Abstract
Microarray technology provides an opportunity to view transcriptions at genomic level under different conditions controlled by an experiment. From an array experiment using a human cancer cell line that is engineered to differ in expression of tumor antigen, integrin α6β4, few hundreds of differentially expressed genes are selected and are clustered using one of several standard algorithms. The set of genes in a cluster is expected to have similar expression patterns and are most likely to be coregulated and thereby expected to have similar function. The highly expressed set of upregulated genes become candidates for further evaluation as potential biomarkers. Besides these benefits, microarray experiment by itself does not help us to understand or discover potential pathways or to identify important set of genes for potential drug targets. In this paper we discuss about integrating protein-to-protein interaction information, pathway information with array expression data set to identify a set of “important” genes, and potential signal transduction networks that help to target and reverse the oncogenic phenotype induced by tumor antigen such as integrin α6β4. We will illustrate the proposed method with our recent microarray experiment conducted for identifying transcriptional targets of integrin α6β4 for cancer progression.
1. Introduction
A micro-array experiment is conducted to study expression profiles of genes in a specimen under different experimental conditions, or over several different time periods. It serves many purposes that include (1) developing a predictive computational model which can be used to predict biomarkers and targets for cancer therapy, (2) gaining some insight on gene regulation when a microarray experiment is conducted in different time points, (3) gaining insight on the genes that may be involved in a situation or disease under investigation, (4) understanding or refining protein-in-protein interaction networks, and (5) annotating uncharacterized genes. In a recent review article on the applications of microarray, Troyanskaya [1] provides some details on the items 2, 4, and 5. Statistical tests are conducted to filter valid signals first and then a subset of genes called differentially expressed genes is selected based on their relative strength or weakness of expression levels with respect to their reference expression values. The differentially expressed probes, which roughly correspond to genes, are reduced to few hundreds while the total number of probes of an experiment is in the order of 20 to 50 thousands.
The set of highly expressed genes are considered to be candidates for biomarkers in a microarray experiment. It is quite difficult to single out the best biomarkers by viewing expression level alone partially due to noise or some association by “guilt.” By integrating microarray expression data with other information pertaining to the protein behavior we can improve the quality of decision on biomarkers as has been proposed by Camargo and Azuaje in [2]. Similarly we vcan gain better insight into gene regulation by associating gene expression with protein interaction network with known cancer related pathways.
A significant volume of works has been done that relates or combines microarray data sets and protein-to-protein interaction networks. Based on the expected outcome, these works may be characterized into (1) annotating uncharacterized genes, (2) refining protein-to-protein interaction network, (3) predicting protein to protein interaction, and (4) refining potential biomarkers from array expression. Integrating protein interaction network information with expression data sets along with other information pertaining to a gene has been used [3–7] for annotating uncharacterized protein. In the recent work of Nariai et al. [6], probabilistic approach has been used to integrate protein to protein interaction, array expression, protein motif, gene knockout phenotype data, and protein localization data for predicting the function of an uncharacterized genes.
Microarray expressions data has also been used for refining protein to protein interaction networks. Zhu et al. [8] have used coexpressed genes from microarray data set to filter the neighbors of protein in an interaction network to enhance the degree of functional consensus among the neighbors.
Array expression data sets are used for predicting protein to protein interaction [9, 10]. Recently Soong et al. [10] have used microarray expression to predict protein to protein interaction. A pair of proteins is represented by a feature vector consisting of a concatenation of expression modes or profiles of those proteins along with the Pearson correlation of the expression profiles of these two proteins. They have demonstrated the predictability of using support vector machine with protein to protein interaction of yeast data sets from DIP [11] and 349 yeast microarray expression data sets from GEO [12].
Camargo et al. [12] have integrated array expression data set with expression data for refining potential biomarkers. Their work has some overlapping with our current approach in selecting hub nodes from interaction network and combining with array expression data sets. Their focus, however, was only on refining the biomarkers derived from array expression as opposed to providing insight into potential signal transduction pathways or any other intermediate activities that are not revealed in an array expression.
We take a different approach that compliments the strength of interaction data sets and array expression data sets. The array data sets capture the expression levels at different experimental conditions (or time points) while the information on interaction networks represents experimentally determined and as well as predicted interaction between pairs of proteins in a two-dimensional space without paying attention to the context, the temporal relations, or the process. By bringing two different types of modalities of information together, we believe we can discover some important genes that may have played important roles in the final observation of the array expression.
Suppose we consider a binary case of studying the expression pattern of a cell line of healthy and sick subjects. Examining the differentially expressed genes provides information on which genes are up-or downregulated, and their expression levels. This information alone does not provide insight into deciding interesting set of genes that are either taking part of the progression or the cause of the disease under consideration. We will show how to integrate gene expression with expression patterns with protein to protein interaction, and known genes in disease pathways to gain insight onto a small subset of interesting genes relevant to the disease under investigation.
To illustrate and to apply the idea of integrating microarray data with protein to protein interaction network, and disease related pathways, we use our recent microarray study for identifying transcriptional targets of integrin α6β4 for cancer progression. Jun Chung and his associates have used the affymetrix HG-U133A_2 to identify transcriptional targets of integrin α6β4. The goal of the study is to identify α6β4 transcriptional targets important for breast cancer progression. The α6β4 integrin, an epithelial-specific integrin, functions as a receptor for the members of the laminin family of extra cellular matrix proteins [13, 14]. While the primary known function of α6β4 is to contribute to tissue integrity through its ability to mediate the formation of hemidesmosomes (HDs), there is growing evidence suggesting that this integrin also plays a pivotal role in functions associated with cancer progression [13, 14]. For example, high expression of this integrin in women with breast cancer has been shown to correlate significantly with mortality and disease states [13, 14]. However, therapeutic targets of breast cancer that overexpress α6β4 are not yet well characterized. For this reason, it is essential to elucidate the mechanism by which α6β4 contributes to breast cancer progression.
We describe the data set, methods, and approaches in Section 2. It is followed by results in Section 3. In Section 4, we summarize and discuss the results.
2. Materials and Methods
2.1. Data
We are focusing on genes of Homo sapiens and their expressions for this experiment. From Affymetrix site at http://www.affymetrix.com/, we have downloaded the annotations (HG-U133A_2.na22.annot) for the genes that are tested in a microarray experiment.
The gene expression data is from our recent microarray experiment using the affymetrix HG-U133A_2 to identify transcriptional targets of integrin α6β4. Our study here describes the gene expression profile obtained from MDA-MB-435 mock transfectants (α6β4 negative human cancer cell line) and MDA-MB-435 β4 integrin transfectants (α6β4 positive human cancer cell lines). Out of oligonucleotide probe sets representing approximately 22 277 genes, expression of β4 integrin in MDA-MB-435 cells up regulated 149 genes by twofold or higher. 193 genes are down regulated by over two fold change. We anticipate that microarray data will lead to not only the identification of α6β4 target genes that are important for breast cancer cell growth, survival, and invasion, but also the discovery of signaling pathways leading to the expression of these genes.
The protein to protein interaction databases include MIPS [15], DIP [11], BIND [16, 17], GRID and I2D [18]. Noise is often a factor in many protein to protein interaction dataset. To minimize the noise and its impacts on the final outcome, we apply ensemble-based method for selecting the interaction. That is, by applying majority voting on interacting pairs from different the database, we can improve the accuracy and minimize the errors in their interaction information. I2D provides experimentally determined and predicted protein to protein interaction with easy to use interface, and thus we have downloaded I2D [18] for homo sapiens genome.
2.2. Data Preprocessing
Suppose we are gathering protein to protein interaction from different sources each with their own accuracy. By combining the results of independent test or source that has prediction accuracy over 50%, we can obtain prediction accuracy better than any one method alone. Suppose we have n independent sources each with some predefined fixed prediction accuracy, say p. Without loss of generality, let us assume n is an odd number. By accepting the decision of majority predictors among n, the combined accuracy is given by the following formula:
| (1) |
where k = ⌈(n/2).
Suppose nine independent predictors each with prediction accuracy 0.65 are combined by majority votes, the combined prediction accuracy becomes 0.83.
I2D [18] collects and maintains protein to protein interaction from various sources and we have downloaded the interaction information pertaining to Homo Sapiens. By applying the majority votes, we have minimized some plausible noise in the data set.
The microarray experiment was repeated three times and in each repetition the expressions of genes under the following two conditions are measured: (1) integrin negative cell line (control), and (2) integrin positive cell line. Out of the 22 277 genes we have selected only 8512 genes that have valid signal in all measurements. The average of the log ratio between the integrin positive and the control expression in all the repetitions is taken as the expression of a gene. From the expressions, we could create different expression patterns based on the values such as up regulated fold changes over 2 to 3, 3 to 4, and over 4. Among the down regulated genes, we may have the similar groups. For simplicity, we have taken only two patterns, namely, up regulated and down regulated genes. The up regulated genes are those that have fold changes (log of the ratio 2) over 1 and the down regulated are those that have the fold changes (log of the ratio 0.5) less than −1.
2.3. Methods
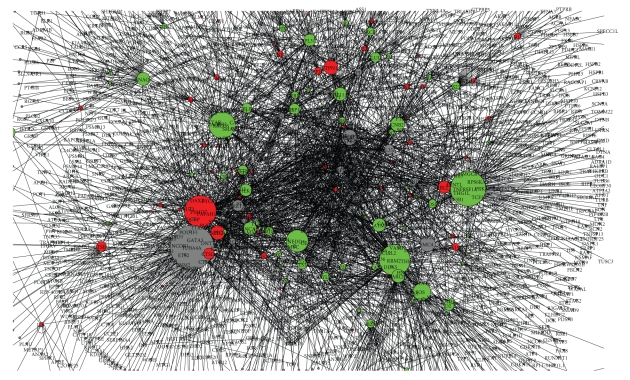
We have downloaded human protein to protein interaction networks from I2D, which have 13 560 genes that have connectivity from 1 to 694. The connectivity or degree of a node is defined as the number of edges connected to the network and we consider each edge as bidirectional connection. As expected, the interaction follows the scale free distribution. For the purpose of integrating the interaction network with the microarray expression data set, we have extracted a subnetworks from the whole networks that interact with the differentially expressed genes from the experiment. The selected sub networks, which we refer to as G, have 2186 genes including the 190 differentially expressed genes, and 3130 edges. A view of Graph G is shown in Figure 1 as created by Navigator [19]. The up and down regulated genes are shown in red and green, respectively, and the size of each node corresponds to the degree of interaction of that node in the graph.
Figure 1.
A view of protein to protein interaction associated with the differentially expressed genes. We refer to this graph as G.
In a typical microarray analysis, the differentially expressed genes are ranked based on their fold changes and the first few of them as taken as important. We feel that using expression fold change alone to determine the importance of a gene is quite weak. We take a different approach in this paper for discovering a set of important genes under a given experimental condition. We create the subgraphs, say G, of protein to protein interaction networks that is associated with the differentially expressed genes from the microarray experiment. It is generally believed that the connectivity of nodes in G roughly reflects the importance of the gene in the interaction [20]. We found that even the network G has the property of a typical scale free network indicating only a small fraction of the node has large connectivity.
2.3.1. Selecting a Set of Important Genes Based on Topological Structure
In the recent work, Jeong et al. [20] and Twe et al. [21] have suggested that essential proteins are over represented among those proteins having high degree of connectivity, which can be attributed to the central role in mediating interactions among numerous, less connected proteins. Hub nodes in an interaction network are defined as a set of nodes with very high degree of interaction with neighbors and the corresponding threshold for connectivity is defined quite arbitrarily. Vallabhajosyula et al. [22] have studied the issue on selecting hub nodes and the impacts on their functional significance, but unfortunately they were unable to provide and prescriptive definition or method on selecting hub nodes. They, however, stated that the nodes with relatively high degree of interaction are likely to have very high functional significance. In the literature, we found that people have applied varying criteria in selecting the threshold for hub nodes; for example, Batada et al. [23] have defined hub nodes as those connect to over 90% or 95% of the nodes in the network. Biasing from the finding in [22] that the top few percentage of nodes with high degree of interaction has better functional significance, we selected the hub nodes; those that are in the top 3% of the nodes ranked based on the decreasing order of connectivity.
We also believe that important genes must also play a role in the stability of the network, that is, removal of such node will break the network into disconnected subnetworks. An articulation node in a graph plays the role of connecting or keeping the graph together and the removal of such node separates the graph into subgraphs. Thus the hub genes that play articulation role in an interaction network seem to have more functional significance.
A minimum spanning tree is acyclic graph that connects all the nodes in a network such that the summation of cost in all the edges is minimal and thus eliminates redundant paths among the nodes. A node with high degree of connectivity in minimum spanning tree will indeed play an important role. In a protein interaction network the edge cost is taken to be 1 and we construct a minimal spanning tree using Kruskal's algorithm [24]. We selected the hub nodes from the minimum spanning tree and consider them as important genes too.
As described above, three set of potentially important nodes can be selected from the following different methods: (1) hub nodes from the interaction networks, (2) hub nodes from the set of articulation nodes, and (3) hub nodes from the minimum spanning tree. The nodes satisfying condition 2 are indeed a subset of those satisfying condition 1 and hence we have only two distinct conditions, namely, 2 and 3. We define a set of important genes; those that satisfy either conditions 2 or 3.
2.3.2. Important Genes Based on Pathways and Interaction
Pandey Lab at the Johns Hopkins University and the Institute of Bioinformatics [25] maintains experimentally determined ten cancer signaling pathways for Homo Sapiens, namely, EGFR1, TGF, beta Receptor, TNF, alpha/NF-kB, α6β4 Integrin, ID, Hedgehog, Notch, Wnt, AR, and Kit Receptor. We have obtained the genes in each of the ten cancer pathways and extracted sub network, say Gp, from the interaction network that interacts with any genes in the cancer pathway. The important nodes of Gp include the ones from the three following methods or sources.
Hub nodes of Gp.
Hub nodes of the articulation nodes of Gp.
Hub nodes of the minimum spanning tree created from Gp.
The nodes satisfying condition 2 are indeed a subset of those satisfying condition 1 and hence we have only two distinct conditions, namely, 2 and 3. The important nodes related to cancer pathway are those that satisfy either condition 2 or 3.
Besides examining the important nodes in each graph, we can examine the cliques or near cliques for similar functional association of genes. Han et al. [26] along with many other researchers have used cliques or near cliques in an interaction network to find functional group of genes. A clique is a fully connected subgraph of a graph and find cliques in a network is computationally intractable. For many practical purposes, near cliques are computed.
3. Results
From the microarray experiment, we have two different expression patterns, namely, up-and downregulated genes. The up regulated genes are those that have valid signal across three trials and have expression level over 2 times that of the reference gene. Similarly the down regulated genes are those that have valid signal across three trials and have inverse expression level over 2 with respect to the reference gene. We list the first 14 up and down regulated genes of our experiment in Table 1. We combined the gene expression with gene interaction by selecting subset of the interaction graph that associates with all the differentially expressed genes. The selected subgraphs, which we refer to as G, have 2186 genes including the 190 differentially expressed genes, and 3130 edges. Note that there is no single hub node among the 14 high ranking up regulated nodes of G. On the other hand, there are 5 hub nodes among the high ranking down regulated nodes. There seems to be no correlation among the hub nodes of an interacting graph with highly up or down regulated genes.
Table 1.
The high ranking 14 up regulated genes based on the fold changes. For each gene in the list the connectivity in the protein interaction network G is given. None of the ranked upregulated genes are hub nodes.
| Genes | Fold changes | Connectivity in G |
|---|---|---|
| IL8 | 5.63 | 11 |
| S100A3 | 4.86 | 4 |
| SOX4 | 4.54 | 2 |
| SLCO4A1 | 4.12 | 2 |
| MAGEH1 | 3.77 | 9 |
| AKR1C1 | 3.72 | 2 |
| MAD1L1 | 3.45 | 21 |
| IL24 | 3.35 | 1 |
| HSPA6 | 3.25 | 13 |
| NRCAM | 3.18 | 10 |
| COL6A1 | 3.07 | 5 |
| ASPH | 3.03 | 2 |
| TUSC3 | 2.98 | 1 |
| PEG10 | 2.87 | 1 |
From the graph G, we select the set of important genes based on topological structure, which involves selecting the hub nodes and following the procedures described in the previous section. The cutoff connectivity for the hub nodes in G is 16 and there are 60 hub genes out of 2186 genes. Out of the 60 hub nodes, 49 are from the differentially expressed genes (12 of them are up regulated and the rest are down regulated). The graph G has 200 articulation genes and out of which 60 satisfy the hub condition (degree 16 or above). The minimum spanning tree of G was constructed assuming the edge cost is 1. The nodes with connectivity 9 or better in the minimum spanning tree satisfy the hub node property. The minimum spanning tree has 77 hub genes and out of which 17 of them are up regulated and 46 are down regulated. In agreement with conditions 2 and 3 in Section 2, 57 genes are selected as important ones out of which 12 are up regulated and 35 are down regulated. These genes are listed in Table 3.
Table 3.
The important set of genes based on topological structure of interaction network. Selecting the nodes that satisfy condition 2 (the articulation nodes among the hub nodes of the network) and condition 3 (the hub nodes of the minimum spanning tree). The inverse of fold changes for down regulated genes is shown. Thus the table includes the genes that are not considered in the experiment or neither up-or downregulated.
| Gene | Regulation | Fold change |
|---|---|---|
| CPNE1 | Up | 2.63 |
| CTSB | Up | 2.78 |
| CTSD | Up | 2.00 |
| MAD1L1 | Up | 3.45 |
| MEF2C | Up | 2.21 |
| PCM1 | Up | 2.01 |
| PRKAR2B | Up | 2.80 |
| PSMD7 | Up | 2.01 |
| PTPN1 | Up | 2.06 |
| RBL2 | Up | 2.62 |
| RGS20 | Up | 2.20 |
| SOD2 | Up | 2.10 |
| ADSL | Down | 2.23 |
| ATM | Down | 2.09 |
| BID | Down | 2.09 |
| CASP1 | Down | 3.07 |
| CBLB | Down | 2.32 |
| CCNB2 | Down | 2.32 |
| CDC7 | Down | 2.17 |
| CHEK1 | Down | 2.06 |
| CTTN | Down | 2.63 |
| DDX17 | Down | 2.13 |
| DGCR14 | Down | 2.04 |
| ETS1 | Down | 2.50 |
| FOS | Down | 2.94 |
| GLUL | Down | 5.13 |
| GNAQ | Down | 2.39 |
| ID2 | Down | 2.86 |
| MACF1 | Down | 2.05 |
| MRE11A | Down | 2.61 |
| MYLK | Down | 4.25 |
| NEDD4 | Down | 2.01 |
| PAFAH1B2 | Down | 2.57 |
| PPFIA1 | Down | 2.41 |
| PRKAB2 | Down | 2.56 |
| PRSS23 | Down | 2.47 |
| RAB27A | Down | 2.74 |
| RAB8B | Down | 2.27 |
| RPL31 | Down | 2.08 |
| RRM2 | Down | 2.32 |
| SMARCB1 | Down | 2.14 |
| SNCA | Down | 5.24 |
| TGFB2 | Down | 2.06 |
| THBS1 | Down | 3.70 |
| TNFRSF1A | Down | 2.13 |
| TPM1 | Down | 4.62 |
| XRCC6 | Down | 2.08 |
| DDX19B | Down | 2.09 |
| *GRINL1B | — | — |
| *HIST2H3PS2 | — | — |
| *NA | — | — |
| *RET | — | — |
| *RPL35A | — | — |
| **SMC2 | — | — |
| **SMC4 | — | — |
| **TPI1 | — | — |
| **TUBA4A | — | — |
*These genes are neither up-or downregulated, nor considered in the experiment.
**These genes are from interaction network that satisfy conditions 2 and 3.
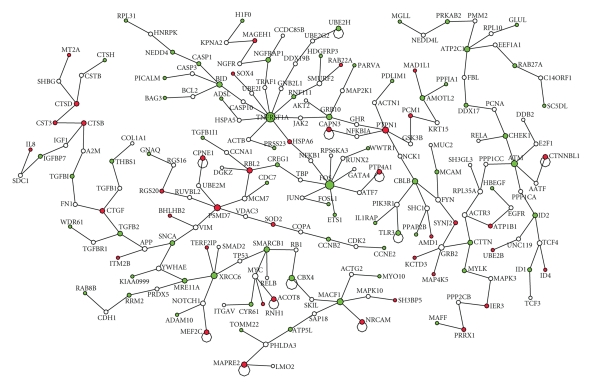
To discover the important genes related to cancer, we have extracted a sub network, which we call Gp, from G such that each node in Gp is directly associating with any one of the genes in cancer pathways that include EGFR1, TGF, beta Receptor, TNF, alpha/NF-kB, Alpha6 Beta4, Integrin, ID, Hedgehog, Notch, Wnt, AR, and Kit Receptor pathways. The genes in these curated pathways for human are downloaded from their web portal [25]. We found 24 nodes in the network with connectivity 12 or better satisfy the hub node property. The pathway related network Gp has 132 articulation genes out of which 23 are hub genes. The minimum spanning tree of Gp is constructed and the backbone of the minimum spanning tree is shown in Figure 2. The minimum spanning tree has 200 genes and 17 out of these genes have connectivity 4 or better satisfy the hub node property. By combining all these three set of hub genes using ensemble method, we have created the important genes related to pathways and are presented in Table 4.
Figure 2.
The minimum spanning tree of the network associated with cancer pathway genes. The backbone of the tree is shown. Up-and down regulated genes are shown in red and green color.
Table 4.
The important genes of network associated with genes in cancer pathways. These genes are obtained by combining three sets of hub genes from interaction network, articulation nodes, and from the minimum spanning tree of Gp. We show the specific pathway a gene is involved with.
| Genes | Regulation | Pathway |
|---|---|---|
| CTSB | Up | |
| CTSD | Up | Tgf_beta,ar |
| PSMD7 | Up | |
| PTPN1 | Up | |
| RBL2 | Up | Tgf_beta |
| SOD2 | Up | Tnf_alpha |
| ATM | Down | |
| BID | Down | Tnf_alpha |
| CASP1 | Down | Tnf_beta |
| CBLB | Down | |
| CCNB2 | Down | Tgf_beta |
| CHEK1 | Down | |
| ETS1 | Down | Tgf_beta,tnf_alpha |
| FOS | Down | Wnt,ar,kit |
| GRB10 | Down | ar |
| ID2 | Down | Tgf_beta,ar |
| MRE11A | Down | |
| NEDD4 | Down | Tgf_beta |
| RRM2 | Down | Egfr1 |
| SNCA | Down | |
| TGFB2 | Down | Egfr1,tgf_beta,tnf_alpha,ar |
| THBS1 | Down | Tgf_beta, tnf_alpha, id,wnt |
| TNFRSF1A | Down | Tgf_beta,notch,kit |
| XRCC6 | Down |
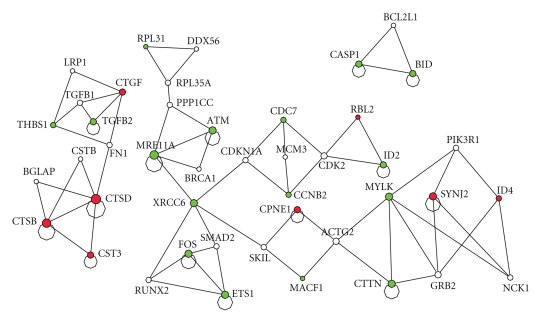
Besides examining the important genes in Gp, the cancer pathway related network, we searched for cliques or near cliques in the network to examine functionally related genes. The cliques from the network Gp is shown in Figure 3.
Figure 3.
The cliques or near cliques from the cancer pathway related network Gp. The up-and downregulated genes are shown in red and green, respectively.
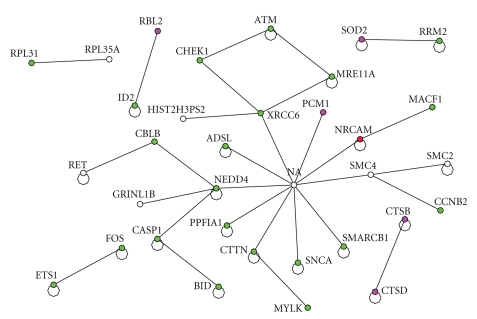
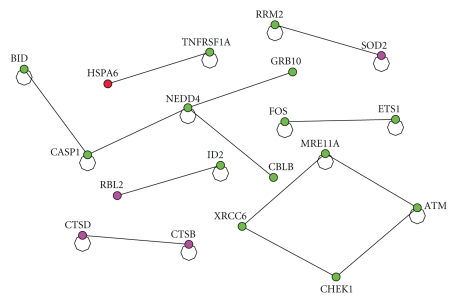
Let us examine the interaction among important genes based on topological structure (from Table 3) and between the highly expressed genes from Table 1. The interaction is shown in Figure 4.
Figure 4.
The interaction between the top 14 up regulated genes from Table 1 with the set of important genes based on network topology (Table 3). The red one represents the gene from Table 1. The green colored ones are down regulated and the red and purple ones are up regulated.
The direct interaction among the genes identified as important nodes due to the known cancer pathways is shown in Figure 5.
Figure 5.
The interaction between the top 14 up regulated genes from Table 1 with the set of important genes based on pathway (Table 4). The red one represents the gene from Table 1. The green colored ones are down regulated and the red and purple ones are up regulated.
4. Summary and Discussion
In this paper we have presented a general method for integrating microarray expression with other complementary information related to gene function so that we can understand and infer information about the set of genes that we are interested. Particularly we focused on integrating protein interaction information and pathway related information with microarray expression. We have applied the proposed general methodology to our recent microarray experiment to discover potential drug target that may lead to novel anticancer therapeutics.
Quite a large body of research works is done in integrating expression data with interaction network and other data sets. Many of the works fall into one or some combination of the following categories: (1) annotating uncharacterized genes, (2) refining protein to protein interaction network, (3) predicting protein to protein interaction, and (4) refining potential biomarkers from array expression. The presented work here has some overlaps with the recent work of Camargo et al. [2], which involved in integrating expression data set with expression data set for refining potential biomarkers of array expression and to annotate uncharacterized genes. They have used hub genes of the interaction network to refine biomarkers of the expression data sets.
The interaction network of Homo sapiens is scale free, that is there are few nodes having very high degree of interaction and facilitate other nodes in mediating their functions. Even the subnetwork of the interaction network that has direct interaction with differentially expressed genes is found to be having the properties of scale free network. Hub nodes in an interaction network are defined as a set of nodes with very high degree of interaction with neighbors and the corresponding threshold for connectivity is defined quite arbitrarily. Biasing from the finding in [22] that the top few percentage of nodes with high degree of interaction has better functional significance, we selected the hub nodes; those that are in the top 3% of the nodes ranked based on the decreasing order of connectivity.
From the Homo sapiens interaction network, we have extracted a sub network called G that is associated with the differential expressed genes of our microarray experiment. Hub nodes in an interaction network are important and we selected the first set of hub nodes from G. A set of articulation nodes, which plays the role of stability of the network, is also important. We selected a set of articulation nodes from G. We have constructed a minimum spanning tree from G and we have selected a set of hub nodes from the minimum spanning tree. We created important set of genes based on topological structure of the interaction network. The hub nodes alone in isolation do not reveal any useful information. Similarly the highly ranked up or down regulated genes by themselves do not provide any clue into any potential signaling pathways either.
On the other hand, when we combine the set of important genes based on the interaction topology from Table 3 and the set of highly expressed genes from Table 1, we started to get some insight into potential signal transduction pattern as shown in Figure 4. The highly expressed gene from the experiment NRCAM, neuronal cell adhesion molecule, is directly interacting with another gene NA (neurocanthocytosis) which is recognized as an important gene from the topology and mediating the down regulation of the following set of tumor suppression genes, CHEK1 [27], XRCC6 [28], SMARCB [29], and ATM [30]. The gene NA acts as a hub gene among the set of important genes and it directly interacts with SMARCB and XRCC4, which directly interacts with CHEK1 which in turn directly interacting with ATM. It is notable that down regulation of these tumor suppressor genes by integrin α6β4 has a significant implication in cancer biology. Poor prognosis has been associated with over expression of integrin α6β4 and our analysis revealed that loss of these tumor suppressor genes could attribute to malignant phenotype of cancer cells.
Impact of this study lies in the identification and targeting molecular aberrations specific to cancer cells. Many recent studies with targeting a single agent turned out to be a disappointment. This could partly be due to the inability to identify signaling network or loop which is positively or negatively regulated around the single target. To meet this important challenge, a number recent studies are analyzing cancer cell lines and tissue samples to measure alterations at the gene, RNA, and protein level to identify markers and targets for the therapy. While these studies will produce a large amount of data whose analysis is critical in order to understand cancer at the molecular level. For example, a similar microarray analysis of MDA-MB-435 cells that are engineered to differ in integrin α6β4 expression by Chen et al. leads to the identification of couple of invasion and metastasis related genes such as ENPP2 [31] and S100A4 [32]. What makes our study unique from these works is that we are in a position to identify genes and proteins that are functionally connected to drive malignant properties rather than focusing a single gene because targeting these sub networks will inhibit cancer cell functions important for progression. For example, we found the potentially important α6β4 target genes associated with cancer pathway as summarized in Table 4. Those genes are associated with TGF-β [33], TNF-α [34], and EGFR1 pathways [35], whose roles in cancer progression have been well established.
In summary, the integration of interaction network with expression of α6β4 integrin in MDA-MB-435 cancer cells reveals the importance of NRCAM, which we would not have discovered with the expression information alone. Further, the interaction network in Figure 4 helps us to understand how the tumor suppression genes CHEK1, XRCC6, SMARCB, ATM, CHEK1 were down regulated by integrin α6β4. Finally, we envision the discovery of interaction network triggered from tumor antigen such as integrin α6β4 will lead to the development of novel anticancer therapeutics by targeting signaling molecules associated with interaction network.
Table 2.
The high ranking 14 downregulated genes. For each gene in the list, the connectivity in the protein interaction network G is given. The 5 hub nodes among the ranked down regulated genes are underlined.
| Genes | Fold change (inverse) | Connectivity in G |
|---|---|---|
| HBE1 | 9.10 | 1 |
| H1F0 | 7.70 | 7 |
| AZGP1 | 7.64 | 3 |
| SNCA | 5.24 | 44 |
| GLUL | 5.13 | 31 |
| TPM1 | 4.62 | 17 |
| IGFBP7 | 4.54 | 10 |
| MYLK | 4.25 | 28 |
| KCNS3 | 4.23 | 1 |
| NGFRAP1 | 4.12 | 15 |
| DGKI | 3.97 | 1 |
| IL1RAP | 3.92 | 14 |
| THBS1 | 3.70 | 36 |
| MAP1B | 3.65 | 1 |
References
- 1.Troyanskaya OG. Putting microarrays in a context: integrated analysis of diverse biological data. Briefings in Bioinformatics. 2005;6(1):34–43. doi: 10.1093/bib/6.1.34. [DOI] [PubMed] [Google Scholar]
- 2.Camargo A, Azuaje F. Identification of dilated cardiomyopathy signature genes through gene expression and network data integration. Genomics. 2008;92(6):404–413. doi: 10.1016/j.ygeno.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Date SV, Marcotte EM. Discovery of uncharacterized cellular systems by genome-wide analysis of functional linkages. Nature Biotechnology. 2003;21(9):1055–1062. doi: 10.1038/nbt861. [DOI] [PubMed] [Google Scholar]
- 5.Lanckriet GR, Deng M, Cristianini N, Jordan MI, Noble WS. Kernel-based data fusion and its application to protein function prediction in yeast. In: Proceedings of the Pacific Symposium on Biocomputing (PSB '04); January 2004; Big Island of Hawaii, Hawaii, USA. pp. 300–311. [DOI] [PubMed] [Google Scholar]
- 6.Nariai N, Kolaczyk ED, Kasif S. Probabilistic protein function prediction from heterogeneous genome-wide data. PLoS ONE. 2007;2(3, article e337) doi: 10.1371/journal.pone.0000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua HN, Sung W-K, Wong L. An efficient strategy for extensive integration of diverse biological data for protein function prediction. Bioinformatics. 2007;23(24):3364–3373. doi: 10.1093/bioinformatics/btm520. [DOI] [PubMed] [Google Scholar]
- 8.Zhu M, Gao L, Guo Z, et al. Globally predicting protein functions based on co-expressed protein-protein interaction networks and ontology taxonomy similarities. Gene. 2007;391(1-2):113–119. doi: 10.1016/j.gene.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Liu M, Chen X-W. Assessing reliability of protein-protein interactions by integrative analysis of data in model organisms. BMC Bioinformatics. 2009;10, article S4 doi: 10.1186/1471-2105-10-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soong T-T, Wrzeszczynski KO, Rost B. Physical protein-protein interactions predicted from microarrays. Bioinformatics. 2008;24(22):2608–2614. doi: 10.1093/bioinformatics/btn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xenarios I, Salwinski L, Duan XJ, Higney P, Kim S-M, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Research. 2002;30(1):303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Research. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercurio AM, Bachelder RE, Rabinovitz I, O'Connor KL, Tani T, Shaw LM. The metastatic odyssey: the integrin connection. Surgical Oncology Clinics of North America. 2001;10(2):313–328. [PubMed] [Google Scholar]
- 14.Lipscomb EA, Mercurio AM. Mobilization and activation of a signaling competent α6β4 integrin underlies its contribution to carcinoma progression. Cancer and Metastasis Reviews. 2005;24(3):413–423. doi: 10.1007/s10555-005-5133-4. [DOI] [PubMed] [Google Scholar]
- 15.Pagel P, Kovac S, Oesterheld M, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21(6):832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 16.Bader GD, Betel D, Hogue CWV. BIND: the biomolecular interaction network database. Nucleic Acids Research. 2003;31(1):248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willis RC, Hogue CW. Searching, viewing, and visualizing data in the Biomolecular Interaction Network Database (BIND) Current Protocols in Bioinformatics. 2006;(chapter 8: unit 8.9) doi: 10.1002/0471250953.bi0809s12. [DOI] [PubMed] [Google Scholar]
- 18.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biology. 2007;8(5, article R95) doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NAViGaTOR 2.0. 2008, http://ophid.utoronto.ca/ href="http://ophid.utoronto.ca/navigator/index.html"> navigator/index.html.
- 20.Jeong H, Mason SP, Barabasi A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 21.Tew KL, Li XL, Tan SH. Functional centrality: detecting lethality of proteins in protein interaction networks. Genome Informatics. 2007;19:166–177. [PubMed] [Google Scholar]
- 22.Vallabhajosyula RR, Chakravarti D, Lutfeali S, Ray A, Raval A. Identifying hubs in protein interaction networks. PLoS ONE. 2009;4(4, article e5344) doi: 10.1371/journal.pone.0005344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batada NN, Reguly T, Breitkreutz A, et al. Stratus not altocumulus: a new view of the yeast protein interaction network. PLoS Biology. 2006;4(10):p. e317. doi: 10.1371/journal.pbio.0040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. 2nd edition. MIT Press; 2001. [Google Scholar]
- 25.NetPath. http://www.netpath.org.
- 26.Han K, Cui G, Chen Y. Identifying functional groups by finding cliques and near-cliques in protein interaction networks. In: Proceedings of the Frontiers in the Convergence of Bioscience and Information Technologies (FBIT '07); 2007; pp. 159–164. [Google Scholar]
- 27.Wang XQ, Stanbridge EJ, Lao X, Cai Q, Fan ST, Redpath JL. p53-dependent Chk1 phosphorylation is required for maintenance of prolonged G2 arrest. Radiation Research. 2007;168(6):706–715. doi: 10.1667/RR1075.1. [DOI] [PubMed] [Google Scholar]
- 28.Willems P, De Ruyck K, Van den Broecke R, et al. A polymorphism in the promoter region of Ku70/XRCC6, associated with breast cancer risk and oestrogen exposure. Journal of Cancer Research and Clinical Oncology. 2009;135(9):1159–1168. doi: 10.1007/s00432-009-0556-x. [DOI] [PubMed] [Google Scholar]
- 29.Roberts CWM, Biegel JA. The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer Biology and Therapy. 2009;8(5):412–416. doi: 10.4161/cbt.8.5.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes and Development. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, O'Connor KL. Integrin α6β4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene. 2005;24(32):5125–5130. doi: 10.1038/sj.onc.1208729. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Sinha M, Luxon BA, Bresnick AR, O'Connor KL. Integrin α6β4 controls the expression of genes associated with cell motility, invasion, and metastasis, including S100A4/metastasin. The Journal of Biological Chemistry. 2009;284(3):1484–1494. doi: 10.1074/jbc.M803997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prud'homme GJ. Pathobiology of transforming growth factor β in cancer, fibrosis and immunologic disease, and therapeutic considerations. Laboratory Investigation. 2007;87(11):1077–1091. doi: 10.1038/labinvest.3700669. [DOI] [PubMed] [Google Scholar]
- 34.Zidi I, Mestiri S, Bartegi A, Amor NB. TNF-α and its inhibitors in cancer. Medical Oncology. 2009:1–14. doi: 10.1007/s12032-009-9190-3. [DOI] [PubMed] [Google Scholar]
- 35.Kopper L. Lapatinib: a sword with two edges. Pathology and Oncology Research. 2008;14(1):1–8. doi: 10.1007/s12253-008-9018-z. [DOI] [PubMed] [Google Scholar]