 2005; Terns and Terns 2006; Eliceiri 2006). The Box H/ACA RNAs E1/U17/snR30, snR10, E2 and E3, are all required for efficient pre-rRNA processing/maturation. In particular, the function of snR30 is essential for cell viability (Morrissey and Tollervey 1993; Mishra and Eliceiri 1997; Atzorn et al. 2004; Eliceiri 2006). Some Box H/ACA RNAs functioning in rRNA maturation adopt a secondary structure completely unique from the other Box H/ACA RNAs. This structural deviation is exemplified by E1/U17. For instance, E1 RNA contains two stem-loops within its 5′ half, a stem-loop structure within the hinge region, and a double stranded 5′ terminus (Selvamurugan et al. 1997, Eliceiri 2006).
Box H/ACA RNAs of unknown function
Within the past decade a new class of Box H/ACA RNA has been defined. Through size fractionation of RNAs and co-immunoprecipitation with antibodies against Box H/ACA core proteins, numerous small RNAs have been identified in mice and humans that are predicted to fold into the typical “hairpin-hinge-hairpin-tail” structure and assemble into RNPs (Hüttenhofer et al. 2001; Vitali et al. 2003; Kiss et al. 2004). Initially these RNPs were presumed to function as site-specific pseudouridine synthases; however, careful inspection of the Box H/ACA RNA guide sequences failed to identify complementarity to any of the known stable non-coding RNAs, i. e., rRNAs, snRNAs, and tRNAs (Hüttenhofer et al. 2001; Vitali et al. 2003; Kiss et al. 2004). Thus, these RNAs have been dubbed “orphan Box H/ACA RNAs”. While it is possible that these orphan RNAs may participate in a function yet to be ascribed to Box H/ACA RNAs, a role for them in directing pseudouridylation of mRNA, or some yet to be discovered noncoding RNA, has not been ruled out. In this regard, it has been reported that HBII-52, a Box C/D RNA (a separate class of small non-coding RNA responsible for guiding RNA 2′-O-methylation using a Box C/D RNA and a distinct set of four core proteins, see Yu et al. 2005 for a discussion of Box C/D RNPs) displays sequence complementarity with the Vb exon of the serotonin receptor 5-HT2CR (Cavaille et al. 2000). Indeed, Kishore and Stamm (2006) were able to detect an interaction between HBII-52 and a 5-HT2CR mini-gene. Strikingly, although it is not yet clear as to whether the target nucleotide in the mRNA is modified, this interaction affected the pattern of alternative splicing for this pre-mRNA transcript (Kishore and Stamm 2006). It is particularly exciting to speculate that some orphan Box H/ACA may play similar roles in the regulation of gene expression.
H/ACA RNP Core Proteins
All Box H/ACA RNAs examined to date, including the telomerase RNA, assemble with an evolutionarily conserved set of four core proteins (Kiss 2002; Meier 2005; Terns and Terns 2006). In S. cerevisiae, these proteins are: Cbf5p (Dyskerin in humans, NAP57 in rats, and Nop60B in drosophila), Gar1p, Nhp2p (L7Ae in archaea) and Nop10p (Henras et al. 1998; Watkins et al. 1998; Dragon et al. 2000; Pogacic et al. 2000; Watanabe and Gray 2000; Rozhdestvensky et al. 2003; Wang and Meier 2004). All core proteins are essential for cell viability (Bousquet-Antonelli et al. 1997; Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998; Dez et al. 2001). To date, the core proteins have been characterized primarily in the context of Box H/ACA RNAs that directs pseudouridylation (See below).
Cbf5p is the Box H/ACA RNP Pseudouridylase
Cbf5p was first identified in S. cerevisiae as a low affinity centromere binding protein that is essential for cell viability (Jiang et al. 1993). The following year, Meier and Blobel identified NAP57 as a protein that interacts with Nopp140, a nucleolar phosphoprotein of 140kD, expected to play a role in ribosome biogenesis (Meier and Blobel 1994). Excitingly, NAP57 was found to be 71% identical and 85% homologous to yeast Cbf5p, as well as having homologues in prokaryotic organisms (p35, E. coli; Bsp35, B. subtilis) (Figure 3). Through immunofluorescence and immunoelectron microscopy NAP57 was localized to the dense fibrillar component of the nucleolus and cajal bodies. Localization of NAP57 to the nucleolus, as well as an interaction with Nopp140, led to the proposition that NAP57 (and yeast Cbf5p) may function as a chaperone during the process of ribosome assembly.
Figure 3.
Yeast Cbf5p exhibits high sequence similarity to other pseudouridine synthases. Regions directly adjacent to the catalytic aspartate residue are strictly conserved within the TruB family of pseudouridine synthases. Grey shaded boxes represent amino acids which are strictly conserved or conservative amino acid substitutions with respect to its column. Black underlined letters (only apparent in the TruB of E. coli row) represent a non-similar amino acid with respect to that column.
A major break in elucidating the function of both Cbf5p and NAP57 occurred when E. coli p35 was identified as TruB, the pseudouridine synthase responsible for Ψ55 formation in virtually all elongator tRNAs (Nurse et al. 1995; Becker et al. 1997). Shortly thereafter, it was shown that Cbf5p was associated with Box H/ACA RNAs, and that point mutations within conserved residues led to a loss of rRNA pseudouridylation (Lafontaine et al. 1998; Zebarjadian et al. 1999). Interestingly, around a similar time it was also shown that the X-linked form of dyskeratosis congenita (X-DC), a rare bone marrow failure syndrome, was the result of a mutation within the DKC1 gene, the human homologue of NAP57 and Cbf5p (Heiss et al 1998; Knight et al. 1999).
The fact that Cbf5p shares high homology with the pseudouridine synthase TruB, and that a point mutation in Cbf5p globally abolishes the formation of pseudouridines in rRNAs strongly suggests that Cbf5p is a pseudouridine synthase associated with Box H/ACA RNPs (Zebarjadian et al. 1999). Detailed analysis indicates that Cbf5p contains a catalytic domain which is common to all known pseudouridine synthases, as well as a carboxyl-terminal PUA (pseudouridine synthase and archaeosine transglycosylase) domain, which is common to most TruB related pseudouridine synthases (Hamma and Ferre-D’Amare 2006; Terns and Terns 2006; Reichow et al. 2007). Mutation of a strictly conserved aspartic acid residue positioned within the active site abolishes the pseudouridine synthase activity of both TruB and Cbf5p (Ramamurthy et al. 1999; Zebarjadian et al. 1999; Reichow et al. 2007)
While the catalytic mechanism appears to be conserved among pseudouridine synthases, structural comparisons between TruB and archeal Cbf5p (aCbf5p) suggest distinct modes of substrate recognition (Hamma et al. 2005; Hamma and Ferre-D’Amare 2006; Manival et al. 2006; Rashid et al. 2006; Terns and Terns 2006; Reichow et al. 2007). In fact, two peptide segments that are involved in TruB’s recognition of tRNA are not present in aCbf5p. The loss of these peptide segments maybe compensated for by a highly electropositive surface potential of both the catalytic face and PUA domain of aCbf5p, which is indicative of their playing a role in substrate RNA binding (Rashid et al. 2006; Reichow et al. 2007). In addition, the recognition of substrate RNAs by Box H/ACA RNPs is aided by the fact that each RNP is associated with a unique Box H/ACA RNA, which is complementary to the substrate RNA.
Additional Core Proteins
While Cbf5p and its homologues are probably the most well studied proteins of the Box H/ACA RNP, three additional core proteins, namely Gar1p, Nhp2p, and Nop10p, are important constituents as well. Gar1p is a small basic protein consisting of two glycine-arginine-rich (GAR) domains flanking either side of a central core domain (Girard et al. 1992). Interestingly, the Gar1p core domain alone is sufficient for cell viability, and is capable of binding Box H/ACA RNAs in vitro (Bagni and Lapeyre 1998; Girad et al. 1994). However, structural analysis of a Box H/ACA RNP suggests that Gar1p does not engage in interactions with the Box H/ACA RNA (Li and Ye 2006) (see below).
Nhp2p (nonhistone protein) is similarly a small basic protein and shares sequence homology with the ribosomal protein L30 and 15.5K/NHP2L1 (Snu13 in yeast), which is present in both Box C/D RNPs and U4 snRNPs (Henras et al. 1998; Watkins et al. 1998, 2000; Nottrott et al. 1999; Leung and Lamond 2002; Meier 2005; Terns and Terns 2006,). While Nhp2p has not been extensively investigated, recent fluorescence data suggests that L7Ae, Nhp2p’s archeal homologue, may aid in the placement of the target uridine within the active site of the Box H/ACA RNP (Liang et al. 2007).
Nop10p is the smallest of the core proteins, consisting of only 64 amino acids (Henras et al 1998). While Nop10p is conserved across species and is essential for cell viability, it contains no recognizable or known motifs. Interestingly, in vitro analysis indicates that Nop10p and Cbf5p are sufficient to achieve basal levels of enzymatic activity when in the presence of a Box H/ACA RNA and a complementary substrate RNA (Charpentier et al. 2005). However, pseudouridylase activity is stimulated in the presence of the other core proteins (Charpentier et al. 2005).
Beyond the Core Components of the Box H/ACA RNP
As discussed earlier, Box H/ACA RNPs are involved in a range of cellular processes. Thus, it is extremely likely that specific functional classes of Box H/ACA RNPs interact with distinct subsets of proteins, in addition to the four core proteins, allowing further diversification of Box H/ACA RNPs (in addition to the diversity imposed by the different Box H/ACA RNAs). In fact, the catalytic subunit of telomerase—reverse transcriptase—is one such protein (Meier 2005; Terns and Terns 2006). Reverse transcriptase, while associated with the telomerase RNP, is associated with neither the Box H/ACA RNPs involved in RNA pseudouridylation, nor the Box H/ACA RNPs involved in rRNA processing. Furthermore, yeast snR30 and Xenopus oocyte E1, both of which are involved in rRNA processing, have been shown to co-purify and crosslink, respectively, with a number of uncharacterized proteins (Lübben et al. 1995; Smith et al. 2005). However, the identification and function of these proteins have remained elusive.
Structural Analysis of the core proteins of Box H/ACA RNP
In recent years a large amount of attention has been devoted to determining the three-dimensional architecture of Box H/ACA RNPs involved in pseudouridylation. This has been due in large part to the development of techniques allowing for the in vitro reconstitution of catalytically active archeal sRNPs (Charpentier et al. 2005). Thus far, structural information has been gathered exclusively on the archeal Box H/ACA RNP core proteins, while their eukaryotic homologues have, for the most part, escaped crystallographic analysis.
The recent crystallographic analyses of the complexes of archaeal Box H/ACA RNP core proteins, including heterodimeric aCbf5p-aNop10p complexes from both Methanococcus jannaschii and Pyrococcus abyssi, as well as a heterotrimeric aCbf5p-aNop10p-aGar1p complex from Pyrococcus furiosus, have provided a wealth of information regarding the molecular interactions that hold the proteins together, as well as how the individual proteins contribute to the process of RNA pseudouridylation (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006; Reichow et al. 2007; Ye 2007).
Structural comparisons between P. furiosus Cbf5p and E. coli TruB revealed structural homology with a root mean square deviation (r.m.s.d.) of 1.3 Å for 191 Cα atoms (Manival et al. 2006; Rashid et al. 2006). The active sites of the two enzymes superimpose closely, and residues involved in catalysis are all present in equivalent locations. However, several differences were observed between aCbf5p and TruB. Most striking was the lack of two stretches of 17 amino acids in aCbf5p. In TruB, these two segments are present in the thumb-loop domain which is involved in binding the substrate tRNA. In place of the thumb-loop domain, aCbf5p contains a β7/β10 hairpin loop, which is positioned within a similar location in the active site (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006; Terns and Terns 2006; Reichow et al. 2007; Ye 2007). Furthermore, several basic residues form two regions flanking the active site, and are also distributed across the catalytic face and PUA domain. These data hinted at distinct modes of substrate recognition for TruB and Cbf5p (as mentioned previously) (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006; Terns and Terns 2006).
aNop10p folds into an elongated two-domain structure when in complex with aCbf5p. The N-terminal half, which consists of a zinc-coordinating ribbon domain, coordinates a single zinc molecule by four conserved cysteines and is connected to a C-terminal alpha helix via a linker region (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006; Terns and Terns 2006; Reichow et al. 2007; Ye 2007). The linker region is the most highly conserved region of aNop10p and engages in several interactions with aCbf5p along a motif required for the stability of pseudouridine synthases (Koonin 1996; Reichow et al. 2007). The ability of aNop10p to form this elongated structure is dependent on its interaction with aCbf5p, as aNop10p alone appears to be intrinsically disordered (Terns and Terns 2006). Interestingly, yeast Nop10p, as well as other eukaryotic Nop10p, lack zinc coordination. This corroborates the fact that mutations within the N-terminal domain of archaeal Nop10p, which are predicated to disrupt zinc binding, do not affect its function in vitro (Charpentier et al. 2005). Furthermore, while yeast Nop10p lacks zinc coordination, it can assemble with archaeal Cbf5p, indicating that zinc coordination does not play a significant role in this interaction and that their mechanism of interaction is conserved from archaea to eukaryotes (Hamma et al. 2006).
Structural characterization of Box H/ACA protein complexes has also provided information regarding Gar1p. aGar1p folds into a compact six-stranded β-barrel structure, and interacts with aCbf5p at the site opposite to that engaged in binding with aNop10p (Li and Ye 2006; Rashid et al. 2006). aGar1p and aNop10p are separated by a distance of ~20Å in the aCbf5p-aGar1p-aNop10p complex. The fact that aGar1p does not make contact with aNop10p is in line with biochemical evidence that indicates independent interactions between aNop10p and aCbf5p, and aGar1p and aCbf5p (Baker et al. 2005). Interestingly, the β7/β10 hairpin loop of aCbf5p, which is thought to be involved in substrate recognition, is disordered when aCbf5p is not bound by aGar1p (Hamma et al. 2006; Terns and Terns 2006). This suggests that while aGar1p does not interact with the substrate RNA, aGar1p’s interaction with aCbf5p promotes structural rearrangements that are critical for the proper recognition of substrate RNAs and provides an explanation for the requirement of aGar1p for full enzymatic activity.
The structure of the complete Box H/ACA RNP
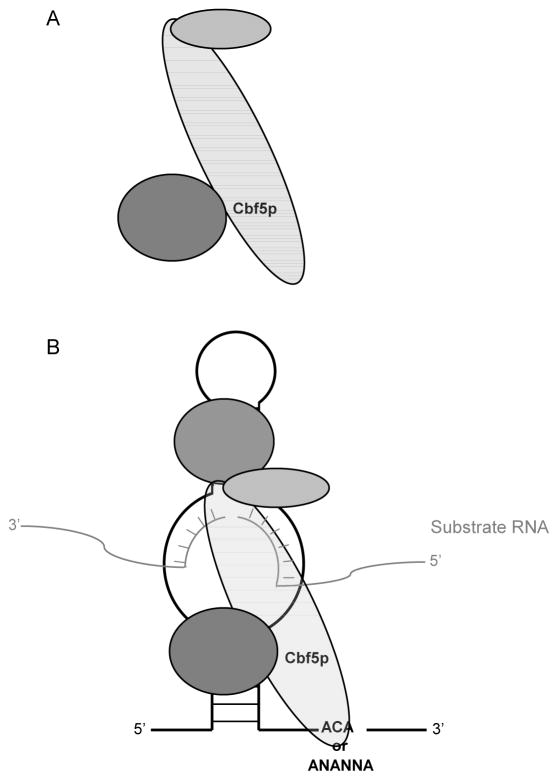
In 2006 Li and Ye published the first crystal structure of a complete Box H/ACA RNP at 2.3 Å resolution (Figure 4b). The structure is comprised of the four core proteins, aCbf5p, aGar1p, aNop10p, and L7Ae from P. furiosus bound to a 65 nucleotide single hairpin Box H/ACA RNA from Archaeoglobus fulgidus. Their structure closely resembles that of the aCbf5p-aNop10p-aGar1p structure, with an r.m.s.d. of 0.69 Å over 420 Cα atoms. The structure of the complete Box H/ACA RNP resembles that of an equilateral triangle with aCbf5p’s catalytic domain positioned in the center. aGar1p, L7Ae (Nhp2p in eukaryotes), and the PUA domain of aCbf5p constitute the three corners. aNop10p is sandwiched between L7Ae and aCbf5p.
Figure 4.
Schematic representation of a Heterotrimeric complex of the core Box H/ACA proteins and complete Box H/ACA RNP. (A) aGar1p, aCbf5p, and aNop10p are capable of forming a trimer in the absence of both a Box H/ACA RNA and Nhp2p. aGar1p interacts with aCbf5p opposite the side of Nop10p (see Rashid et al. 2006 for detail). (B) A schematic composite of the two structures provided by Li and Ye (2006) and Liang et al. (2007). The complete Box H/ACA RNP consists of the four core proteins, a Box H/ACA RNA, and substrate RNA. Nhp2p, Nop10p and the PUA domain of Cbf5p, are responsible for anchoring Cbf5p to the Box H/ACA RNA, and positioning Cbf5p’s catalytic core over the target uridine. (see Li and Ye 2006, and Liang et al. 2007 for atomic structures).
All the core components, excluding aGar1p, are observed making contacts with the Box H/ACA RNA. The upper stem of the hairpin is bound simultaneously by aNop10p, L7Ae, and aCbf5p, while the lower stem and the ACA motif are recognized by the PUA domain of aCbf5p. The two conserved adenines of the ACA motif are involved in extensive interactions, both aromatic and hydrogen bonding, with aCbf5p. Furthermore, numerous interactions are observed between the minor groove of the lower stem and the PUA domain, providing an explanation for the importance of maintaining a lower stem for the proper functioning of a Box H/ACA RNP. The tethering of aCbf5p by aNop10p and L7Ae to the apical stem, as well as the interactions of the aCbf5p PUA domain with the ACA motif and lower stem, positions the catalytic domain of aCbf5p over the pseudouridylation pocket. Thus, aNop10p, L7Ae and the PUA domain of aCbf5p can be seen as serving the purpose of forming a molecular bracket (Reichow et al. 2007; Ye 2007). This provides a structural explanation for the distance of 14–16nt from the Box H or Box ACA motifs to the nucleotide targeted for pseudouridylation (see above).
As mentioned earlier, crystallographic data suggests aGar1p plays a role in substrate recognition through the stabilization of the β7/β10 hairpin of aCbf5p (Rashid et al. 2006). In the crystal structure reported by Li and Ye, aGar1p makes a series of additional contacts with aCbf5p which are not observed in the heterotrimeric aCbf5p-aNop10p-aGar1p complex. Interestingly, when a substrate RNA was modeled into the complete Box H/ACA RNP structure, the additional contacts observed between aGar1p and aCbf5p orient the β7/β10 hairpin of aCbf5p in such a way as to prevent an interaction with the substrate (Li and Ye 2006, Reichow et al. 2007). However, if the β7/β10 hairpin is situated as in the heterotrimeric complex, it is positioned ideally for substrate recognition. Thus, it seems plausible that aGar1p is involved in regulating substrate loading and release through the stabilization of alternate conformations (Reichow et al. 2007).
In 2007 Liang et al. reported the crystal structure of a P. furiosus Box H/ACA RNP in complex with a substrate RNA, in the absence of L7Ae (Figure 4b). The structure indicates that upon the formation of the duplex between the Box H/ACA RNA and substrate RNA, several interactions are established between the duplex and conserved residues of aCbf5p, two of which, His63 and His80, are strictly conserved. Interestingly, a S121G mutation in dyskerin associated with X-DC, affects the amino acid directly adjacent to the amino acid corresponding to Pf Cbf5p His80.
While the structure reported by Liang et al. contained a substrate RNA, the uridine nucleotide targeted for pseudouridylation was ~11Å from the catalytic aspartate residue. Thus, the authors suggest that L7Ae plays a role in target RNA placement within active site, as L7Ae is missing from their structure and the target nucleotide is too far for modification. In line with this notion, fluorescence studies aimed at addressing local conformational changes near the target uridine indicate that upon titration of L7Ae into a preformed aCbf5p-aNop10p-aGar1p-guide RNA-substrate complex, significant changes in fluorescence intensity were observed indicating substantial conformational changes near the target uridine (Liang et al 2007).
Dyskeratosis Congenita (DC)
The functional importance of Box H/ACA RNPs is exemplified by the fact that mutations within core components of the RNP are associated with dyskeratosis congenita (DC). DC is a both clinically and genetically heterogeneous disease (Kirwan and Dokal 2007). Patients often display a variety of features, which include, but are not limited to, mucocutaneous abnormalities, bone marrow failure and an increased risk for the development of specific cancers (Kirwan and Dokal 2007; Vulliamy and Dokal 2007). Genetically, DC can be inherited in three fashions (Meier 2005; Kirwan and Dokal 2007). The first is an X-linked manner and is the most frequent and severe form of the disease. A positional cloning approach identified the gene responsible for X-DC as dyskerin (DKC1) (Heiss et al. 1998). DC can also be transmitted genetically in an autosomal dominant pattern, and mutations within the RNA component of telomerase (TR) have been identified as being responsible for this form of the disease (Vulliamy et al. 2001). Lastly, DC can be inherited in an autosomal recessive fashion. While there are likely several loci responsible for this form, recently Walene et al. (2007) identified mutations within Nop10p as being responsible for a specific subtype.
While a variety of mutations exist which result in DC, most occur within DKC1. The vast majority of mutations are missense mutations; however, a C-terminal truncation, intronic mutations, and mutations within the promoter have all been reported (Knight et al. 1999, 2001; Vulliamy et al. 1999; Salowsky 2002; Meier 2005). Strikingly, mutations in the protein-coding sequences tend to cluster within two locations: an N-terminal region outside of any conserved motifs, and in the PUA domain. Three mutations have been mapped within the catalytic domain as well (Li and Ye et al 2006). While explaining how mutations outside of conserved domains result in DC is rather difficult, mutations within the catalytic domain would be expected to result in a decrease, or even loss of, the ability to carry out the pseudouridylation reaction. Likewise, mutations in the PUA domain would be expected to alter binding of dyskerin to the Box H/ACA RNA, resulting in a similar defect in pseudouridylation. Puzzlingly, however, when mutations within the PUA domain of dyskerin are modeled into the structure provided by Li and Ye, they do not overlap with regions of Cbf5p involved in binding to the lower stem or the ACA motif (Li and Ye 2006). However, one X-DC mutation, S121G, affects an amino acid adjacent to a conserved histidine engaged in interactions with the substrate RNA/target RNA duplex. Whether these mutations overlap with the binding sites for dyskerin will have to await structural characterization of the eukaryotic Box H/ACA RNP. On the other hand, given that dyskerin/Cbf5p is also a component of telomerase and a component of the Box H/ACA RNPs involved in rRNA processing, mutations in dyskerin may affect telomere maintenance and rRNA maturation, respectively, thus linking the telomerase defect and rRNA processing defect to DC (see below). A crystal structure of the core proteins of the Box H/ACA RNP complexed with the telomerase RNA or a Box H/ACA RNA involved in rRNA maturation should help to clarify the effects of dyskerin mutations on telomere maintenance and rRNA maturation.
Molecular Mechanism of DC remains unclear
Analyses of patient cell lines as well as mouse models of DC have identified defects in numerous cellular processes including rRNA modification, rRNA processing, as well as in the maintenance of telomeric DNA (Mitchell et al. 1999b; Ruggero et al. 2003; Mochizuki et al. 2004). The initial discovery that dyskerin is a component of the telomerase RNP led to the general belief that DC was primarily a result of telomere dysfunction (Mitchell et al. 1999,a,b; Vulliamy et al 2001; Meier 2005). However, as discussed previously, Box H/ACA RNPs participate in a range of cellular processes including the pseudouridylation of rRNA and rRNA processing. Furthermore, it is becoming increasingly clear that defects in rRNA pseudouridylation and rRNA processing have profound effects on protein synthesis (King et al. 2003; Liang et al 2007). In line with the notion that rRNA modification and processing defects contribute to disease pathology, a hypomorphic dyskerin mouse model exhibits the hallmark symptoms of DC in the first generation, while no effect on telomere length was observed (Ruggero et al. 2003). However, these mice did display a significant reduction in rRNA pseudouridylation, as well as defects in
rRNA processing (Ruggero et al 2003). Furthermore, using an unbiased proteomic approach, Yoon et al. (2006) showed in the same hypomorphic dyskerin mouse model that internal ribosome entry site (IRES) meditated translation was impaired. Importantly, they showed that impaired cap-independent translation reduced cellular levels of the tumor suppressor p27(Kip1), offering some explanation for the observed predisposition to cancer in DC individuals (Yoon et al. 2006). Lastly, Mochizuki et al. (2004) introduced two dyskerin mutations, G402E and A353V, which are commonly observed in X-DC, into murine embryonic stem (ES) cells. Both cell lines exhibited reduced levels of specific Box H/ACA RNAs, as well as a reduction in rRNA pseudouridylation and processing.
It is interesting to note that Box H/ACA RNPs are also responsible for the formation of pseudouridines in the vertebrate spliceosomal U snRNAs, and that the pseudouridylation of snRNAs is known to contribute to their function in pre-mRNA splicing (Yu et al. 2005). However, to date, no studies have investigated whether any defects in pre-mRNA splicing exist in DC. Thus, it is still an open question as to whether telomere shortening is the main contributor to disease pathology. It is more likely that all of the defects observed (e.g. reduced pseudouridylation and processing of rRNA, telomere shorting, and probably defects in pre-mRNA splicing) contribute to disease pathology.
Concluding Remarks
Box H/ACA RNPs are incredibly diverse and complex macromolecular machines involved in a variety of cellular processes. Within the past decade remarkable progress has been made on defining the structure and mechanism of action of Box H/ACA RNPs. These data have shed light on the detailed molecular interactions that occur between the core protein components of the Box H/ACA RNP, and have also provided information regarding Box H/ACA RNA-protein binding and Box H/ACA RNP-substrate recognition.
While a tremendous amount of information has been gathered, numerous issues remain unresolved. For instance, a complete understanding of how a Box H/ACA RNP recognizes its substrate RNA can only be provided by a structural characterization of a complete Box H/ACA RNP-substrate complex. Furthermore, our understanding of the structure and function of Box H/ACA RNPs involved in rRNA processing and telomere maintenance is still premature. Also, an entire class of Box H/ACA RNPs, the “orphan Box H/ACA RNPs”, is uncharacterized both functionally and structurally. Eukaryotic Box H/ACA RNPs pose an even more challenging situation, as currently there are no enzymatically active reconstitution systems from purified recombinant proteins. Thus, any structure determined will be accompanied by serious doubts as to the biological authenticity of the structure. The structural and functional characterization of the eukaryotic H/ACA RNPs will help to unravel how their unique features are accommodated within the RNP as well as offer insight into the nature of DC mutations.
Acknowledgments
We would like to thank the members of the Yu lab for insightful discussions regarding Box H/ACA RNPs, and especially Chao Huang for help with preparation of the figures. Our work was supported by grant GM62937 (to Yi-Tao Yu) from the National Institute of Health.
References
- ATZORN V, FRAGAPANE P, KISS T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol. 2004;24:1769–78. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAGNI C, LAPEYRE B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem. 1998;273:10868–73. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- BAKER DL, YOUSSEF OA, CHASTKOFSKY MI, DY DA, TERNS RM, TERNS MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005;19:1238–48. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALAKIN AG, SMITH L, FOURNIER MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–34. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- BECKER HF, MOTORIN Y, PLANTA RJ, GROSJEAN H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–9. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUSQUET-ANTONELLI C, HENRY Y, G’ELUGNE JP, CAIZERGUES-FERRER M, KISS T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. Embo J. 1997;16:4770–6. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVAILLE J, BUITING K, KIEFMANN M, LALANDE M, BRANNAN CI, HORSTHEMKE B, BACHELLERIE JP, BROSIUS J, HUTTENHOFER A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97:14311–6. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHARPENTIER B, MULLER S, BRANLANT C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–44. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLLINS K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–83. doi: 10.1016/s0955-0674(00)00103-4. [DOI] [PubMed] [Google Scholar]
- DENNIS PP, OMER A. Small non-coding RNAs in Archaea. Curr Opin Microbiol. 2005;8:685–94. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- DEZ C, HENRAS A, FAUCON B, LAFONTAINE D, CAIZERGUES-FERRER M, HENRY Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001;29:598–603. doi: 10.1093/nar/29.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAGON F, POGACIC V, FILIPOWICZ W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol. 2000;20:3037–48. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELICEIRI GL. The vertebrate E1/U17 small nucleolar ribonucleoprotein particle. J Cell Biochem. 2006;98:486–95. doi: 10.1002/jcb.20821. [DOI] [PubMed] [Google Scholar]
- FU D, COLLINS K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell. 2003;11:1361–72. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- FU D, COLLINS K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006;20:531–6. doi: 10.1101/gad.1390306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GANOT P, BORTOLIN ML, KISS T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997a;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- GANOT P, CAIZERGUES-FERRER M, KISS T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997b;11:941–56. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- GIRARD JP, LEHTONEN H, CAIZERGUES-FERRER M, AMALRIC F, TOLLERVEY D, LAPEYRE B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. Embo J. 1992;11:673–82. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIRARD JP, BAGNI C, CAIZERGUES-FERRER M, AMALRIC F, LAPEYRE B. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J Biol Chem. 1994;269:18499–506. [PubMed] [Google Scholar]
- HAMMA T, REICHOW SL, VARANI G, FERRE-D’AMARE AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol. 2005;12:1101–7. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- HAMMA T, FERRE-D’AMARE AR. Pseudouridine synthases. Chem Biol. 2006;13:1125–35. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- HEISS NS, KNIGHT SW, VULLIAMY TJ, KLAUCK SM, WIEMANN S, MASON PJ, POUSTKA A, DOKAL I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–8. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- HENRAS A, HENRY Y, BOUSQUET-ANTONELLI C, NOAILLAC-DEPEYRE J, GELUGNE JP, CAIZERGUES-FERRER M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. Embo J. 1998;17:7078–90. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUTTENHOFER A, KIEFMANN M, MEIER-EWERT S, O’BRIEN J, LEHRACH H, BACHELLERIE JP, BROSIUS J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. Embo J. 2001;20:2943–53. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JADY BE, BERTRAND E, KISS T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–52. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG W, MIDDLETON K, YOON HJ, FOUQUET C, CARBON J. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol Cell Biol. 1993;13:4884–93. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG W, LIM MY, YOON HJ, THORNER J, MARTIN GS, CARBON J. Overexpression of the yeast MCK1 protein kinase suppresses conditional mutations in centromere-binding protein genes CBF2 and CBF5. Mol Gen Genet. 1995;246:360–6. doi: 10.1007/BF00288609. [DOI] [PubMed] [Google Scholar]
- KING TH, LIU B, MCCULLY RR, FOURNIER MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11:425–35. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- KIRWAN M, DOKAL I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2007 doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- KISHORE S, STAMM S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–2. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- KISS AM, JADY BE, DARZACQ X, VERHEGGEN C, BERTRAND E, KISS T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–9. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISS AM, JADY BE, BERTRAND E, KISS T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24:5797–807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISS T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–8. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- KNIGHT SW, HEISS NS, VULLIAMY TJ, GRESCHNER S, STAVRIDES G, PAI GS, LESTRINGANT G, VARMA N, MASON PJ, DOKAL I, POUSTKA A. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50–8. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIGHT SW, VULLIAMY TJ, MORGAN B, DEVRIENDT K, MASON PJ, DOKAL I. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum Genet. 2001;108:299–303. doi: 10.1007/s004390100494. [DOI] [PubMed] [Google Scholar]
- KOONIN EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–5. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAFONTAINE DL, BOUSQUET-ANTONELLI C, HENRY Y, CAIZERGUES-FERRER M, TOLLERVEY D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–37. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGE TS, EZROKHI M, AMALDI F, GERBI SA. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol Biol Cell. 1999;10:3877–90. doi: 10.1091/mbc.10.11.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEUNG AK, LAMOND AI. In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles. J Cell Biol. 2002;157:615–29. doi: 10.1083/jcb.200201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI L, YE K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–7. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- LIANG B, XUE S, TERNS RM, TERNS MP, LI H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol. 2007;14:1189–95. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- LIANG XH, LIU Q, FOURNIER MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–77. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- LUBBEN B, FABRIZIO P, KASTNER B, LUHRMANN R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:11549–54. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- LUKOWIAK AA, NARAYANAN A, LI ZH, TERNS RM, TERNS MP. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. Rna. 2001;7:1833–44. [PMC free article] [PubMed] [Google Scholar]
- MANIVAL X, CHARRON C, FOURMANN JB, GODARD F, CHARPENTIER B, BRANLANT C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 2006;34:826–39. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATTICK JS, MAKUNIN IV. Non-coding RNA. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- MEIER UT, BLOBEL G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–14. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIER UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MISHRA RK, ELICEIRI GL. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci U S A. 1997;94:4972–7. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITCHELL JR, CHENG J, COLLINS K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999a;19:567–76. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITCHELL JR, WOOD E, COLLINS K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999b;402:551–5. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- MOCHIZUKI Y, HE J, KULKARNI S, BESSLER M, MASON PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc Natl Acad Sci U S A. 2004;101:10756–61. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRISSEY JP, TOLLERVEY D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–77. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NARAYANAN A, LUKOWIAK A, JADY BE, DRAGON F, KISS T, TERNS RM, TERNS MP. Nucleolar localization signals of box H/ACA small nucleolar RNAs. Embo J. 1999;18:5120–30. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI J, TIEN AL, FOURNIER MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–73. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- NOTTROTT S, HARTMUTH K, FABRIZIO P, URLAUB H, VIDOVIC I, FICNER R, LUHRMANN R. Functional interaction of a novel 15.5kD [U4/U6. U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. Embo J. 1999;18:6119–33. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NURSE K, WRZESINSKI J, BAKIN A, LANE BG, OFENGAND J. Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. Rna. 1995;1:102–12. [PMC free article] [PubMed] [Google Scholar]
- POGACIC V, DRAGON F, FILIPOWICZ W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028–40. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMAMURTHY V, SWANN SL, PAULSON JL, SPEDALIERE CJ, MUELLER EG. Critical aspartic acid residues in pseudouridine synthases. J Biol Chem. 1999;274:22225–30. doi: 10.1074/jbc.274.32.22225. [DOI] [PubMed] [Google Scholar]
- RASHID R, LIANG B, BAKER DL, YOUSSEF OA, HE Y, PHIPPS K, TERNS RM, TERNS MP, LI H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell. 2006;21:249–60. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- REICHOW SL, HAMMA T, FERRE-D’AMARE AR, VARANI G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–64. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHARD P, DARZACQ X, BERTRAND E, JADY BE, VERHEGGEN C, KISS T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. Embo J. 2003;22:4283–93. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROZHDESTVENSKY TS, TANG TH, TCHIRKOVA IV, BROSIUS J, BACHELLERIE JP, HUTTENHOFER A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–77. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUGGERO D, GRISENDI S, PIAZZA F, REGO E, MARI F, RAO PH, CORDON-CARDO C, PANDOLFI PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–62. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- RUHL DD, PUSATERI ME, ELICEIRI GL. Multiple conserved segments of E1 small nucleolar RNA are involved in the formation of a ribonucleoprotein particle in frog oocytes. Biochem J. 2000;348(Pt 3):517–24. [PMC free article] [PubMed] [Google Scholar]
- SALOWSKY R, HEISS NS, BENNER A, WITTIG R, POUSTKA A. Basal transcription activity of the dyskeratosis congenita gene is mediated by Sp1 and Sp3 and a patient mutation in a Sp1 binding site is associated with decreased promoter activity. Gene. 2002;293:9–19. doi: 10.1016/s0378-1119(02)00725-4. [DOI] [PubMed] [Google Scholar]
- SELVAMURUGAN N, JOOST OH, HAAS ES, BROWN JW, GALVIN NJ, ELICEIRI GL. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–6. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH JL, WALTON AH, ELICEIRI GL. UV-crosslinking of E1 small nucleolar RNA to proteins in frog oocytes. J Cell Physiol. 2005;203:202–8. doi: 10.1002/jcp.20223. [DOI] [PubMed] [Google Scholar]
- TANG TH, BACHELLERIE JP, ROZHDESTVENSKY T, BORTOLIN ML, HUBER H, DRUNGOWSKI M, ELGE T, BROSIUS J, HUTTENHOFER A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–41. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERNS MP, TERNS RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- TERNS M, TERNS R. Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- VITALI P, ROYO H, SEITZ H, BACHELLERIE JP, HUTTENHOFER A, CAVAILLE J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–51. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VULLIAMY TJ, KNIGHT SW, HEISS NS, SMITH OP, POUSTKA A, DOKAL I, MASON PJ. Dyskeratosis congenita caused by a 3′ deletion: germline and somatic mosaicism in a female carrier. Blood. 1999;94:1254–60. [PubMed] [Google Scholar]
- VULLIAMY T, MARRONE A, GOLDMAN F, DEARLOVE A, BESSLER M, MASON PJ, DOKAL I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- VULLIAMY TJ, DOKAL I. Dyskeratosis congenita: The diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2007 doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- WALNE AJ, VULLIAMY T, MARRONE A, BESWICK R, KIRWAN M, MASUNARI Y, AL-QURASHI FH, ALJURF M, DOKAL I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum Mol Genet. 2007;16:1619–29. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG C, MEIER UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. Embo J. 2004;23:1857–67. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE Y, GRAY MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28:2342–52. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATKINS NJ, GOTTSCHALK A, NEUBAUER G, KASTNER B, FABRIZIO P, MANN M, LUHRMANN R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. Rna. 1998;4:1549–68. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATKINS NJ, SEGAULT V, CHARPENTIER B, NOTTROTT S, FABRIZIO P, BACHI A, WILM M, ROSBASH M, BRANLANT C, LUHRMANN R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–66. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- YE K. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol. 2007;17:287–92. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- YOON A, PENG G, BRANDENBURGER Y, ZOLLO O, XU W, REGO E, RUGGERO D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–6. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- YU Y-T, TERNS RM, TERNS MP. Mechanisms and functions of RNA-guided RNA modifications. In: Grosjean H, editor. Fine-tuning of RNA functions by modification and editing, Topics in Current Genetics series. Vol. 12. Springer-Verlag Press; New York, USA: 2005. pp. 223–262. [Google Scholar]
- ZEBARJADIAN Y, KING T, FOURNIER MJ, CLARKE L, CARBON J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–72. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Y, TOMLINSON RL, LUKOWIAK AA, TERNS RM, TERNS MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]