Abstract
Cancer cell membrane proteins are released into the plasma/serum by exterior protein cleavage, membrane sloughing, cellular secretion or cell lysis, and represent promising candidates for interrogation. Because many known disease biomarkers are both glycoproteins and membrane bound, we chose the hydrazide method to specifically target, enrich, and identify glycosylated proteins from breast cancer cell membrane fractions using the LTQ Orbitrap mass spectrometer. Our initial goal was to select membrane proteins from breast cancer cell lines and then to use the hydrazide method to identify the N-linked proteome as a prelude to evaluation of plasma/serum proteins from cancer patients. A combination of steps facilitated identification of the glycopeptides and also defined the glycosylation sites. In MCF-7, MDA-MB-453 and MDA-MB-468 cell membrane fractions, use of the hydrazide method facilitated an initial enrichment and site mapping of 27 N-linked glycosylation sites in 25 different proteins. However, only three N-linked glycosylated proteins, galectin-3 binding protein, lysosome associated membrane glycoprotein 1, and oxygen regulated protein, were identified in all three breast cancer cell lines. In addition, MCF-7 cells shared an additional 3 proteins with MDA-MB-453. Interestingly, the hydrazide method isolated a number of other N-linked glycoproteins also known to be involved in breast cancer including, epidermal growth factor receptor (EGFR), CD44, and the breast cancer 1, and early onset isoform 1 (BRCA1) biomarker. Analyzing the N-glycoproteins from membranes of breast cancer cell lines highlights the usefulness of the procedure for generating a practical set of potential biomarkers.
Keywords: breast cancer, biomarkers, n-linked glycosylation, glycoprotein, proteomics, mass spectrometry, post-translational modification, hydrazide, HER2, EGFR
Introduction
Breast tumors are the most common malignancy in women and the second leading cause of cancer death in American females. Until cancer prevention becomes a reality, early detection and better treatment are the two most effective means to reduce cancer mortalities1. Although mammography screening is successful for detecting many breast tumors, it often fails to identify malignancies in young women, as well as in women with dense breasts, implants or lobular tumors2. In addition, mammography is unaffordable to many women not only in the USA, but also in the overwhelming majority of the developing world 3. Protein cancer biomarkers provide a promising approach that may advance the field in disease detection and treatment1. To achieve this clinical goal, discovery of more specific, sensitive and reliable biomarkers is essential 4. A patients blood plasma/serum is considered to be the most desirable clinical specimen for biomarker research because it is attainable non-invasively, extraction is simple and affordable, and it is likely to contain tumor markers, albeit in significantly low abundance. It is well documented that not only do tumors leak or secrete proteins into the circulation, but also the surrounding stroma releases proteases and other mediators of cancer growth 5 6. The current panel of known tumor markers include carcinoma antigen 27.29 [CA 27.29] for breast cancer 7, prostrate-specific antigen [PSA] for prostrate cancer 8, cancer antigen 125 [CA125] for ovarian cancer 9, carcinoembryonic antigen [CEA] for a wide spectrum of carcinomas 7, 10, and alpha-fetoprotein [AFP] for liver 11 and testicular cancers 10. These proteins are all examples of useful circulation-borne cancer biomarkers that probably arise from the primary tumor or the surrounding stroma 7, 8. However, the discovery of new cancer biomarkers in serum is hindered by two major technical hurdles: the low abundance of disease related proteins in sera and the interference of the 20 or so highly abundant normal serum proteins that make up 99% of the circulating proteome12–14. Serum protein concentrations cover more than 10 orders of magnitude. This enormous range exceeds the dynamic range of any current analytical method or instrument and further complicates the discovery of new cancer biomarkers. Therefore, direct protein profiling of tumor cells may be the most effective way to identify novel cancer signatures, before screening human sera.
However, extracts secreted by or extracted from breast cancer cell lines, proximal fluid, and breast tumor biopsies have highly complex proteomes that must be extensively and reproducibly fractionated in order to identify candidate cancer biomarkers with clinical relevance. A number of fractionation strategies are currently used to reduce sample complexity. These include chromatography, often with the combination of ion exchange and reverse phase modalities such as the multidimensional protein identification technology (MUDPIT)15, fractionations based on molecular weight cut-off filters14, fractionations based on hydrophobicity such as that developed using reverse phase separations16, the capture of cysteine-containing peptides with biotinylated thiol reagents17, the capture of phosphorylated peptides using immobilized metal affinity chromatography18 or dendrimer conjugation chemistry 19, and glycopeptide capture 20. Since most clinically useful tumor markers such as PSA21, CA12522, 23, CEA24, 25, or AFP26, are both N-glycosylated and O-glycosylated we take advantage of the hydrazide method’s utility to isolate, enrich, and identify the N-glycoproteome. In addition, important proteins such as fibroblast growth factor (FGF)27, epidermal growth factor receptor (EGFR)28, insulin receptor (IR)29, and insulin-like growth factor 1 (IGF-1)30 that are all N-glycosylated may also be enriched, identified, and site-mapped by the hydrazide method. In this study we investigated cell membrane glycoproteins of several breast cancer cell lines with the goal of detecting new potential cancer biomarkers.
For the past 30 years the hydrazide method has been used to isolate and enrich glycoproteins 31. More recently Aebersold’s group described its use for isolating and site mapping N-linked glycoproteins from serum32. However many of the most abundant proteins are also N-linked glycosylated molecules precluding the discovery of any low abundance glycoproteins that are released from tissue into the serum. The use of the hydrazide method would be more effective in the search of potential biomarkers if applied to the source of disease. Loo’s group recently reported the hydrazide method was effective in characterizing N-linked glycoproteins in salivary fluids 33. The hydrazide method is initiated by first oxidizing the sugar residues in the glycosylated peptides with periodate before reacting with a hydrazide resin34. Covalent attachment of glycopeptides to the resin allows the samples to be thoroughly washed free of non-covalently-attached peptides and other contaminates before enzymatic release of the N-linked peptides with PNGase F leaving a signature of deaminated asparagine residues converted to aspartic acid. These samples may then be processed by data-dependent LC-MS/MS so that the glycopeptide is identified and the glycosylation site mapped.
Since excised human tumors are both limited in quantity, heterogeneous for cell type and variable for cancer sub-type, we chose to evaluate the utility of the proposed method with human breast cancer cell lines that are homogenous for cell-type, available in reasonable quantities and represent different subtypes of breast cancer. The names and characteristics of the cell lines are: MCF-7, hormone receptor positive and HER2 negative; MDA-MB-453, hormone receptor negative and HER2 positive; and MDA-MB-468, hormone receptor (estrogen receptor (ER) and progesterone receptor (PR)) and HER2 negative (triple negative). We selected N-linked glycosylated proteins in our concentrated membrane extracts of breast cancer cell lines using the hydrazide system since many of the membrane proteins released into the plasma/serum are frequently glycosylated. The identified N-glycoproteins in cancer cells then can guide further detection of candidate biomarkers in serum/plasma from breast cancer patients and lead to the development of clinically useful diagnostic assays.
Materials and Methods
Cell Culture
MCF-7, MDA-MB-453 and MDA-MB-468 human breast cancer cell lines were maintained in DMEM culture medium supplemented with L-glutamine and sodium pyruvate, 10% FBS, and 1% penicillin and streptomycin. Cells were grown to 70% confluency in 10 cm petri dishes. Media was removed and cells were washed with PBS, harvested by centrifugation (1,000 × g, 2 min 4 °C) and the pellets stored at −80 °C until needed.
Membrane preparation
The frozen cell pellets were homogenized in ice cold 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM PMSF, and protease cocktail (Roche Cat. No. 04693132001) using an ultrasonic cell disrupter (Fisher Scientific, sonic dismembrater model 100, at setting 4 for 2 × 10 seconds at 30 seconds intervals) on ice. Samples were centrifuged at 1,500 × g for 5 min at 4 °C to remove large debris, and the supernatant re-centrifuged at (100,000 × g, 1 hr, 4 °C). The pellet was solubilized in 40 mM Tris-HCl pH 8.3, 6 M guanidine HCl, 0.2% Rapigest™ (Waters), 5 mM DTT, centrifuged (15,000 × g, 2 min, RT), and supernatant diluted to <1 M guanidine HCl with 40 mM Tris-HCl pH 8.3. The sample was sequentially treated with DTT, iodoacetamide, and trypsin (overnight, 37 °C) according to manufacturers protocol (Promega). The pH was adjusted to pH 3 with TCA, the samples were centrifuged (15,000 × g, 2 min, RT) and the supernatant was passed through an activated, washed C18 spin column (The Nest Group, Inc.) according to manufacturers protocol. The eluate was vacuum dried and a fraction (2%) was reserved for LC-MS/MS without further treatment.
Hydrazide method
The remainder of dried eluates in microcentrifuged tubes were dissolved in coupling buffer (100 mM sodium acetate, 150 mM NaCl, pH 5.5), centrifuged (15,000 × g, 2 min, RT), and the supernatant was treated with sodium periodate (15 mM final concentration, 30 min, RT) by end over end rotation in the dark. The oxidation reaction was quenched with 20 mM sodium sulfite (20 mM final concentration, 15 min, RT) and the sample was added to the hydrazide resin (200 µl wet resin, Biorad, previously washed with coupling buffer following manufacturers instructions). The coupling reaction was performed overnight at 37 °C with end over end rotation. The resin was washed (1 mL) sequentially twice each with H2O, 1.5 M NaCl, methanol, acetonitrile, and finally with 50mM NH4CO3 pH 8.0. The slurry was then treated with PNGase F (New England BioLabs Inc.), centrifuged (15,000 × g, 2 min, RT), the supernatant removed and the pellet washed with water/acetonitrile (50/50, 1 mL) and re-centrifuged. The pooled samples were vacuum dried.
LC-MS/MS analysis
Dried samples were re-dissolved in Buffer A (H2O/acetonitrile/formic acid, 98.9/1/0.1, typically 100 µl), separated by nanospray LC (Eskigent technologies, Inc. Dublin, CA), and analyzed by online tandem mass spectrometry (LTQ Orbitrap, Thermo Fisher). Aliquots were injected (10 µl) onto a reverse phase column (New Objective C18, 15 cm, 75 µM diameter, 5 µm particle size equilibrated in Buffer A) and eluted (300 nL/min) with an increasing concentration of Buffer B (acetonitrile/water/formic acid, 98.9/1/0.1; min 0/5, 10/10, 112/40, 130/60, 135/90, 140/90). The effluent from the column was passed directly into an integrated nanospray emitter tip connected to the LTQ Orbitrap mass spectrometer. Eluted peptides were analyzed by MS and data-dependent MS/MS acquisition (collision-induced dissociation CID), previously optimized for samples, selecting the 7 most abundant precursor ions for MS/MS with a dynamic exclusion duration of 15.0 seconds.
Data base analysis
The mass spectra were searched against a human trypsin indexed database (two trypsin missed cleavages), with variable modifications of carboxyamidomethylation, methionine oxidation, and deamination of asparagine residues using the Bioworks software (Thermo Fisher) based on the SEQUEST algorithm. Positive protein identification was accepted for peptides with a Xcorr for doubly charged ions ≥ 2.0 and for triply charged ions ≥ 2.5.
Quantitative data analysis was performed using the Scaffold (Proteome Software, Inc.) software program. The Bioworks search results were uploaded into the scaffold software program and a filter with a 99% minimum protein ID probability (calculated probability of correct protein identification), with a minimum number of 2 unique peptides for one protein, and with a minimum peptide ID probability of 99.9% was set. Note that this filter setting affects not only which proteins are shown but also the report values shown for the Number of Peptides, Number of Identified Spectra, and Number of Unique Spectra. Scaffold verifies peptide identification derived from MS/MS sequencing results using X! Tandem35 ProteinProphet computer algorithms36. Scaffold validates these peptide identifications using PeptideProphet37 and derives corresponding protein probabilities using ProteinProphet36. Scaffold normalizes MS/MS data between samples with similar total protein amounts allowing a comparison of protein abundance between samples. Each of the breast cancer cell line samples is a combination of three replicate experiments. Normalization entails averaging the spectral counts for all the samples and then multiplying the spectral counts in each sample by the average divided by the individual sample’s sum. Since we are analyzing relatively equivalent amounts of proteins in each MS/MS experiment, combining three replicate experiments per breast cancer cell line sample and only looking at the most abundantly expressed proteins in each sample, the abundance differences between each breast cancer cell line is significant.
Results
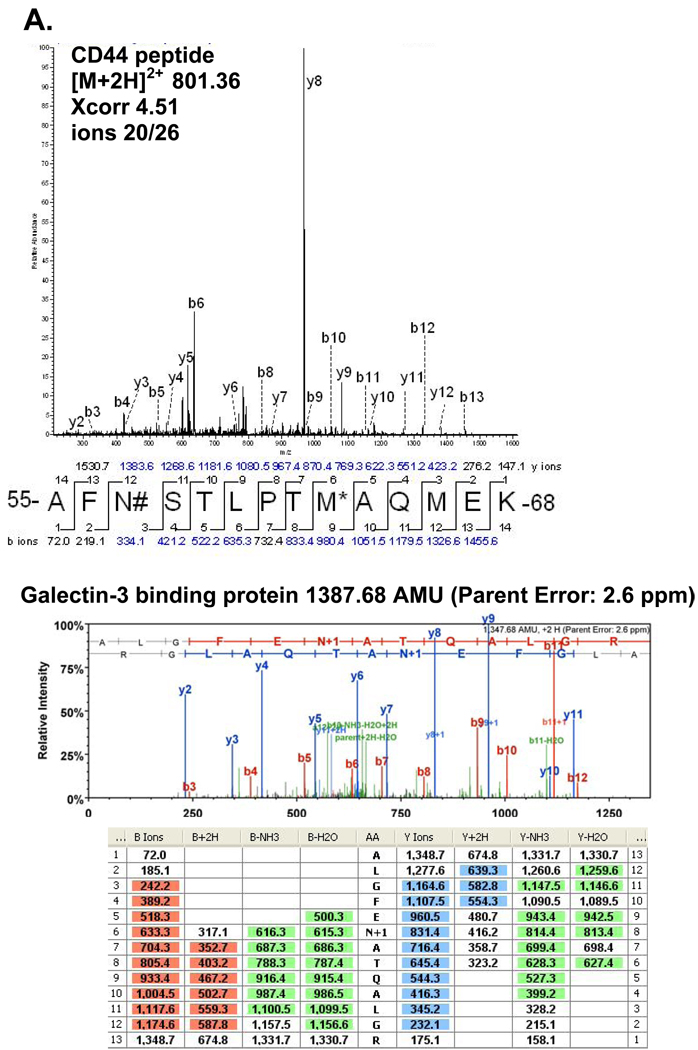
The aim of this study was to isolate, identify and site-map N-linked glycosylated membrane proteins as a more direct way to define new tumor related proteins, since most of the cancer biomarkers used in clinical practice today are glycosylated cell membrane proteins. Isolation of N-linked glycoproteins provides an opportunity to study a unique subset of proteins in the cell membrane of breast cancers. Hydrazide beads were used to bind periodate oxidized sugar residues of glycosylated peptides after trypsin cleavage of cancer cell membrane proteins. A total of 27 N-glycosylated sites in 25 glycoproteins (haptoglobin and intercellular adhesion molecule 1 had two N-glycosylated sites) with the consensus site NXS/T (where X may be any amino acid except proline) emerged from the analysis of the three cell lines (Tables 1), with MCF-7 yielding 15 total proteins, MDA-MB-453 yielding 13 total proteins, and MDA-MB-468 yielding 6 total proteins for a combined total of 25 different N-glycosylated proteins. In addition, 12 other N-glycosylated proteins were site mapped without the conventional NXS/T motif (Supplemental Table 1). In Table 1 only three N-glycosylated NXS/T consensus site proteins were shared between all three different cell types. These are galectin-3 binding protein, oxygen regulated protein precusor, and lysosome-associated membrane glycoprotein 1 (LAMP1). The MCF-7 and MDA-MB-453 cell lines were found to share three proteins, cathepsin D, pigment epithelium-derived factor, and clusterin isofrom 2. Interestingly, of the 25 site mapped NXS/T motif N-linked glycoproteins, this analysis revealed a breast cancer stem cell marker CD44 (Figure 1A). An example of the Scaffold display of MS/MS data indicating the N-linked glycosylation site mapped to the galectin-3 binding protein peptide is shown in Figure 1B. Surprisingly, we also found a well-known breast cancer biomarker: breast cancer 1, early onset (BRCA1) a suppressor of uncontrolled tumor cell proliferation (Supplemental Figure 1). While interesting, some caution should be attached to this identification because the peptide identified had two additional deaminated asparagines outside the NXS/T motif, possibly due to novel N-glycosylations, or deamination during the oxidative process of the hydrazide method, or SNPs. The MSMS spectrum also revealed a surprisingly intense high m/z b- ladder, unusual for tryptic peptides. Confirmation of the identification would require high-resolution accurate m/z assignments on the fragment ions that is not possible at this time. In addition, spontaneous deamination of asparagine and glutamine is commonly found in mass spectrometry samples and the parent ion for the BRCA1 N-linked glycopeptide had a mass error less than 2 ppm. A number of other proteins, varyingly implicated in breast cancer, were also identified, including galectin-3 binding protein, cathepsin D, and keratin 18 (supplemental table 1 and supplemental figure 2). Although the MS/MS spectra of the non-NXS/T motif N-glycoproteins were purified by covalently attaching the N-glycan to the hydrazide resin and subjected to stringent washing conditions we could not rule out contamination. In addition, most of these non-NXS/T motif N-glycoproteins had high xcorr factors, ion matching ratios, and were subjected to manual inspection (supplemental table 1 and supplemental figure 2).
Table 1.
A compilation of all the site mapped N-linked glycosylation sites within the consensus sequence NXS/T found on proteins in membrane fractions of MCF-7, MDA-MB-453, and MDA-MB-468 breast cancer cell lines (#=deaminated asparagine, @=carboxyamidomethylation, *=oxidation, z=charge state). Only peptides with good Xcorrelation numbers (good XC: +1=1.5, +2=2.0, +3=2.5 and rigorous XC +1=1.9, +2=2.5, +3=3.5) were chosen and manually inspected.
| N-linked glycosylated proteins | Breast cancer cell lines | MH+ | z | Peptide | XC | Ions |
|---|---|---|---|---|---|---|
| galectin 3 binding protein | MCF-7, MDA-MB-453, MDA-MB-468 | 1348.68556 | 2 | R.ALGFEN#ATQALGR.A | 4.17 | 20/24 |
| oxygen regulated protein | MCF-7, MDA-MB-453, MDA-MB-468 | 1819.92586 | 2 | R.LSALDNLLN#HSSM*FLK.G | 3.50 | 16/30 |
| lysosome associated membrane protein 1 | MCF-7, MDA-MB-453, MDA-MB-468 | 1160.6059 | 2 | R.GHTLTLN#FTR.N | 2.66 | 15/18 |
| cathepsin D preproprotein | MCF-7, MDA-MB-453 | 1110.57896 | 2 | K.GSLSYLN#VTR.K | 3.75 | 15/18 |
| clusterin isoform 2 | MCF-7, MDA-MB-453 | 1684.81769 | 2 | R.LAN#LTQGEDQYYLR.V | 4.94 | 22/26 |
| pigment epithelium-derived factor | MCF-7, MDA-MB-453 | 2944.51497 | 4 | K.VTQN#LTLIEESLTSEFIHDIDRELK.T | 5.45 | 44/144 |
| breast cancer 1, early onset isoform 1 | MCF-7 | 1392.61251 | 2 | K.EEN#QGKN#ESN#IK.P | 2.16 | 13/22 |
| Integrin alpha-2 | MCF-7 | 1646.79483 | 2 | R.YFFN#VSDEAALLEK.A | 4.23 | 22/26 |
| leucine-rich alpha-2-glycoprotein 1 | MCF-7 | 1553.94138 | 2 | R.KLPPGLLAN#FTLLR.T | 2.64 | 13/26 |
| solute carrier family 3 | MCF-7 | 2323.14523 | 2 | -.DIENLKDASSFLAEWQN#ITK.- | 5.38 | 22/38 |
| Transforming growth factor beta-1 | MCF-7 | 2761.44069 | 3 | R.LASPPSQGEVPPGPLPEAVLALYN#STR.D | 4.17 | 29/104 |
| thrombospondin 1 | MCF-7 | 1367.69137 | 2 | K.VVN#STTGPGEHLR.N | 2.21 | 13/24 |
| Na+/K+ -ATPase beta 1 subunit | MCF-7 | 2383.22138 | 2 | K.YLQPLLAVQFTN#LTM*DTEIR.I | 3.25 | 16/38 |
| purinergic receptor P2X4 | MCF-7 | 1106.60920 | 2 | K.AAEN#FTLLVK.N | 3.19 | 15/18 |
| chromosome 20 open reading frame 3 | MDA-MB-453 | 1424.70562 | 2 | R.AGPN#GTLFVADAYK.G | 3.86 | 18/26 |
| lysosomal-associated membrane protein 2 | MDA-MB-453 | 1883.96905 | 2 | K.IAVQFGPGFSWIAN#FTK.A | 4.81 | 24/32 |
| prosaposin | MDA-MB-453 | 2783.35430 | 3 | K.DVVTAAGDM*LKDN#ATEEEILVYLEK.T | 4.01 | 27/96 |
| zinc finger protein 92 (HTF12) isoform 2 | MDA-MB-453 | 1668.77584 | 2 | -.C@GNAFN#QSSNLTARK.- | 2.61 | 10/28 |
| haptoglobin | MDA-MB-453 | 1795.99526 | 3 | K.VVLHPN#YSQVDIGLIK.L | 3.58 | 24/60 |
| haptoglobin | MDA-MB-453 | 2680.37631 | 3 | K.MVSHHN#LTTGAYLINEQWLLTTAK.N | 4.40 | 36/92 |
| clusterin | MDA-MB-453 | 1684.81769 | 2 | R.LAN#LTQGEDQYYLR.V | 4.74 | 22/26 |
| tissue inhibitor of metalloproteinases 1 | MDA-MB-453 | 1753.87554 | 2 | K.FVGTPEVN#QTTLYQR.Y | 3.73 | 20/28 |
| zinc-alpha-2-glycoprotein | MDA-MB-453 | 2067.92540 | 3 | K.DIVEYYN#DSN#GSHVLQGR.F | 2.76 | 20/68 |
| zinc-alpha-2-glycoprotein | MDA-MB-453 | 3417.59034 | 4 | K.AREDIFM*ETLKDIVEYYN#DSN#GSHVLQGR.F | 4.71 | 41/168 |
| CD44 antigen (PGP-1) | MDA-MB-468 | 1601.71857 | 2 | K.AFN#STLPTM*AQM*EK.A | 4.51 | 20/26 |
| human epidermal growth factor receptor | MDA-MB-468 | 1176.61066 | 2 | K.DSLSIN#ATNIK.H | 3.42 | 18/20 |
| intercellular adhesion molecule 1 ICAM-1 | MDA-MB-468 | 1614.76459 | 2 | R.LNPTVTYGN#DSFSAK.A | 3.99 | 20/28 |
| intercellular adhesion molecule 1 ICAM-1 | MDA-MB-468 | 999.61971 | 2 | R.AN#LTVVLLR.G | 2.41 | 12/16 |
| oxygen regulated protein | MDA-MB-468 | 1194.63648 | 2 | R.VFGSQN#LTTVK.L | 3.85 | 16/20 |
Figure 1.
A) MS/MS data of CD44 peptide demonstrating the N-linked glycosylation site (#=deaminated asparagines; *=oxidized methionine). B) Scaffold MS/MS data of galectin-3 binding protein peptide demonstrating the N-linked glycosylation site (1387.68 AMU, parent error measurement: 2.6 ppm).
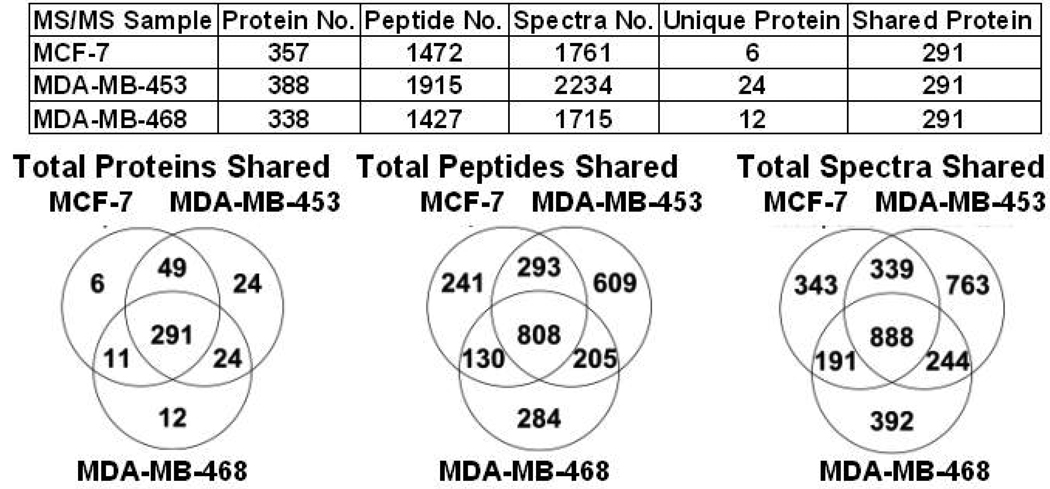
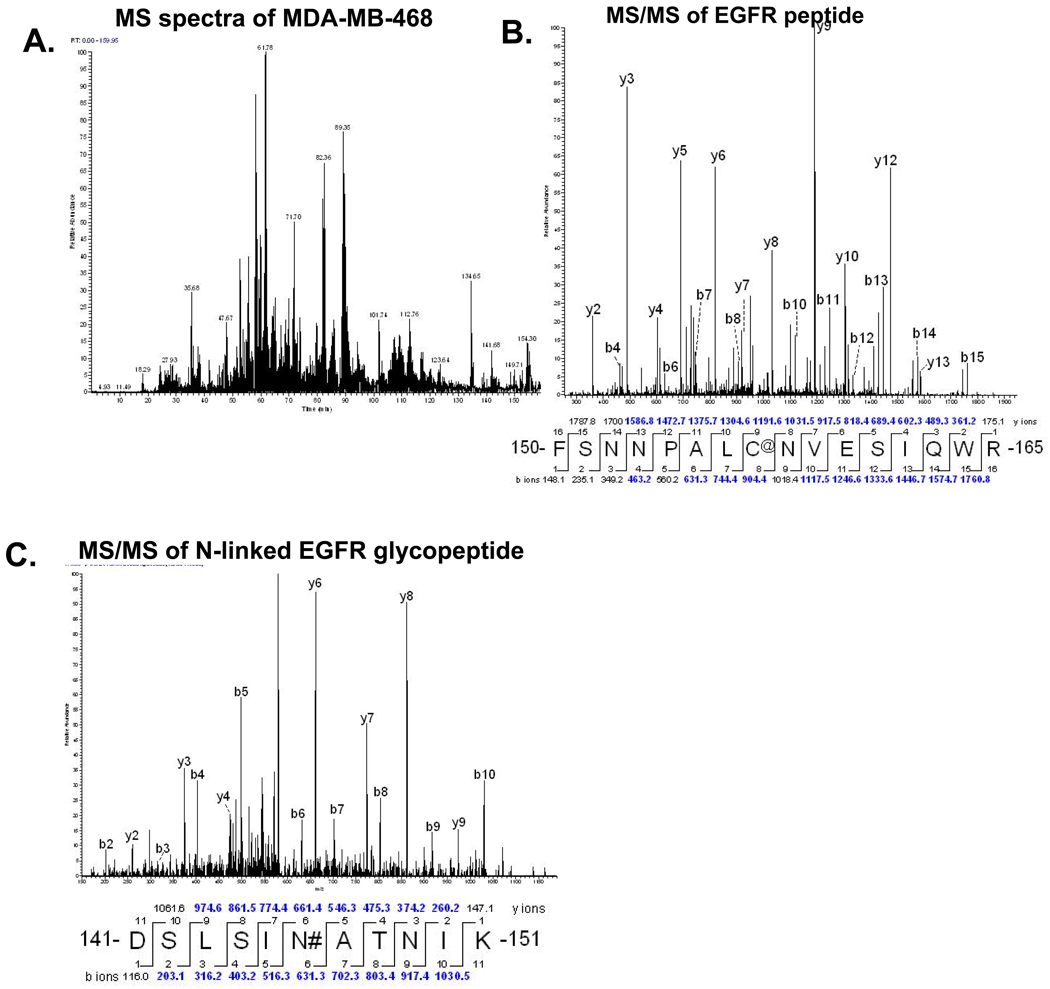
Since N-linked proteins were isolated from crude membrane fractions we compared the overlap of N-linked glycoproteins to the membrane sources to determine selectivity of the hydrazide enrichment method. First we analyzed the starting material before hydrazide treatment using total membrane proteins from three separate isolation experiments for each of the three breast cancer cell lines. We found that MCF-7, MDA-MB453, and MDA-MB-468 had over 291 proteins in common (Figure 2). In addition, by comparing the total peptides in common and total spectra shared we observed that our starting material had relatively equivalent amounts of protein present. An example of the base peak MS chromatogram of MDA-MB-468 membrane fractions and MS/MS of the signature EGFR found in MDA-MB-468 breast cancer cell lines is shown in Figure 3A and 3B. The hydrazide method clearly enriched for an EGFR N-linked glycopeptide that had not been detectable in the crude membrane fractions (Figure 3C). A representative 2D plot (ion map) for each crude membrane fraction from each breast cancer cell is shown in Supplemental Figure 3.
Figure 2.
A chart and Venn diagrams outlining the total membrane proteins, total membrane peptides, total membrane spectra, unique proteins and shared proteins between MCF-7, MBA-MD-453, and MDA-MB-468 breast cancer cell lines.
Figure 3.
Figure 6. A) Total ion chromatogram of the whole membrane fraction of MDA-MB-468 cells. B) MS/MS data of EGFR peptide from whole membrane fraction (@=carboxyamidomethylation). C) MS/MS data of EGFR N-linked glycopeptide enriched by the hydrazide method from MDA-MB-468 whole membrane fraction (#=deaminated asparagines).
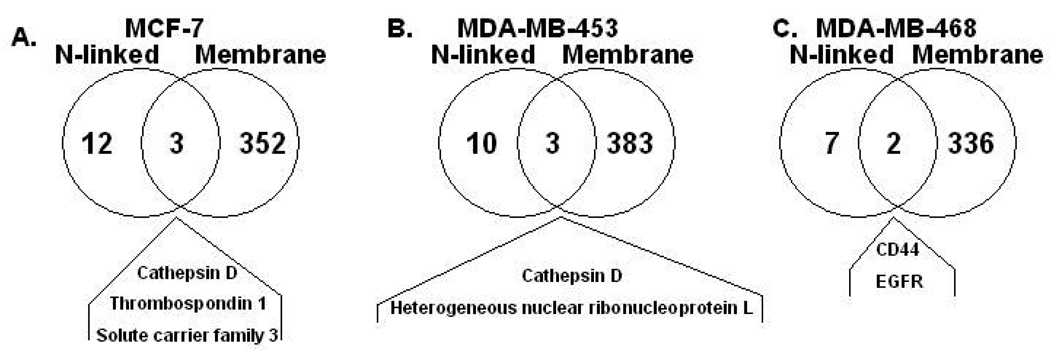
When the 15 site mapped N-linked glycosylated proteins in MCF-7 membrane fractions were compared to the total protein of MCF-7 membrane fractions by a Venn diagram (Figure 4A) only three proteins including cathepsin D, solute carrier family 3 member 2, and thrombospondin 1, were present in both preparations. Only one, solute carrier family 3 member 2 isoform (101 spectral counts), of the three proteins had high abundance spectral counts in the crude membrane fractions. While cathepsin D (13 spectral counts) and thrombospodin 1 (0 spectral counts) had either low or no spectral counts, respectively. Two of the non-NXS/T motif N-glcoproteins found also had high abundant spectral counts, keratin 18 (150 spectral counts) and H2B histone family member C (163 spectral counts). This data indicates that the fractionation of N-linked glycoproteins from crude membrane fractions using the hydrazide method both significantly enriched for low abundant glycoproteins and identified glycoproteins from high abundant membrane proteins. The same comparison of 13 site mapped N-linked glycosylated proteins in the MDA-MB-453 relative to total membrane proteins is shown in Figure 4B. Only two proteins, cathepsin D (7 spectral counts) and heterogeneous nuclear ribonucleoprotein L (22 spectral counts), were found in both preparations. The two non-NXS/T glycoproteins, H2B histone family member C (192 spectral counts) and keratin 18 (29 spectral counts) were also found in the MDA-MB-453 cell line. The 6 N-linked glycoproteins site mapped in MDA-MB-468 membranes had only two shared proteins, CD44 (33 spectral counts) and EGFR (22 spectral counts), also seen in the total cell membrane fraction. Since the majority of the site mapped N-linked glycoproteins were not detected in the starting material of the crude membrane fractions, it appears that the hydrazide method did in fact significantly enriched the glycoproteome in all three breast cancer cell lines.
Figure 4.
A) A Venn diagram comparison of N-linked glycosylated site mapped proteins to total membrane proteins in MCF-7 cells. B) A Venn diagram comparison of N-linked glycosylated site mapped proteins to total membrane proteins in MDA-MB-453 cells. C) A Venn diagram comparison of N-linked glycosylated site mapped proteins to total membrane proteins in MDA-MB-468 cells.
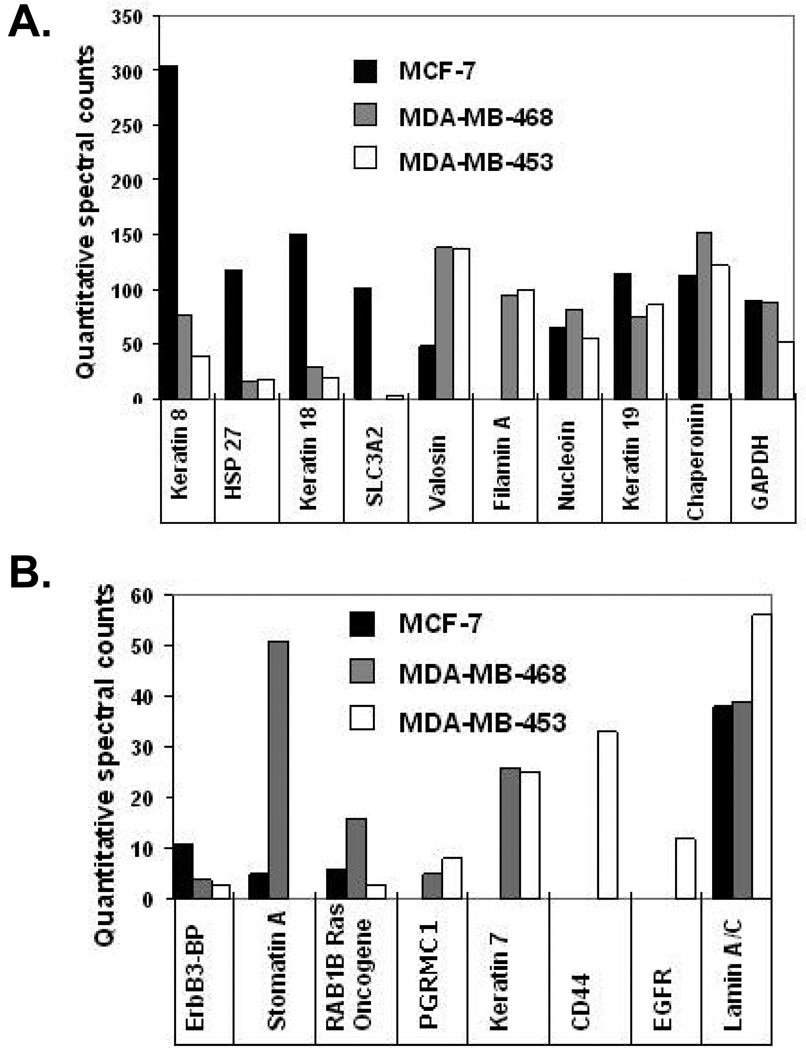
After we identified a set of glycoprotein potential biomarkers from membrane fractions we also analyzed the quantitative expression of proteins between the three human breast cancer cell line membranes. Using the Scaffold program we quantitatively compared the spectral counts of three combined experiments for each of the three breast cancer cell lines (Supplemental Table 2), with over 400 proteins quantified. To simplify comparisons among the large number of proteins from the 3 breast cancer cell lines, a subset of the data was analyzed in charts highlighting quantitatively distinct proteins between the three cell lines (Figure 5A and 5B). A number of proteins that are unique or elevated in each cell line are readily appreciated. Erb-binding protein, solute carrier protein 3, keratin 8, keratin 18, and heat shock protein 27 (HSP27) are increased in hormone receptor positive and HER2 negative MCF-7 cells (black bar). MDA-MB-453 (grey bar), a HER2 positive cell line, demonstrates an elevation of enolase 1, RAB1B Ras oncogene, stomatin, keratin 18, filamin A, and valosin. Triple negative breast cancer cell line MDA-MB-468 (white bar) has a distinctive set of elevated proteins including CD44, EGFR, filamin A, and valosin. A number of elevated proteins were in high quantity in all three breast cancer cell lines such as stress-induced-phosphoprotein-1 (STIP-1), keratin 19, lamin A/C, chaperonin, and glyceraldehydes-3-phospate dehydrogenase (GAPDH) to name a few. The expression of distinct protein profiles in the three types of breast cancer cell line membrane fractions may be useful in the further stratification of the major types of breast cancer. In addition, the high expression of these proteins may enhance detection after release in sera or plasma from cancer patients and could represent potential cancer biomarkers.
Figure 5.
Compilation of proteins highlighting the relative quantitative spectral counts of distinct proteins found in the membrane fractions of MCF-7 (black bar), MDA-MB-453 (grey bar), and MDA-MB-468 (white bar) breast cancer cell lines. A) Spectral count scale is 0–350 counts for proteins. B) Spectral count scale is 0–60 counts for proteins. Each bar represents quantitatively normalized spectral counts of 3 separate experiments for each breast cancer cell line complied in the software program scaffold.
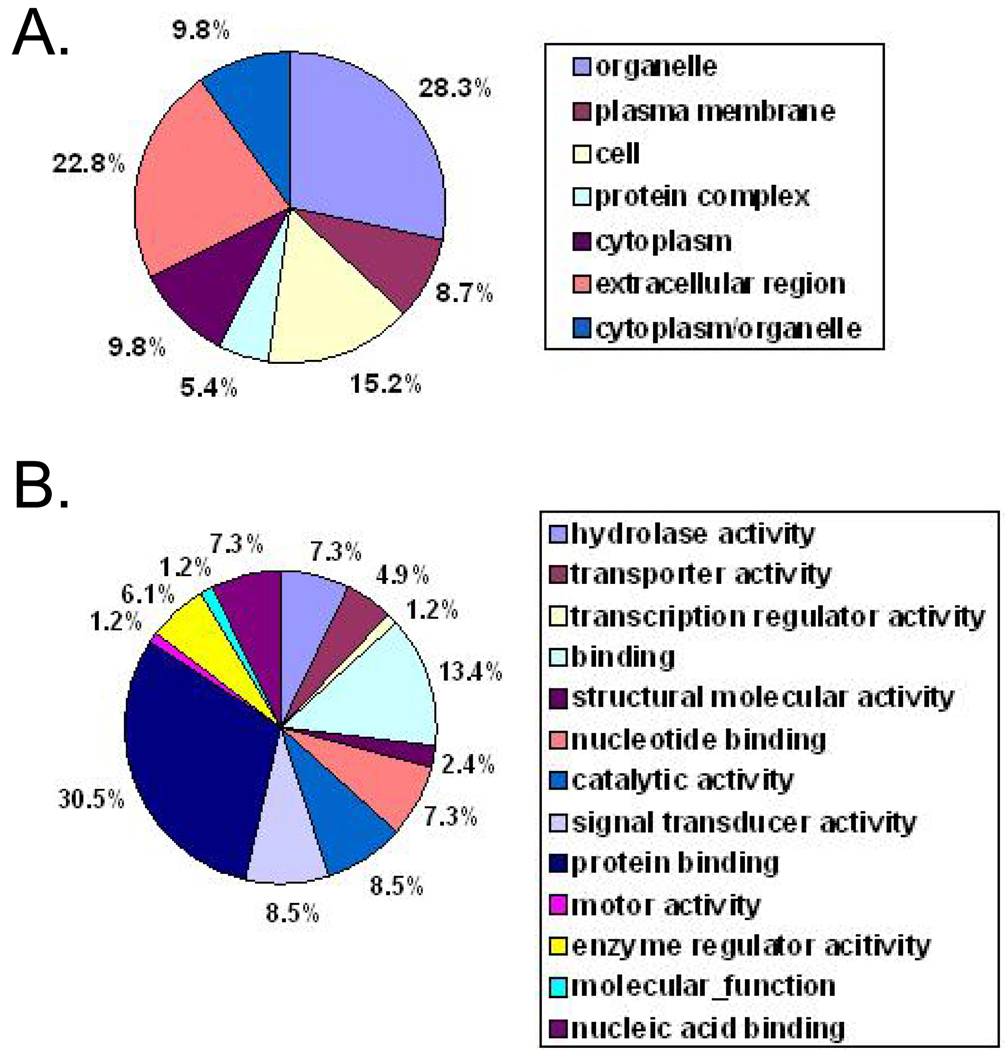
Since N-linked glycoproteins clearly were enriched from crude cell membrane fractions using the hydrazide method, it was of interest to assess the location (Figure 6A) and cellular function (Figure 6B) of these defined proteins. N-linked proteins were classified into a pie chart of differential cell compartments and function using an in-house developed protein sorting software program (UCLA Bioinformatics Department). As expected, this analysis revealed a heavy representation of membrane-associated proteins (Figure 6A) with diverse functions (Figure 6B).
Figure 6.
A) Pie graph of the percentage of all N-linked glycosylated proteins found in different cell compartments of breast cancer cell lines MCF-7, MDA-MB-453, and MDA-MB-468. B) Pie graph of the percentage of proteins of all N-linked glycosylated proteins found in different cellular functions of breast cancer cell lines MCF-7, MDA-MB-453, and MDA-MB-468.
Discussion
The current study explored the hydrazide method as an effective way to preselect N-linked glycoproteins from cell membranes before applying the resolving power of coupled microbore chromatography-mass spectrometry to identify the proteins. The results indicate this approach was successful in enriching, identifying and site-mapping 27 N-linked glycosylation sites in 25 glycoproteins from crude membrane samples. The method allowed us to dig deeper into the glycosylated membrane proteome than is possible with conventional cellular fractionation. This can be clearly seen in all three human breast cancer cell lines examined. For example, in MCF-7 cell membrane isolates there was only an overlap of 3 N-linked glycoproteins, with the 357 total membrane proteins detected by LC-MS/MS alone, while 12 N-linked glycoproteins were uncovered only after N-linked glycoprotein hydrazide enrichment. Similarly, only 3 glycoproteins were detectable by LC-MS/MS in MDA-MB-453 among the 388 total membrane proteins but N-linked glycoprotein enrichment revealed an additional 10 N-linked glycoproteins after hydrazide enrichment. Finally in MDA-MB-468 cells only 2 N-linked glycoproteins were seen by LC-MS/MS among the 338 total membrane proteins identified. Both these N-linked glycoproteins are signature proteins of the MDA-MB-468 cell line. Clearly there was significant enrichment of the N-linked glycoprotein proteome using the hydrazide system. Recently published was a modification of the original hydrazide method32 that significantly increased the selection and identification of cell surface glycoproteins by chemically tagging glycoproteins on intact living cells38. The application of this method would significantly reduce the contamination of proteins from intracellular membranes found in crude membrane extracts.
Traditionally N-glycosylation typically occurs at the NXS/T motif on glycoproteins, where X is any amino acid except proline. Interestingly, we also detected a number of non-NXS/T motif glyco-sites with the hydrazide method. We can not rule these peptides out as contaminating peptides that underwent deamination through the hydrazide process nor could we dismiss them due to the covalent isolation of the glyco-residues and stringent washing. Therefore we included them in this manuscript (Supplemental Table 1). We also included representative MS/MS data (Supplemental Figure 2). Some of these glco-sites have serine or threonine located right next to the asparagines (NS/T) or two amino acids to the right (NXXS/T).
The identified N-linked glycoprotein differences and similarities between the three cell lines are especially interesting, since out of 291 shared membrane proteins there were only 3 glycoproteins in common. In addition, these three proteins were not detectable in the crude membrane fraction, providing further support for the utility of selective glycoprotein enrichment by the hydrazide method. MCF-7 and MDA-MB-453 cells shared three NXS/T motif glycoproteins in common. Cathepsin D is reported to be secreted in more aggressive breast cancer cell lines39 and clusterin when elevated promotes tumor cell survival by having anti-apoptotic effects on chemotherapy treated breast cancer40. On the other hand pigment epithelium-derived factor (PEDF) expression was found to be dramatically decreased in breast cancer 41. PEDF is one of the most potent inhibitors of angiogenesis and is a candidate tumor suppressor in a variety of cancers 42. Further study of increased expression of PEDF in hormone and receptor positive breast cancer cell lines compared to triple negative cell lines may correlate with the prognosis of this cancer. We also detected another angiogenesis inhibitor protein, thrombospondin-1, in MCF-7 cells. Interestingly, the expression of thrombospondin-1 is inversely correlated with vascularization in breast cancer.43 Further study in HER2 positive and HER2 negative cell lines will be necessary to determine the significance of the presence of these glycoproteins in breast cancer cell lines. Three other interesting MCF-7 glycoproteins that did not have the traditional NXS/T motif where keratin and hypothetical protein LOC9768 isoform 1, KIAA0101 or p15(PAF). Keratin KRT8/18 expression is known to differentiate distinct subtypes of grade 3 invasive ductal carcinoma of the breast 44. We also see an abundant amount of keratin 8 in the MCF-7 breast cancer cell lines membrane fraction, an apparent candidate biomarker for that particular breast cancer cell type. Hypothetical protein LOC9768 isoform 1, KIAA0101 or p15(PAF), is a 15-kDa protein containing a conserved proliferating cell nuclear antigen (PCNA)-binding motif found to be involved in the regulation of DNA repair, cell cycle progression, and cell proliferation45, 46. KIAA0101 is over-expressed in tumors of the breast, uterine cervix, brain, kidney, esophagus, lungs and colon45, 46.
A single non-NXS/T motif glycoprotein, tankyrase 1, was shared by the triple negative MDA-MB-468 breast cancer cells and the HER2 negative MCF-7 breast cancer cells. Tankyrase 1 is known to be involved in the positive regulation of telomeres which play a critical role in cellular senescence, apoptosis and tumorigenesis47, 48. Increased expression of tankyrase 1 interacts with and releases a negative regulator of telomerase, telomere repeat binding factor 1 (TRF1), inducing telomere elongation47. Tankyrase 1 also is elevated in tumor tissues and its expression is higher in estrogen and progesterone negative tumors49. The discovery of a N-linked glycosylation site may be important in the regulation and function of tankyrase 1 as well as being a useful post-translational modification (PTM) for biomarker detection.
After hydrazide enrichment, LC-MS/MS identified breast cancer 1, early onset isoform 1 (BRCA 1) as a N-linked glycoprotein in MCF-7 cells. Point mutations in BRCA 1 and BRCA 2 have been well documented in 30% of hereditary breast cancers50. Previously (based on the “Keil rule”) it was believed trypsin cleaves at lysine or arginine but not before proline, however Rodriguez et al. reported that many peptides are actually cleaved with trypsin before proline residues51. In addition, one of the N-linked sites on the BRCA1 glycopeptide is a predicted N-linked site (https://db.systemsbiology.net) based on the N-linked glycosylation consensus sequence NXS/T at amino acid site 913 (EENQGKN*ESNIK). The enrichment and identification of BRCA 1 as a N-linked glycoprotein also provides support for the utility of the hydrazide method in discovering additional glycoproteins as potential breast cancer biomarkers.
N-linked glycosylation plays a key role in the expression levels and receptor activity of several receptor tyrosine kinases (RTK) such as EGFR and HER252. N-linked glycosylation of the EGFR positively effects the conformation necessary for receptor-receptor self-dimerization. Four hypothetical N-linked glycosylation sites located in domain III of EGFR have been proposed as potential key contributors to EGFR self-dimerization which is necessary for ligand activation of the receptor53. We have identified and site-mapped a N-linked glycosylation site in the domain III region of EGFR in MDA-MB-468 breast cancer cells. The EGFR peptide, 347DSLSINATNIK357, contains a N-linked glycosylation consensus site (NXS/T) that was also detected in purified EGFR from A431 epidermal cancer cell line54. In addition, the hydrazide method identified a N-linked glycosylation at N57 of CD44 which matched one of five N-linked glycosylation sites necessary for CD44 conformational interaction with hyaluronic acid leading to CD44 activation55. Both O-glycosylation and N-glycosylation of CD44 play a critical role in its function55. Interestingly the presence of CD44 cell surface receptors and the absence of CD24 is a phenotype found in breast cancer stem cells and basal-like breast tumors otherwise known as triple negative cancers (without PR, without ER and without HER2 receptor) such as the MDA-MB-468 breast cancer cell line56–58. The CD44+/CD24- phenotype correlates with metastasis and invasiveness59. Identification of key post-translational modifications (PTMs) that play key roles in receptor function and activity may be important as targets for drug development as well as for the development of PTM specific biomarkers.
The high sensitivity of LC-MS/MS allowed detection of progesterone receptor membrane component 1 (PGRMC1) in both of the estrogen receptor (ER) negative breast cancer cell lines tested and not in the ER positive MCF-7 cell line (Supplemental Table 2). PGRMC1 is found to be highly expressed in ER negative cell lines compared to ER positive cell lines60. This agreement between our mass spectrometry data with previously published reports confirms the sensitivity and reliability of the LTQ Orbitrap as a biomarker discovery tool.
A primary goal of this study was to enrich for N-linked glycoproteins in crude cancer cell membrane fractions as a prelude to discover of potential biomarkers similar to the currently known biomarkers HER2, MUC1, ER, and PR. Since we were able to isolate 25 N-linked glycoproteins with many of the proteins playing major roles in cancer we also analyzed the proteins differentially expressed in the membrane fraction that may also be secreted or sloughed off into the interstitial fluid of tumors in vivo. Significant overlaps and differences between the membrane proteins were seen in each tumor cell line, but quantitatively each cell line appeared to have a unique proteome signature. Comparison of the normalized spectral counts of membrane proteins in MCF-7, MDA-MB-453, and MDA-MB-468 cells using the scaffold software program revealed signature differences between the three breast cancer cell lines. MCF-7 had high quantities keratin 8 and 18, solute carrier protein 3, HSP 27 and ErbB-binding protein, while MDA-MB-453 had high quantities of enolase 1, nucleolin, RAB1B Ras oncogene, stomatin, filamin A, valosin and cytokeratin 7. MDA-MB-468 cells had high quantities of EGFR, CD44, filamin A, progesterone receptor component 1, and valosin. All three cancer cell lines also share high expression of STIP1, keratin 19, chaperonin, and HSP-alpha. Any one of these proteins could represent signature new candidate biomarkers of cancer for a blood assay. Further studies of breast cancer cell lines, proximal fluid, tissue samples, and patient matched serum samples using affinity-based assays will be necessary to validate reproducibility and functionality of each of these candidate biomarkers.
This preliminary study of N-linked glycoprotein proteome in breast cancer cell lines has led to the identification of 27 glycosylation sites in 25 proteins. An important extension of this work will be to document the correlation between protein quantity and breast cancer type and severity. Already an overlap in the glycoproteins found in cancer membrane fractions and secreted or released proteins has been detected in the breast cell line culture media (data not shown). Another important extension of this work will be to compare both the conventional O-linked glycoproteins of the cell membrane and the monosacharide O-linked N-acetylglucosamine (O-GlcNAc)61 of the nucleocytoplasmic fraction between the different breast cancer cell lines, tissue and the interstitial fluid around the tumor sites. The discovery of unique candidate biomarkers will help with the stratification of breast tumor types. Future development of antibodies that specifically recognize changes in glycosylation PTMs of candidate biomarkers would further the stratification of breast cancer. In addition, discovery of high abundant proteins secreted into the proximal fluid of breast cancer cell lines or interstitial fluid of tumor tissue may lead to a plausible affinity-based assay capable of identifying these trace level biomarkers in the large volume of circulating blood proteins of breast cancer patients.
Supplementary Material
Acknowledgments
This work was supported in part by the California Breast Cancer Research Program (6JB-0013), the Department of Defense (DAMD17-01-1-0179), the National Institute of Health (1RO1CA93736), the Gonda Foundation, the EIF-Women Cancer Research Fund and Friends of the Breast Program at UCLA.
References
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3(4):243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Porter PL, El-Bastawissi AY, Mandelson MT, Lin MG, Khalid N, Watney EA, Cousens L, White D, Taplin S, White E. Breast tumor characteristics as predictors of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst. 1999;91(23):2020–2028. doi: 10.1093/jnci/91.23.2020. [DOI] [PubMed] [Google Scholar]
- 3.Anderson BO, Braun S, Lim S, Smith RA, Taplin S, Thomas DB. Early detection of breast cancer in countries with limited resources. Breast J. 2003;9 Suppl 2:S51–S59. doi: 10.1046/j.1524-4741.9.s2.4.x. [DOI] [PubMed] [Google Scholar]
- 4.Bertucci F, Goncalves A. Clinical proteomics and breast cancer: strategies for diagnostic and therapeutic biomarker discovery. Future Oncol. 2008;4(2):271–287. doi: 10.2217/14796694.4.2.271. [DOI] [PubMed] [Google Scholar]
- 5.Chen ST, Pan TL, Juan HF, Chen TY, Lin YS, Huang CM. Breast tumor microenvironment: proteomics highlights the treatments targeting secretome. J Proteome Res. 2008;7(4):1379–1387. doi: 10.1021/pr700745n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5(11):845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 7.Bast RC, Jr, Ravdin P, Hayes DF, Bates S, Fritsche H, Jr, Jessup JM, Kemeny N, Locker GY, Mennel RG, Somerfield MR. 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol. 2001;19(6):1865–1878. doi: 10.1200/JCO.2001.19.6.1865. [DOI] [PubMed] [Google Scholar]
- 8.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 9.Bast RC, Jr, Klug TL, St John E, Jenison E, Niloff JM, Lazarus H, Berkowitz RS, Leavitt T, Griffiths CT, Parker L, Zurawski VR, Jr, Knapp RC. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N Engl J Med. 1983;309(15):883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- 10.Bates SE, Longo DL. Tumor markers: value and limitations in the management of cancer patients. Cancer Treat Rev. 1985;12(3):163–207. doi: 10.1016/0305-7372(85)90037-4. [DOI] [PubMed] [Google Scholar]
- 11.Debruyne EN, Delanghe JR. Diagnosing and monitoring hepatocellular carcinoma with alpha-fetoprotein: new aspects and applications. Clin Chim Acta. 2008;395(1–2):19–26. doi: 10.1016/j.cca.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Domon B, Aebersold R. Challenges and opportunities in proteomics data analysis. Mol Cell Proteomics. 2006;5(10):1921–1926. doi: 10.1074/mcp.R600012-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 14.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2(10):1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24(3):413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 17.Schrimpf SP, Meskenaite V, Brunner E, Rutishauser D, Walther P, Eng J, Aebersold R, Sonderegger P. Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics. 2005;5(10):2531–2541. doi: 10.1002/pmic.200401198. [DOI] [PubMed] [Google Scholar]
- 18.Corthals GL, Aebersold R, Goodlett DR. Identification of phosphorylation sites using microimmobilized metal affinity chromatography. Methods Enzymol. 2005;405:66–81. doi: 10.1016/S0076-6879(05)05004-4. [DOI] [PubMed] [Google Scholar]
- 19.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2(8):591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4(2):144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Holmes EH, Greene TG, Tino WT, Boynton AL, Aldape HC, Misrock SL, Murphy GP. Analysis of glycosylation of prostate-specific membrane antigen derived from LNCaP cells, prostatic carcinoma tumors, and serum from prostate cancer patients. Prostate Suppl. 1996;7:25–29. [PubMed] [Google Scholar]
- 22.Lloyd KO, Yin BW, Kudryashov V. Isolation and characterization of ovarian cancer antigen CA 125 using a new monoclonal antibody (VK-8): identification as a mucin-type molecule. Int J Cancer. 1997;71(5):842–850. doi: 10.1002/(sici)1097-0215(19970529)71:5<842::aid-ijc24>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Kui Wong N, Easton RL, Panico M, Sutton-Smith M, Morrison JC, Lattanzio FA, Morris HR, Clark GF, Dell A, Patankar MS. Characterization of the oligosaccharides associated with the human ovarian tumor marker CA125. J Biol Chem. 2003;278(31):28619–28634. doi: 10.1074/jbc.M302741200. [DOI] [PubMed] [Google Scholar]
- 24.Comegys MM, Lin SH, Rand D, Britt D, Flanagan D, Callanan H, Brilliant K, Hixson DC. Two variable regions in carcinoembryonic antigen-related cell adhesion molecule1 N-terminal domains located in or next to monoclonal antibody and adhesion epitopes show evidence of recombination in rat but not in human. J Biol Chem. 2004;279(33):35063–35078. doi: 10.1074/jbc.M404431200. [DOI] [PubMed] [Google Scholar]
- 25.Paxton RJ, Mooser G, Pande H, Lee TD, Shively JE. Sequence analysis of carcinoembryonic antigen: identification of glycosylation sites and homology with the immunoglobulin supergene family. Proc Natl Acad Sci U S A. 1987;84(4):920–924. doi: 10.1073/pnas.84.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson PJ, Poon TC, Hjelm NM, Ho CS, Ho SK, Welby C, Stevenson D, Patel T, Parekh R, Townsend RR. Glycan composition of serum alpha-fetoprotein in patients with hepatocellular carcinoma and non-seminomatous germ cell tumour. Br J Cancer. 1999;81(7):1188–1195. doi: 10.1038/sj.bjc.6690828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J Biol Chem. 2006;281(37):27178–27189. doi: 10.1074/jbc.M601248200. [DOI] [PubMed] [Google Scholar]
- 28.Stroop CJ, Weber W, Gerwig GJ, Nimtz M, Kamerling JP, Vliegenthart JF. Characterization of the carbohydrate chains of the secreted form of the human epidermal growth factor receptor. Glycobiology. 2000;10(9):901–917. doi: 10.1093/glycob/10.9.901. [DOI] [PubMed] [Google Scholar]
- 29.Sparrow LG, Gorman JJ, Strike PM, Robinson CP, McKern NM, Epa VC, Ward CW. The location and characterisation of the O-linked glycans of the human insulin receptor. Proteins. 2007;66(2):261–265. doi: 10.1002/prot.21261. [DOI] [PubMed] [Google Scholar]
- 30.Firth SM, Baxter RC. Characterisation of recombinant glycosylation variants of insulin-like growth factor binding protein-3. J Endocrinol. 1999;160(3):379–387. doi: 10.1677/joe.0.1600379. [DOI] [PubMed] [Google Scholar]
- 31.Gemeiner P, Viskupic E. Stepwise immobilization of proteins via their glycosylation. J Biochem Biophys Methods. 1981;4(5–6):309–319. doi: 10.1016/0165-022x(81)90071-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5(6):1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 34.Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6(1):141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17(20):2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 36.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 37.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 38.Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of Nlinked cell surface glycoproteins. Nat Biotechnol. 2009;27(4):378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mbeunkui F, Metge BJ, Shevde LA, Pannell LK. Identification of differentially secreted biomarkers using LC-MS/MS in isogenic cell lines representing a progression of breast cancer. J Proteome Res. 2007;6(8):2993–3002. doi: 10.1021/pr060629m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranney MK, Ahmed IS, Potts KR, Craven RJ. Multiple pathways regulating the anti-apoptotic protein clusterin in breast cancer. Biochim Biophys Acta. 2007;1772(9):1103–1111. doi: 10.1016/j.bbadis.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai J, Parr C, Watkins G, Jiang WG, Boulton M. Decreased pigment epithelium-derived factor expression in human breast cancer progression. Clin Cancer Res. 2006;12(11 Pt 1):3510–3517. doi: 10.1158/1078-0432.CCR-06-0094. [DOI] [PubMed] [Google Scholar]
- 42.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, Cornwell ML, Pins MR, Borensztajn J, Crawford SE. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat Med. 2003;9(6):774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 43.Chandrasekaran S, Guo NH, Rodrigues RG, Kaiser J, Roberts DD. Pro-adhesive and chemotactic activities of thrombospondin-1 for breast carcinoma cells are mediated by alpha3beta1 integrin and regulated by insulin-like growth factor-1 and CD98. J Biol Chem. 1999;274(16):11408–11416. doi: 10.1074/jbc.274.16.11408. [DOI] [PubMed] [Google Scholar]
- 44.Walker LC, Harris GC, Holloway AJ, McKenzie GW, Wells JE, Robinson BA, Morris CM. Cytokeratin KRT8/18 expression differentiates distinct subtypes of grade 3 invasive ductal carcinoma of the breast. Cancer Genet Cytogenet. 2007;178(2):94–103. doi: 10.1016/j.cancergencyto.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Yuan RH, Jeng YM, Pan HW, Hu FC, Lai PL, Lee PH, Hsu HC. Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2007;13(18 Pt 1):5368–5376. doi: 10.1158/1078-0432.CCR-07-1113. [DOI] [PubMed] [Google Scholar]
- 46.Yu P, Huang B, Shen M, Lau C, Chan E, Michel J, Xiong Y, Payan DG, Luo Y. p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues. Oncogene. 2001;20(4):484–489. doi: 10.1038/sj.onc.1204113. [DOI] [PubMed] [Google Scholar]
- 47.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282(5393):1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 48.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579(4):859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 49.Poonepalli A, Banerjee B, Ramnarayanan K, Palanisamy N, Putti TC, Hande MP. Telomere-mediated genomic instability and the clinico-pathological parameters in breast cancer. Genes Chromosomes Cancer. 2008;47(12):1098–1109. doi: 10.1002/gcc.20608. [DOI] [PubMed] [Google Scholar]
- 50.Lynch HT, Silva E, Snyder C, Lynch JF. Hereditary breast cancer: part I. Diagnosing hereditary breast cancer syndromes. Breast J. 2008;14(1):3–13. doi: 10.1111/j.1524-4741.2007.00515.x. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez J, Gupta N, Smith RD, Pevzner PA. Does trypsin cut before proline? J Proteome Res. 2008;7(1):300–305. doi: 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- Contessa JN, Bhojani MS, Freeze HH, Rehemtulla A, Lawrence TS. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008;68(10):3803–3809. doi: 10.1158/0008-5472.CAN-07-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandes H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association: a study with an aberrant epidermal growth factor receptor (EGFRvIII/DeltaEGFR) expressed in cancer cells. J Biol Chem. 2001;276(7):5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 54.Wu SL, Kim J, Hancock WS, Karger B. Extended Range Proteomic Analysis (ERPA): a new and sensitive LC-MS platform for high sequence coverage of complex proteins with extensive post-translational modifications-comprehensive analysis of beta-casein and epidermal growth factor receptor (EGFR) J Proteome Res. 2005;4(4):1155–1170. doi: 10.1021/pr050113n. [DOI] [PubMed] [Google Scholar]
- 55.Bartolazzi A, Nocks A, Aruffo A, Spring F, Stamenkovic I. Glycosylation of CD44 is implicated in CD44-mediated cell adhesion to hyaluronan. J Cell Biol. 1996;132(6):1199–1208. doi: 10.1083/jcb.132.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dontu G. Breast cancer stem cell markers - the rocky road to clinical applications. Breast Cancer Res. 2008;10(5):110. doi: 10.1186/bcr2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponti D, Zaffaroni N, Capelli C, Daidone MG. Breast cancer stem cells: an overview. Eur J Cancer. 2006;42(9):1219–1224. doi: 10.1016/j.ejca.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 58.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, Grabau D, Ferno M, Borg A, Hegardt C. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R, Jr, Badve S, Nakshatri H. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neubauer H, Clare SE, Wozny W, Schwall GP, Poznanovic S, Stegmann W, Vogel U, Sotlar K, Wallwiener D, Kurek R, Fehm T, Cahill MA. Breast cancer proteomics reveals correlation between estrogen receptor status and differential phosphorylation of PGRMC1. Breast Cancer Res. 2008;10(5):R85. doi: 10.1186/bcr2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whelan SA, Hart GW. Proteomic approaches to analyze the dynamic relationships between nucleocytoplasmic protein glycosylation and phosphorylation. Circ Res. 2003;93(11):1047–1058. doi: 10.1161/01.RES.0000103190.20260.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.