Abstract
In our previous study, we utilized a 26-residue amphipathic α-helical antimicrobial peptide L-V13K (Chen et al., Antimicrob Agents Chemother 2007, 51, 1398−1406) as the framework to study the effects of peptide hydrophobicity on the mechanism of its antimicrobial action. In this study, we explored the effects of net charge and the number of positively charged residues on the hydrophilic/polar face of L-V13K on its biological activity (antimicrobial and hemolytic) and biophysical properties (hydrophobicity, amphipathicity, helicity, and peptide self-association). The net charge of V13K analogs at pH 7 varied between −5 and +10 and the number of positively charged residues varied from 1 to 10. The minimal inhibitory concentrations (MIC) against six strains of Pseudomonas aeruginosa as well as other gram-negative and gram-positive bacteria were determined along with the maximal peptide concentration that produces no hemolysis of human red blood cells (MHC). Our results show that the number of positively charged residues on the polar face and net charge are both important for both antimicrobial activity and hemolytic activity. The most dramatic observation is the sharp transition of hemolytic activity on increasing one positive charge on the polar face of V13K i.e., the change from +8 to +9 resulted in greater than 32-fold increase in hemolytic activity (250 μg/ml to <7.8 μg/ml, respectively).
Keywords: antimicrobial peptide, antimicrobial activity, hemolytic activity, hydrophobicity, amphipathicity, helicity, peptide self-association
INTRODUCTION
The widespread use of classical antibiotics has resulted in the emergence of many antibiotic-resistant strains. Therefore, the development of a new class of antibiotics is critical. Cationic antimicrobial peptides (AMPs) have become important candidates as potential therapeutic agents. Cationic AMPs are found in organisms that are evolutionarily quite distant, ranging from plants and insects to birds, animals (including molluscs, crustaceans, amphibians, fish, and mammals), and humans.1-3 Cationic AMPs have unusually broad spectra of “antimicrobial” activity, which include an ability to kill or neutralize bacteria (gram-positive and gram-negative), fungi (including yeasts), parasites (including planaria and nematodes), cancer cells, and even enveloped viruses like HIV and herpes simplex virus.4
Although the exact mode of action of AMPs has not been established, it is generally accepted that the cytoplasmic membrane is the main target of many AMPs, whereby peptide accumulation in the membrane causes increased permeability and a loss of barrier function, resulting in the leakage of cytoplasmic components and cell death. The development of resistance to membrane active peptides whose sole target is the cytoplasmic membrane is thought to be considerably reduced when compared with that of many current antibiotics, which have more specific molecular targets.3 This prediction has been substantiated in several studies.5-7 However, the major barrier for the use of AMPs as antibiotics is their toxicity or ability to lyse eukaryotic cells, normally expressed as hemolytic activity (toxicity to human red blood cells). This is the main reason preventing their applications as injectable therapeutics.
Cationic AMPs of the α-helical class have two unique features: a net positive charge of at least +2 and an amphipathic character, with a nonpolar face and a polar/charged face.8 Factors believed to be important for antimicrobial activity have been identified, including peptide hydrophobicity, the presence of positively charged residues, an amphipathic nature that segregates basic and hydrophobic residues, and secondary structure. It has also been shown that linearity of amphipathic α-helical AMPs is not a prerequisite for lytic activity but rather affects selectivity between gram-negative and gram-positive bacteria and between mammalian cells and bacteria.9
To investigate what features of α-helical AMPs could be changed to control specificity between prokaryotic and eukaryotic cells, we chose a peptide, V681, which had an excellent antimicrobial activity but also exhibited high toxicity to human red blood cells as measured by hemolytic activity.10 We showed that a single valine to lysine substitution in the center of the nonpolar face dramatically reduced toxicity and increased the therapeutic index.10 This was the first report that a single substitution could act as a specificity determinant in α-helical AMPs that controls activity between eukaryotic and prokaryotic cells. We then showed that the sole target of this peptide was the membrane by comparing the l- and d-enantiomers of peptide V13K. These peptides had equal activities suggesting that the antimicrobial mechanism did not involve a stereoselective interaction with a chiral enzyme, lipid, or protein receptor.11 In addition, the all d-peptide was resistant to proteolytic enzyme degradation, which enhances the potential of D-V13K as a clinical therapeutic. We subsequently investigated the role of hydrophobicity on the nonpolar face and showed that there was an optimum hydrophobicity on the nonpolar face required to obtain the best therapeutic index.12 Increases in hydrophobicity beyond the optimum resulted in a dramatic reduction in antimicrobial activity, which correlated with increase peptide self-association ability.12 We have now increased the list of factors important for antimicrobial activity to include: (1) the importance of lack of secondary structure in benign medium, but inducible structure in the presence of the hydrophobic environment of the membrane; (2) the presence of a positively-charged residue in the center of the nonpolar face of amphipathic cyclic β-sheet and α-helical peptides as a determinant of locating the peptides to the interface region of prokaryotic membranes and decreasing transmembrane penetration into eukaryotic membranes; and (3) importance of peptide self-association in an aqueous environment to the biological activities of these peptides.10-14
In this study, to investigate the effects of net charge and the number of positively charged residues in the mechanism of action of α-helical AMPs, we either systematically decreased or increased the net charge of peptide L-V13K on the polar face to show that net charge and the number of positively charged residues have significant effects on the biological activities in prokaryotic and eukaryotic cells.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Purification
Synthesis of the peptides was carried out by solid-phase peptide synthesis using t-butyloxycarbonyl chemistry and 4-methylbenzhydrylamine resin (0.97 mmol/g) and purification by reversed-phase high-performance liquid chromatography (RP-HPLC), as described previously.10-12 Peptide purification was performed on a Zorbax 300 SB-C8 column (250 mm × 9.4 mm I.D.; 6.5 μm particle size, 300 Å pore size; Agilent Technologies, Little Falls, DE) with a linear AB gradient (0.1% acetonitrile/min) at a flow rate of 2 ml/min, where eluent A was 0.2% aqueous trifluoroacetic acid (TFA), pH 2, and eluent B was 0.2% TFA in acetonitrile, where the shallow 0.1% acetonitrile/min gradient started 12% below the acetonitrile concentration required to elute the peptide on injection of analytical sample using a gradient of 1% acetonitrile/min.15 The purity of the peptides was verified by analytical RP-HPLC as described later and was further characterized by mass spectrometry and amino acid analysis.
Analytical RP-HPLC and Temperature Profiling of Peptides
Crude and purified peptides were analyzed on an Agilent 1100 series liquid chromatograph (Little Falls, DE). Runs were performed on a Zorbax 300 SB-C8 column (150 mm × 2.1 mm I.D.; 5 μm particle size, 300 Å pore size) from Agilent Technologies using a linear AB gradient (1% acetonitrile/min) and a flow rate of 0.25 ml/min, where eluent A was 0.2% aqueous TFA, pH 2, and eluent B was 0.2% TFA in acetonitrile. Temperature profiling analyses were performed on the same column in 3°C increments, from 5 to 80°C using a linear AB gradient of 0.5% acetonitrile/min, as described previously.10-13 Peptide −5 (see later) does not dissolve at pH 2; thus, no retention time (tR) and no self-association parameter (PA) values could be determined.
Characterization of Helical Structure
The mean residue molar ellipticities of peptides were determined by circular dichroism (CD) spectroscopy, using a Jasco J-720 spectro-polarimeter (Easton, MD) at 5°C under benign (nondenaturing) conditions (50 mM NaH2PO4/Na2HPO4/100 mM KCl, pH 7.0), hereafter referred to as KP buffer, as well as in the presence of an α-helix inducing solvent, 2,2,2-trifluoroethanol, TFE, (50 mM NaH2PO4/Na2HPO4/100 mM KC1, pH 7.0 buffer/50% TFE). A 10-fold dilution of an approximately 500 μM stock solution of the peptide analogs was loaded into a 0.1 cm quartz cell and its ellipticity scanned from 195 to 250 nm.
Determination of Peptide Amphipathicity
Amphipathicity of peptide analogs was determined by the calculation of hydrophobic moment16 using the software package Jemboss version 1.2.1,17 modified to include a hydrophobicity scale determined in our laboratory. The hydrophobicity scale used in this study is as follows: Trp, 33.0; Phe, 30.1; Leu, 24.6; Ile, 22.8; Met, 17.3; Tyr, 16.0; Val, 15.0; Pro, 10.4; Cys, 9.1; His, 4.7; Ala, 4.1; Arg, 4.1; Thr, 4.1; Gln, 1.6; Ser, 1.2; Asn, 1.0; Gly, 0.0; Glu, −0.4; Asp, −0.8; and Lys, −2.0.18 These hydrophobicity coefficients were determined from RP-HPLC at pH 7 (10 mM Na2HPO4 buffer containing 50 mM NaCl) of a model random coil 10-residue peptide sequence, Ac-X-G-A-K-G-A-G-V-G-L-amide, where position X was substituted by all 20 naturally occurring amino acids. We propose that this HPLC-derived scale reflects the relative differences in hydrophilicity/hydrophobicity of the 20 amino acid side chains more accurately than previously determined scales.
Pseudomonas aeruginosa Strains Used in this Study
Strain PAO1 was isolated from a human wound in 1955 in Australia19; strain WR5 was isolated from a burn patient at Walter Reed Army Hospital, Washington, DC, in 1976 and is a natural toxA mutant isolate but is virulent in experimental mouse models20,21; strain PAK was originally isolated at Memorial University, St. John's, Newfoundland, Canada, and is widely used in the analysis of pili22,23; strain PA14 was originally isolated as a clinical isolate in 1995 at the Massachusetts General Hospital, Boston, and is virulent in a variety of plant and animal models of infection24; strain M2 was originally isolated in 1975 from the gastrointestinal tract of a healthy CF1 mouse, University of Cincinnati College of Medicine, and Shriners Burns Institute, Cincinnati, OH, and is virulent in a burn mouse model of P. aeruginosa infection25; and strain CP204 was isolated from a cystic fibrosis patient in 1989 at the National Jewish Medical and Research Center, Denver, CO. All strains have been maintained at −80°C in the laboratory of Michael Vasil.
Measurement of Antimicrobial Activity (MIC)
Minimal inhibitory concentrations (MICs) were determined by a standard microtiter dilution method in brain heart infusion (BHI) medium. Briefly, cells were grown overnight at 37°C in BHI broth and were diluted in the same medium. Serial dilutions of the peptides were added to the microtiter plates in a volume of 50 μl, followed by the addition of 50 μl of bacteria to give a final inoculum of 5 × 105 colony-forming units (CFU)/ml. The plates were incubated at 37°C for 24 h, and the MICs were determined as the lowest peptide concentration that inhibited growth. However, for MIC determination of microorganisms other than P. aeruginosa clinical isolates, Mueller Hinton Broth (MHB) medium was used instead of BHI medium. In addition, serial dilutions of the 10× compound were added to the microtiter plates in a volume of 10 μl followed by 90 μl of bacteria to give a final inoculum of 5 × 105 CFU/ml.
Measurement of Hemolytic Activity (MHC)
Peptide samples were added to 1% human erythrocytes in phosphate-buffered saline (100 mM NaCl, 80 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4) and the reaction mixtures were incubated at 37°C for 18 h in microtiter plates. Twofold serial dilutions of the peptide samples were carried out in order to determine the concentration that produced no hemolysis. This determination was made by withdrawing aliquots from the hemolysis assays and removing unlysed erythrocytes by centrifugation (800g). Hemoglobin release was determined spectrophotometrically at 570 nm. The hemolytic activity was determined as the maximal peptide concentration that caused no hemolysis of erythrocytes after 18 h. The control for no release of hemoglobin was a sample of 1% erythrocytes without any peptide added. Since erythrocytes were in an isotonic medium, no detectable release (<1% of that released upon complete hemolysis) of hemoglobin was observed from this control during the course of the assay.
Calculation of Therapeutic Index (MHC/MIC Ratio)
The therapeutic index is a widely accepted parameter to represent the specificity of antimicrobial reagents. It is calculated by the ratio of MHC (hemolytic activity) and MIC (antimicrobial activity); thus, larger values of therapeutic index indicate greater antimicrobial specificity. It should be noted that both the MHC and MIC values are carried out by serial twofold dilutions; thus, for individual bacteria and individual peptides, the therapeutic index could vary as much as fourfold if the peptide is very active in both hemolytic and antimicrobial activities; of course, if a peptide has poor or no hemolytic activity, the major variation in the therapeutic index comes from the variation in the MIC value (as much as twofold).
RESULTS
Peptide Design
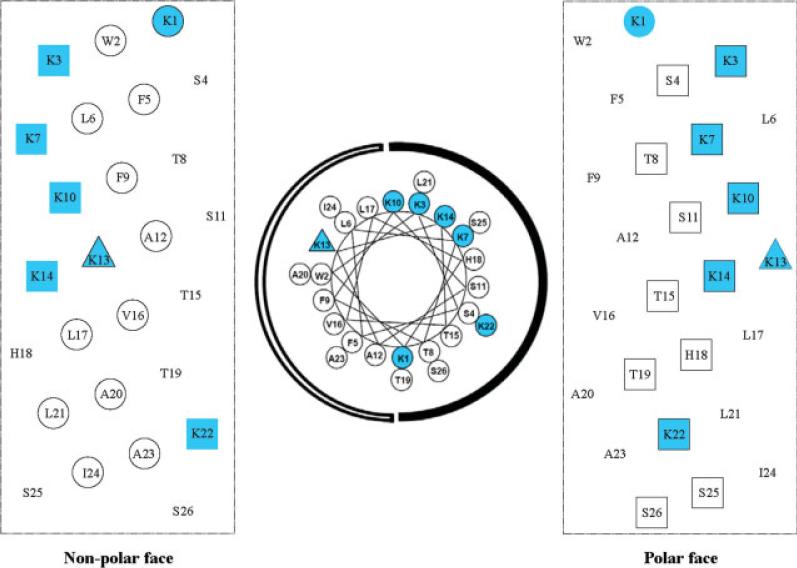
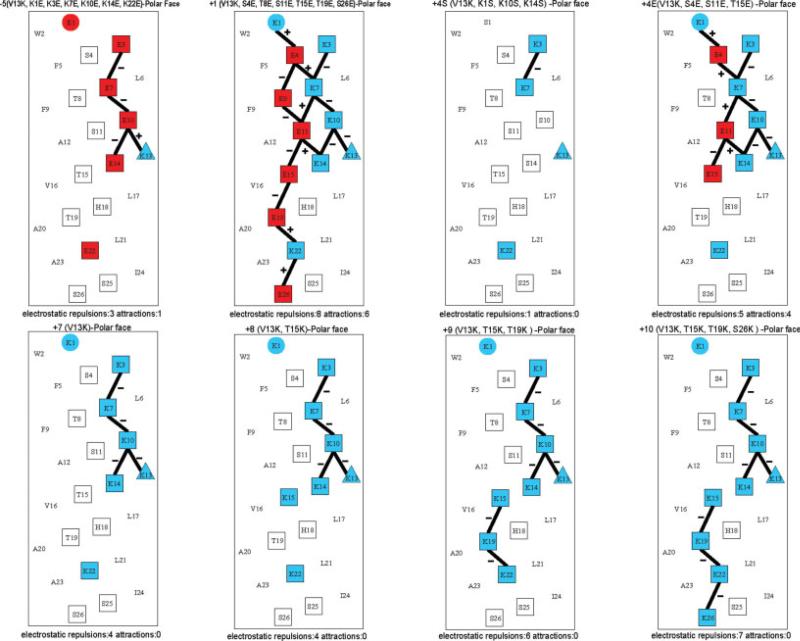
Peptide V13K is a 26-residue amphipathic peptide, which adopts an α-helical conformation in a hydrophobic environment and contains a hydrophilic positively charged lysine residue in the center of the nonpolar face (position 13) (Figures 1 and 2).10-12 In this study, we used peptide V13K as a framework to systematically alter net positive charge and the number of positively charged residues on the polar face of the helix, while keeping the hydrophobic/nonpolar face unchanged. The peptide sequences are shown in Table I and named according to their net charge at pH 7. Figure 2 shows the peptides analogs represented as helical nets. Decreasing net charge was achieved by decreasing the number of positively charged residues or increasing the number of negatively charged residues on the polar face (e.g., peptide −5, peptide +1, peptide +4S, peptide +4E). Peptide +4S, with three substitutions in the polar face of V13K, resulted in a net charge decrease from +7 to +4 when lysine residues were converted to polar noncharged serine residues. Peptide +4E also had three substitutions in the polar face of V13K. The same net charge shift was achieved by substitution of serine or threonine residues with glutamic acid residues (negatively charged at pH 7). Similarly, peptide +1 had six glutamic acid substitutions while maintaining the seven lysine residues to decrease the net charge to +1. All six lysine residues other than the lysine at position 13 were replaced by glutamic acid residues to give a peptide with a net charge of −5. Increasing the net charge was achieved by increasing the number of positively charged residues in the V13K sequence by systematically replacing one, two, and three serine and threonine residues by lysine residues to generate peptides with a net charge of +8, +9, and +10 at pH 7. In summary, the net charge of V13K analogs at pH 7 varied between −5 and +10 and the number of positively charged residues varied from 1 to 10. The electrostatic interactions along the helix (i → i + 3 and i → i + 4) varied between 1 and 7 (see Figure 2). It has been previously shown that intrachain electrostatic attractions and repulsions can affect folding and stability of monomeric α-helices.
FIGURE 1.
Representation of the lead compound V13K as helical nets/helical wheel showing the polar/hydrophilic face and nonpolar/hydrophobic face. In the helical wheel, the nonpolar face is indicated as an open arc and the polar face is shown as a solid arc. In the helical net presentations, the residues making up the nonpolar face of the amphipathic peptide are circled; residues making up the polar face are boxed. The lysine residue at position 13 of the sequence is denoted by a triangle. Positively charged residues are colored blue.
FIGURE 2.
Helical net representation of the sequences of lead compound V13K and analogs. The amino acid residues on the polar faces are boxed. The positively charged residues are colored blue; the negatively charged residues are colored red. The i → i + 3 and i → i + 4 potential electrostatic interactions along the helix are shown as black bars, − denotes repulsions, + denotes attractions. The numbers of electrostatic interactions (repulsions and attractions) on the polar face are indicated at the bottom of each helical net. The one-letter code is used for amino acid residues. The sequences are shown in Table I.
Table I.
Peptides Used in this Study
| Sequencea |
Number of Positively Charged Residues | Number of Negatively Charged Residues | ||||||
|---|---|---|---|---|---|---|---|---|
| Peptide Name | 1 | 13 | 26 | Substitution | Net Chargeb | Amphipathicityc | ||
| –5 | Ac–E–W–E–S–F–L–E–T–F–E–S–A–K–E–T–V–L–H–T–A–L–E–A–I–S–S–amide | V13K, K1E, K3E, K7E, K10E, K14E, K22E | 1 | 6 | –5 | 4.79 | ||
| +1 | Ac–K–W–K–E–F–L–K–E–F–K–E–A–K–K–E–V–L–H–E–A–L–K–A–I–S–E–amide | V13K, S4E, T8E, S11E, T15E, T19E, S26E | 7 | 6 | +1 | 5.38 | ||
| +4S | Ac–S–W–K–S–F–L–K–T–F–S–S–A–K–S–T–V–L–H–T–A–L–K–A–I–S–S–amide | V13K, K1S, K10S, K14S | 4 | 0 | +4 | 4.87 | ||
| +4E | Ac–K–W–K–E–F–L–K–T–F–K–E–A–K–K–E–V–L–H–T–A–L–K–A–I–S–S–amide | V13K, S4E, S11E, T15E | 7 | 3 | +4 | 5.18 | ||
| V13K | Ac–K–W–K–S–F–L–K–T–F–K–S–A–K–K–T–V–L–H–T–A–L–K–A–I–S–S–amide | V13K | 7 | 0 | +7 | 4.92 | ||
| +8 | Ac–K–W–K–S–F–L–K–T–F–K–S–A–K–K–K–V–L–H–T–A–L–K–A–I–S–S–amide | V13K, T15K | 8 | 0 | +8 | 5.11 | ||
| +9 | Ac–K–W–K–S–F–L–K–T–F–K–S–A–K–K–K–V–L–H–K–A–L–K–A–I–S–S–amide | V13K, T15K, T19K | 9 | 0 | +9 | 5.18 | ||
| +10 | Ac–K–W–K–S–F–L–K–T–F–K–S–A–K–K–K–V–L–H–K–A–L–K–A–I–S–K–amide | V13K, T15K, T19K, S26K | 10 | 0 | +10 | 5.26 | ||
| +7b | Ac–K–W–K–S–F–L–S–T–F–K–S–A–K–K–T–V–L–H–K–A–L–S–A–I–S–K–amide | V13K, K7S, T19K, K22S, S26K | 7 | 0 | +7 | 4.85 | ||
| +7c | Ac–S–W–K–S–F–L–K–T–F–K–S–A–K–K–K–V–L–H–T–A–L–K–A–I–S–S–amide | V13K, K1S, T15K | 7 | 0 | +7 | 5.08 | ||
| +7d | Ac–S–W–S–S–F–L–K–T–F–S–S–A–K–K–K–V–L–H–K–A–L–K–A–I–S–K–amide | V13K, K1S, K3S, K10S, T15K, T19K, S26K | 7 | 0 | +7 | 5.26 | ||
| +6 | Ac–S–W–K–S–F–L–K–T–F–S–S–A–K–K–T–V–L–H–T–A–L–K–A–I–S–K–amide | V13K, K1S, K10S, S26K | 6 | 0 | +6 | 4.99 | ||
| +5 | Ac–S–W–K–S–F–L–K–T–F–S–S–A–K–K–T–V–L–H–T–A–L–K–A–I–S–S–amide | V13K, K1S, K10S | 5 | 0 | +5 | 4.92 | ||
Peptide sequences are shown using the one-letter code for amino acid residues; Ac- denotes Nα-acetyl and -amide denotes Cα-terminal amide. The blue letters denote positively charged residues and the red letters denote negatively charged residues. Histidine is considered neutral at pH 7.
The net charges are calculated at pH 7.
Amphipathicity of peptide analogs was determined by calculation of hydrophobic moment using the software package Jemboss version 1.2.1, modified to include a hydrophobicity scale determined in our laboratory.18 (See Experimental Procedures).
Secondary Structure of Peptides
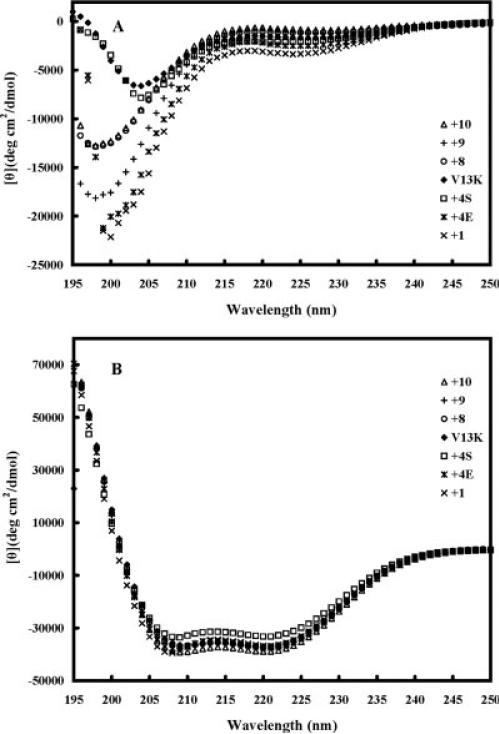
Figure 3 shows the CD spectra of the peptide analogs in different environments, i.e., under benign conditions (in KP buffer) (Figure 3A) and in KP buffer with 50% TFE to mimic the hydrophobic environment of the membrane (Figure 3B). All the peptides exhibited negligible secondary structure in benign buffer (Figure 3A, Table II) but were fully folded α-helical structures in the presence of 50% TFE (Figure 3B, Table II): all the peptide analogs showed a typical α-helix spectrum with double minima at 208 nm and 222 nm. The helicities of the peptides in benign buffer and in 50% TFE relative to that of peptide +10 in 50% TFE were determined (Table II).
FIGURE 3.
Circular dichroism (CD) spectra of peptide V13K and its analogs in benign buffer (100 mM KCl, 50 mM NaH2PO4/Na2HPO4 (A) at pH 7.0, 5°C and (B) in the presence of buffer-TFE (1:1, v/v).
Table II.
Biophysical Data of V13K Analogs
| Benignb |
50% TFE |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptide Name | Number of Positively Charged Residues | Number of Negatively Charged Residues | Net Charge | Amphipathicitya | Hydrophobicity tRd | [θ]222b | % Helixe | [θ]222b | % Helixe | PAc |
| +1 | 7 | 6 | +1 | 5.38 | 85.2 | −3250 | 8 | −36,400 | 94 | 4.07 |
| +4S | 4 | 0 | +4 | 4.87 | 83.6 | −2100 | 5 | −32,750 | 85 | 4.12 |
| +4E | 7 | 3 | +4 | 5.18 | 85.8 | −2400 | 6 | −37,100 | 96 | 3.39 |
| V13K | 7 | 0 | +7 | 4.92 | 77.2 | −1600 | 4 | −37,050 | 96 | 2.44 |
| +8 | 8 | 0 | +8 | 5.11 | 76.5 | −1150 | 3 | −36,600 | 95 | 2.37 |
| +9 | 9 | 0 | +9 | 5.18 | 73.2 | −1000 | 3 | −37,000 | 96 | 1.87 |
| +10 | 10 | 0 | +10 | 5.26 | 71.9 | −750 | 2 | −38,550 | 100 | 1.91 |
Amphipathicity of peptide analogs was determined by calculation of hydrophobic moment using the software package Jemboss version 1.2.1, modified to include a hydrophobicity scale determined in our laboratory.18
The mean residue molar ellipticities [θ]222 (deg cm2/dmol) at wavelength 222 nm were measured at 5°C in benign conditions (100 mM KCl, 50 mM NaH2PO4/Na2HPO4, pH 7.0) or in benign buffer containing 50% TFE by circular dichroism spectroscopy.
PA denotes dimerization parameter of each peptide during RP-HPLC temperature profiling, which is the maximal retention time difference of (tRt – tR5 for peptide analogs) – (tRt – tR5 for control peptide C) within the temperature range; tRt – tR5 is the retention time difference of a peptide at a specific temperature (tRt) compared with that at 5°C (tR5). The sequence of control peptide C is Ac-ELEKGGLEGEKGGKELEK-amide.
tR denotes retention time in RP-HPLC at pH 2 and room temperature, which is a measure of overall hydrophobicity.
The helical content (in percentage) of a peptide relative to the molar ellipticity value of peptide +10 in the presence of 50% TFE.
The continuous hydrophobic surface on the nonpolar face of amphipathic α-helices has been shown to stabilize α-helical structure in benign conditions. However, all peptides in this study contain a lysine residue in the center of this hydrophobic surface, which would as observed (Table II), disrupt α-helix formation in benign conditions. The same lysine residue also prevents self-association, as discussed later. In addition, the peptides that vary in the number of positively charged residues from 4 to 10 decrease in helicity in benign conditions as the net charge is increased (Table II) which could be related to the increase in destabilizing electrostatic repulsions (see Figure 2).
Peptide Self-Association
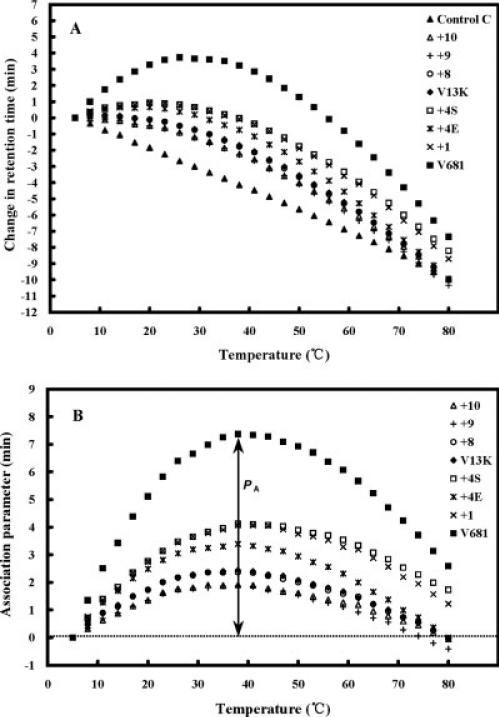
Peptide self-association (i.e. the ability to oligomerize/dimerize) in aqueous solution is a very important parameter for antimicrobial activity.10-13 It is well accepted that the amphipathicity of AMPs is necessary for their mechanism of action, because the positively charged polar face will help the molecules to reach the biomembrane through electrostatic attraction with the negatively charged head groups of phospholipids, and then the nonpolar face of the peptides will allow insertion into the membrane through hydrophobic interactions, causing increased permeability and loss of barrier function of target cells.1 If the self-association ability of a peptide in aqueous media is too strong (e.g., forming stable folded dimers and burying the nonpolar face), it could decrease the ability of the peptide to dissociate and pass through the capsule and cell wall of microorganisms and, hence, prevent penetration into the cytoplasmic membrane to kill target cells. The ability of peptides to self-associate was determined by the technique of RP-HPLC temperature profiling at pH 2.13,26,27 The reason pH 2 is used to determine self-association of cationic AMPs is that highly positively charged peptides are not eluted from reversed-phase columns at pH 7 because of nonspecific binding to negatively charged silanols on the column matrix. This is not a problem at pH 2 since the silanols are protonated and nonspecific interactions are eliminated. At pH 2 the interactions between the peptide and the reversed-phase matrix involve ideal behavior, i.e., only hydrophobic interactions between the preferred binding domain (nonpolar face) of the amphipathic molecule and the hydrophobic surface of the column matrix.28 Interestingly, most native amphipathic cationic peptides (approximately 73%) do contain Asp/Glu residues, which would be negatively charged at pH 7 and might be expected to decrease dimerization at pH 7 compared to pH 2. However, the majority of the peptides in this study do not contain these residues and the dimerization results at pH 2 reflect the conditions at pH 7 where most biological and other biophysical studies are carried out. Figure 4A shows the retention behavior of the peptides after normalization to the retention times at 5°C. Control peptide C shows a linear decrease in the retention behavior with increasing temperature and is representative of most peptides which have no ability to self-associate during RP-HPLC. Control peptide C is a monomeric random coil peptide in both aqueous and hydrophobic media; thus, its retention behavior within the temperature range of 5 to 80°C represents only the general effects of temperature on peptide retention behavior, i.e., a linear decrease in peptide retention time with increasing temperature due to greater solute diffusivity and enhanced mass transfer between the stationary and mobile phase at higher temperatures.29 This method has been used with amphipathic cyclic β-sheet peptides13 and amphipathic α-helical peptides10-12 to measure the ability of peptides to self-associate. How the method works has been described in detail in other publications.10,13,26,27 To allow for the general temperature effects, the data for the control peptide was subtracted from each temperature profile as shown in Figure 4B. Thus, the peptide self-association parameter, PA, represents the maximum change in peptide retention time relative to the random coil peptide C. Note that the higher the PA value, the greater the self-association.
FIGURE 4.
Peptide self-association ability as monitored by RP-HPLC temperature profiling. In panel (A), the retention time of peptides are normalized to 5°C through the expression (tRt – tR5), where tRt is the retention time at a specific temperature of an antimicrobial peptide or control peptide C, and tR5 is the retention time at 5°C. In panel (B), the retention behavior of the peptides was normalized to that of control peptide C through the expression (tRt – tR5 for V13K analogs) – (tRt – tR5 for control peptide C). The maximum change in retention time from the control peptide C defines the peptide association parameter, denoted PA.
In our case, we have a histidine residue at position 18, which is deprotonated at pH 7 (uncharged), but would be fully charged at pH 2 and, hence, add an extra positive charge to the polar face of V13K. Thus, peptides denoted +4S, V13K, +8, +9, and +10, reflecting their net charge at pH 7, will have a net charge of +5, +8, +9, +10, and +11, respectively, at pH 2 during temperature profiling. Interestingly, 38% of native amphipathic α-helical AMPs do contain histidine residues.
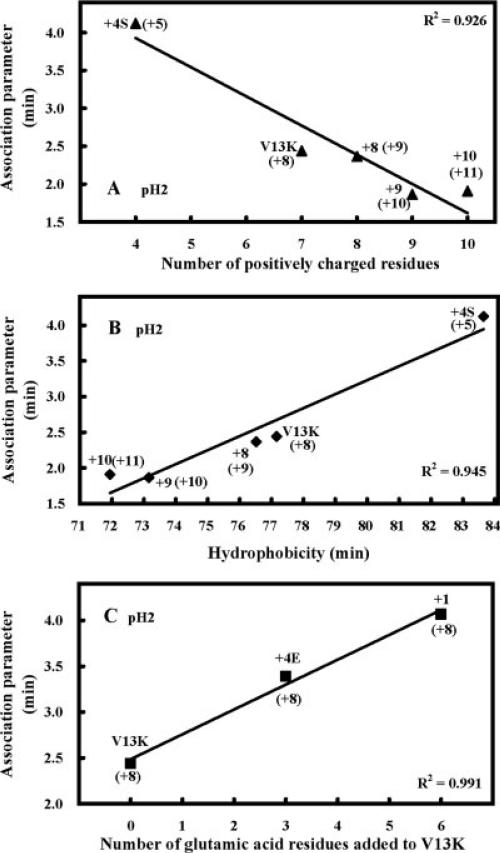
Peptide self-association dramatically decreases with an increase in positive charge even though the nonpolar face of these analogs remained unchanged (Figure 5A, Table II). For example, an increase in net charge from +5 to +11 decreased peptide self-association from 4.1 to 1.9. Interestingly, this decrease directly correlated with the decrease in overall hydrophobicity of the analogs (Figure 5B, Table II). This result shows that overall hydrophobicity decreases with increasing positive charge on the polar face, which in turn decreases peptide self-association. In contrast, adding glutamic acid residues to the polar face of V13K increased peptide self-association from 2.4 for V13K to 4.1 for peptide +1 (Figure 5C, Table II). This can be explained by the fact that these residues are protonated at pH 2 and peptides V13K, +4E, and +1, which have net charges of +7, +4, and +1, respectively, at pH 7 will all have a net charge of +8 at pH 2. Thus, the increase in self-association is due to the increase in hydrophobicity of protonated glutamic acid residues compared to the hydrophobicity of serine and threonine residues.18
FIGURE 5.
Correlation of the number of positively charged residues (A), hydrophobicity (B), and number of glutamic acid residues added to V13K (C) with RP-HPLC association parameter, PA. The numbers in brackets indicate the net charge on the peptide at pH2. Calculation of PA is shown in Figure 4.
Amphipathicity
The sequence of our lead compound, V13K, even with a lysine residue in the center of the nonpolar face is still very amphipathic with a value of 4.92. To place this value in perspective, the sequence can be shuffled at all positions other than lysine 13 to give a amphipathic value of 0.14 (Ac-SHAVLTLKLKKTKKFSWKSAFKATSI-amide) or a maximum value of 5.26 while maintaining the nonpolar face of V13K and shuffling the six positively charged residues on the polar face (Table I) or a maximum value of 6.74 (Ac-AWSKLLKSFTKHKKKFAKALTSIVST-amide) when residues are shuffled to optimized amphipathicity.
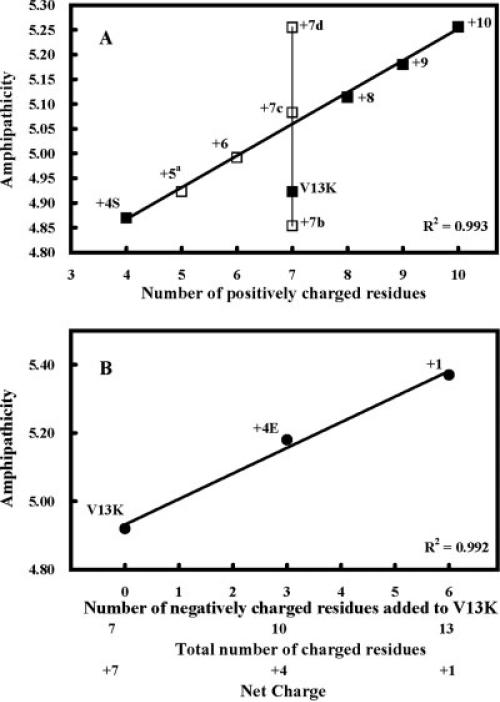
There is an increase in amphipathicity as we increase the net positive charge from +4 (peptide +4S) to +8, +9, and +10 (Figure 6A). Peptide +4S has an amphipathicity of 4.87 and peptide +10 has an amphipathicity of 5.26 for a change of 0.39, which is small compared to the change in amphipathicity from a nonamphipathic molecule (see earlier). This same change in amphipathicity can be observed with +7 peptides where the six positively charged residues are shuffled on the polar face. Amphipathicity changes from 4.85 for peptide +7b to 5.26 for peptide +7d (change 0.41) (Table I, Figure 6A). Thus, though there is a linear relationship with amphipathicity and increasing net charge (Figure 6A), neither amphipathicity nor net charge is linearly related with hemolytic activity (Figure 7B). On the other hand, increasing net charge, which increases amphipathicity does increase antimicrobial activity (Figure 7B).
FIGURE 6.
Correlation of the number of charged residues and net charge with amphipathicity. Panel A shows the number of positively charged residues correlated with amphipathicity. The closed squares denote peptides used in this study. The open squares show the amphipathicity of peptide sequence shown in Table I. Panel B shows the effect of negatively charged residues on amphipathicity.
FIGURE 7.
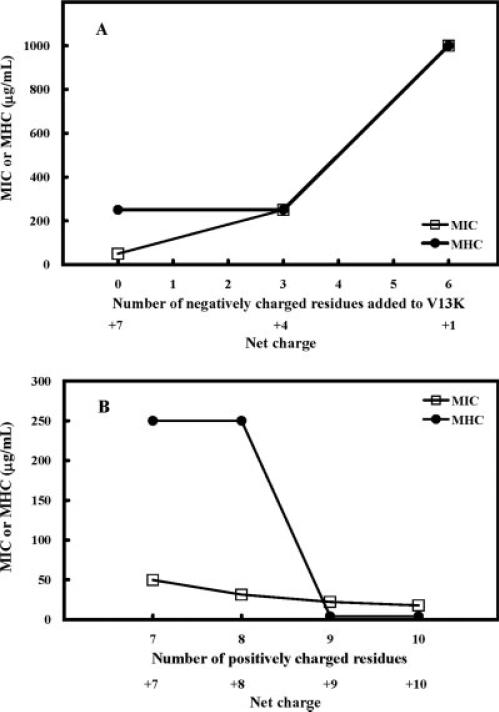
Relationships of number of negatively charged residues added to V13K or net charge (A), number of positively charged residues or net charge (B) and antimicrobial activity (MIC, open symbols), hemolytic activity (MHC, closed symbols).
Amphipathicity increases linearly with increasing number of negatively charged residues added to the polar face (3 or 6 glutamic acid residues) while maintaining the seven positively charged residues (peptides denoted V13K, +4E, and +1, Table I, Figure 6B). This increase in amphipathicity with the addition of six negatively charged residues is small varying from 4.92 for V13K (zero negatively charged residues) to 5.38 for peptide +1 (six negatively charged residues) for a change of 0.46 in amphipathicity. In this case, the increase in amphipathicity is directly correlated with a decrease in net charge from +7 (V13K) to +4 (peptide +4E) to +1. However, though the decrease in net charge results in a dramatic loss of antimicrobial activity and hemolytic activity there is no linear correlation with amphipathicity or net charge and biological activity (Figure 7A).
Overall, increasing amphipathicity is the outcome of increasing the number of charged residues on the polar face or changing the location of the charged residues, but not the key parameter to explain biological activity. The key parameter seems to be net charge where increasing net charge on the polar face of an amphipathic α-helix increases biological activity, both hemolytic and antimicrobial activity (Figures 7A and 7B, see discussion later).
Peptide Hemolytic Activity
The hemolytic activities of the peptides against human erythrocytes were determined as a measure of peptide toxicity toward higher eukaryotic cells.30-32 The MHC is the maximal peptide concentration that produces no hemolysis after 18 h of incubation at 37°C. The lead compound, V13K, exhibited very low hemolysis against human red blood cells with a MHC value of 250 μg/ml after 18 h of incubation at 37°C (Table III), which decreased hemolytic activity more than 32-fold compared to the original peptide, V681 (MHC = 7.8 μg/ml). In Table III, reducing the net charge of V13K to +1 or −5, totally prevented hemolysis as well as eliminated antimicrobial activity. Comparing peptides +4E, V13K, +8, +9, and +10, the hemolytic activity was very low (MHC = 250.0 μg/ml) for peptides with a wide range of net charge (from +4 to +8), but peptide +9, and +10, exhibited very high levels of hemolytic activity (MHC < 7.8 μg/ml). The MHC values decreased by more than 32-fold from the +8 peptide to the +9 peptide, which involved a single T19K substitution on the polar face. Thus, it seems that hemolytic activity is very sensitive to the net charge once the optimum is passed and possibly to the location of the charged residues on the α-helix.
Table III.
Biological Activity of V13K Analogs Against Pseudomonas aeruginosa Strains
| Hemolytic Activity |
Antimicrobial Activity |
Therapeutic Indexa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml)d |
|||||||||||||||
| Peptide Name |
Number of Positively Charged Residues |
Number of Negatively Charged Residues |
Net Charge |
MHC (μg/ml)b |
Foldc | PAO 1 | PAK | CP 204 | PA 14 | WR 5 | M 2 | GMg | Folde | MHC/MIC | Foldf |
| V681 | 6 | 0 | +6 | 7.8 | 0.03 | 15.6 | 31.3 | 15.6 | 62.5 | 62.5 | 15.6 | 27.8 | 1.8 | 0.3 | 0.06 |
| −5 | 1 | 6 | −5 | >1000.0 | 8.0 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | 1000.0 | 0.05 | 2.0 | 0.4 |
| +1 | 7 | 6 | +1 | >1000.0 | 8.0 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | 1000.0 | 0.05 | 2.0 | 0.4 |
| +4S | 4 | 0 | +4 | 125.0 | 0.5 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | >500.0 | 1000.0 | 0.05 | 0.1 | 0.02 |
| +4E | 7 | 3 | +4 | 250.0 | 1.0 | 500.0 | 250.0 | 250.0 | 125.0 | 250.0 | 250.0 | 250.0 | 0.2 | 1.0 | 0.2 |
| V13K | 7 | 0 | +7 | 250.0 | 1.0 | 31.3 | 125.0 | 7.8 | 62.5 | 250.0 | 31.3 | 49.6 | 1.0 | 5.0 | 1.0 |
| +8 | 8 | 0 | +8 | 250.0 | 1.0 | 31.3 | 62.5 | 7.8 | 31.3 | 62.5 | 31.3 | 31.3 | 1.6 | 8.0 | 1.6 |
| +9 | 9 | 0 | +9 | <7.8 | 0.02 | 7.8 | 31.3 | 15.6 | 15.6 | 62.5 | 31.3 | 22.1 | 2.2 | 0.2 | 0.04 |
| +10 | 10 | 0 | +10 | <7.8 | 0.02 | 15.6 | 15.6 | 7.8 | 15.6 | 31.3 | 31.3 | 17.5 | 2.8 | 0.2 | 0.04 |
Therapeutic index is the ratio of the MHC value (μg/ml) over the geometric mean MIC value (μg/ml). Large values indicate greater antimicrobial specificity.
MHC is the maximal peptide concentration that produces no hemolysis of human red blood cells after 18 h in the standard microtiter dilution method. When no detectable hemolytic activity was observed at 1000 μg/ml, a value of 2000 μg/ml was used for calculation of the therapeutic index and fold improvement. When hemolytic activity was still observed at 7.8 μg/ml, a value of 3.9 μg/ml was used for calculation of the therapeutic index and fold improvement.
The fold improvement in hemolytic activity compared to that of lead compound, V13K.
MIC is minimal inhibitory concentration that inhibited growth of six P. aeruginosa strains in brain heart infusion (BHI) medium at 37°C after 24 h. MIC is given based on three sets of determinations. When no detectable antimicrobial activity was observed at 500 μg/ml, a value of 1000 μg/ml was used for calculation of the therapeutic index and fold improvement. The relative susceptibilities of P. aeruginosa strains to ciprofloxacin, with the number 1 denoting the most susceptible strain, are as follows: PAOl, 4; WR5, 1; PAK, 4; PA14, 8; M2, 8; CP204, 64.
The fold improvement in antimicrobial activity (geometric mean data) compared to that of lead compound, V13K.
The fold improvement in therapeutic index compared to that of lead compound, V13K.
GM, geometric mean of the MICs for the six P. aeruginosa clinical isolates.
Peptide Antimicrobial Activity Against Pseudomonas aeruginosa Strains
Pseudomonas is a genus of gram-negative bacteria with high intrinsic resistance to traditional antibiotics; thus, it is one of the most severe threats to human health. Resistance levels have been steadily increasing in recent years, and P. aeruginosa is also known to produce proteolytic enzymes that make it even less susceptible to AMPs.33 P. aeruginosa strains used in this study are a diverse group of clinical isolates from different places in the world (see Experimental Procedures). Antibiotic susceptibility tests show that these P. aeruginosa strains share similar susceptibility to most antibiotics except that there is about a 64-fold difference for the range of ciprofloxacin susceptibility (Table III). The P. aeruginosa strain most resistant to ciprofloxacin was CP204, a clinical isolate from cystic fibrosis patients, was, in contrast, the strain that was most susceptible to our AMPs. Antimicrobial activities of peptide analogs against P. aeruginosa strains are shown in Table III. The geometric means of MICs for six P. aeruginosa strains were calculated to provide an overall evaluation of the antimicrobial activities of the peptides with a different net charge and a different number of positively charged residues. The fold improvement was calculated by comparing the geometric mean MIC of each peptide to that of the lead compound, V13K. In general, reducing the net charge to +4 or less made the peptide analogs inactive (peptide −5, +1, +4S, +4E); increasing the net charge from +7 to +10 (peptide V13K, +8, +9, and +10), the antimicrobial activity increased stepwise (Geometric mean of MIC = 49.6, 31.3, 22.1, 17.5 μg/ml, respectively). Thus, the number of positively charged residues on the polar face and net charge are both important for antimicrobial activity and hemolytic activity. Comparison of peptide +4S and peptide +4E, both with the same net charge of +4 but a different number of positively charged residues, showed that the number of positively charged residues on the polar face is also important. Peptide +4E had seven positively charged residues and had significantly higher antimicrobial activity than peptide +4S with four positively charged residues.
Peptide Antimicrobial Activity Against Other Microorganisms
The antimicrobial activities against four gram-negative bacteria strains and three gram-positive bacteria strains are compared in Table IV. Geometric mean of MIC was calculated to provide an overall view of antimicrobial activity of different analogs. It is clear that our peptides were effective in killing the microorganisms tested, while gram-negative bacteria were more sensitive than gram-positive bacteria. Decreasing the net charge to +4 significantly reduced antimicrobial activity of V13K for both gram-positive and gram-negative bacteria. Increasing net charge on the polar face of V13K analogs from +7 to +10 had little effect on antimicrobial activity. The most active analog was peptide +8 and the least active analog was peptide +4S (Table IV). The MIC values reported for P. aeruginosa PAO1 were different between Tables III and IV, since the assay procedures used are different (see Experimental Procedures).
Table IV.
Biological Activity of V13K Analogs Against Different Gram-Negative (A) and Gram-Positive (B) Bacteria
| Antimicrobial Activity |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemolytic Activity |
MICc (μg/ml) |
Therapeutic Index |
||||||||||
| Peptide Name |
Number of Positively Charged Residues |
Net Charge |
MHCa (μg/ml) |
Foldb |
P. aeruginosa PAOl |
E. coli C498 UB1005 |
E. coli C857 DH5α |
S. typhimurium C587 14208S |
GMg | Foldd | MHC/MICe | Foldf |
| A. Gram-negative bacteria | ||||||||||||
| +4S | 4 | +4 | 125.0 | 0.5 | >64 | 32 | 2 | >64 | 32.0 | 0.1 | 3.9 | 0.1 |
| V13K | 7 | +7 | 250.0 | 1.0 | 8 | 8 | 1 | 4 | 4.0 | 1.0 | 62.5 | 1.0 |
| +8 | 8 | +8 | 250.0 | 1.0 | 4 | 8 | 1 | 4 | 3.4 | 1.2 | 74.3 | 1.2 |
| +9 | 9 | +9 | <7.8 | 0.02 | 8 | 8 | 1 | 4 | 4.0 | 1.0 | 1.0 | 0.02 |
| +10 | 10 | +10 | <7.8 | 0.02 | 4 | 8 | 2 | 4 | 4.0 | 1.0 | 1.0 | 0.02 |
| Antimicrobial Activity |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemolytic Activity |
MICc (μg/ml) |
Therapeutic Index |
|||||||||
| Peptide Name |
Number of Positively Charged Residues |
Net Charge |
MHCa (μg/ml) |
Foldb |
S. aureus C622 ATCC3923 |
S. epidermidis C621 |
B. subtilus C971 ATCC6633 |
GMg | Foldd | MHC/MICe | Foldf |
| B. Gram-positive bacteria | |||||||||||
| +4S | 4 | +4 | 125.0 | 0.5 | >64 | 64 | 32 | 80.6 | 0.5 | 1.6 | 0.3 |
| V13K | 7 | +7 | 250.0 | 1.0 | >64 | 16 | 32 | 40.3 | 1.0 | 6.2 | 1.0 |
| +8 | 8 | +8 | 250.0 | 1.0 | 32 | 8 | 32 | 20.2 | 2.0 | 12.4 | 2.0 |
| +9 | 9 | +9 | <7.8 | 0.02 | >64 | 8 | 32 | 32.0 | 1.3 | 0.1 | 0.02 |
| +10 | 10 | +10 | <7.8 | 0.02 | >64 | 8 | 16 | 25.4 | 1.6 | 0.2 | 0.02 |
MHC is the maximal peptide concentration that produces no hemolysis of human red blood cells after 18 h in the standard microtiter dilution method. When hemolytic activity was still observed at 7.8 μg/ml, a value of 3.9 μg/ml was used for calculation of the therapeutic index and fold improvement.
The fold improvement in hemolytic activity compared to that of lead compound, V13K.
MIC is minimal inhibitory concentration that inhibited growth of different strains in Mueller-Hinton (MH) medium at 37°C after 24 h. MIC is given based on four sets of determinations. When no detectable antimicrobial activity was observed at 64 μg/ml, a value of 128 μg/ml was used for calculation of the therapeutic index and fold improvement.
The fold improvement in antimicrobial activity (geometric mean data) compared to that of lead compound, V13K.
Therapeutic index is the ratio of the MHC value (μg/ml) over the geometric mean MIC value (μg/ml). Large values indicate greater antimicrobial specificity.
The fold improvement in therapeutic index compared to that of lead compound, V13K.
GM, geometric mean of the MIC values.
Peptide Therapeutic Index
The therapeutic index is a widely accepted parameter to characterize the specificity of antimicrobial agents. It is calculated by the ratio of MHC (hemolytic activity) and MIC (antimicrobial activity); thus, larger values of therapeutic index indicate greater antimicrobial specificity. The therapeutic indices of the peptides for P. aeruginosa clinic isolates are shown in Table III. An increase of one positive charge from +8 to +9 dramatically increased hemolytic activity more than 32-fold (250 μg/ml to <7.8 μg/ml) and decreased the therapeutic index by 40-fold (Table III), 74-fold for gram-negative bacteria (Table IVA), and 124-fold for gram-positive bacteria (Table IVB). Our overall conclusion is that there are an optimum number of positively charged residues of seven on the polar face (net charge +8) for an optimum therapeutic index. Reducing the net charge decreased antimicrobial activity, while increasing the net charge increased both antimicrobial activity and hemolytic activity.
DISCUSSION
We previously proposed a “membrane discrimination” mechanism10-12 of action for AMPs whose sole target is the cytoplasmic membrane to explain the peptide specificity between prokaryotic and eukaryotic membranes based on a “barrel-stave” mechanism34 in eukaryotic cells and a “carpet” mechanism35 in prokaryotic cells. This specificity is based upon a compositional difference in the lipids between the two types of membranes. Eukaryotic cell membranes are generally characterized as zwitterionic phospholipids, a relatively large amount of cholesterol and sphingomyelin, while prokaryotic membrane are composed negatively charged phospholipids. Hemolysis of eukaryotic cells requires the peptides to insert into the hydrophobic core of the membrane, perpendicular to the membrane surface, and interaction of the nonpolar face of the amphipathic α-helix with the hydrophobic lipid core of the bilayer. The peptide may thus form transmembrane channels/pores and the hydrophilic surfaces point inward, producing an aqueous pore (“barrel-stave” mechanism).34 Antimicrobial activity in prokaryotic cells, while maintaining specificity, requires the peptide to lie at the membrane interface parallel with the membrane surface and interaction of the nonpolar face of the amphipathic α-helix with the hydrophobic component of the lipid and interaction of the positively charged residues with the negatively charged head groups of the phospholipid, described as detergent-like manner (“carpet” mechanism).35
The cell wall of the microorganisms is another factor that may affect antimicrobial activity. We have proposed that peptides without secondary structure will behave as unstructured random-coil monomers and can more easily pass through the cell wall of microorganisms. In contrast, amphipathic α-helical peptides can dimerize in benign conditions and the folded structure will hinder the molecule from reaching the cytoplasmic membrane.12 In the presence of a hydrophobic environment of the bilayer the unstructured monomeric peptides will be induced into α-helix, which is essential for antimicrobial activity. In this study, all our peptides have a lysine residue in the center of the nonpolar face as a specificity determinant to maintain the peptides as unstructured monomers in benign conditions, but still allow the peptides to be fully induced into α-helix in the hydrophobic environment of the bilayer (see Figure 3).
To date, almost 300 α-helical AMPs have been reported (Antimicrobial Sequences Database, http://www.bbcm.units.it/~tossi/pag2.htm), which vary markedly in positive charge. For example, the peptide ovispirin-1 (KNLRRIIRKIIHIIKKYG) has the highest charge density of any amphipathic α-helical antimicrobial peptide with positively charged residues representing 7/18 or 38.9% and hydrophobes representing 8/18 or 44.4% at pH 7.36 However, this peptide is highly cytotoxic. The majority of native AMPs have a net charge ranging form +4 to +6,37 which may represent an optimal charge for activity.38 According to Han and Kang, two positively charged amino acids (at pH7) lysine and arginine were composed of approximately 20% of the first 20 residues of α-helical AMPs among 221 representative peptides; while the frequencies of two negatively charged amino acids (at pH 7) glutamic acid and aspartic acid were both below 2.3%.39
Various studies on individual cationic α-helical AMPs have shown that net charge can modulate the antimicrobial specificity and efficacy of these peptides.38 However, regression analyses40 showed that there were no apparent general correlations between net charge and MICs (P > 0.05). For example, Giangaspero et al. used an artificial lead AMP to show that decreasing the net charge reduced antimicrobial activity and increasing the net charge increased antimicrobial activity.37 Dathe et al. showed that increased net charge of magainin 2 from +4 to +5 increased antimicrobial activity; further increase to +7 did not alter the maximal activity observed at +5, however, the hemolytic activity increased.41 Scott et al. modulated the charge of cecropin/melittin hybrids from +5 to +9, and found no statistically significant correlation between net charge and the MIC.42 Moreover, some studies further suggested that differences between the localization of positive charge on arginine and lysine side-chains, may be linked to membrane lipid selectivity and thereby contribute to differences in the antimicrobial action of cationic α-helical AMPs.43
In this study, we have clearly shown that net charge and positively charged residues on polar face significantly affects α-helical AMPs’ antimicrobial activity and hemolytic activity. Decreasing the net charge to a lower level (<+4) made V13K analogs totally inactive, both antimicrobial activity and hemolytic activity. Systematically increasing the net charge from +4 to +8 made V13K analogs more active for antimicrobial activity, and maintained low level hemolytic activity (MHC = 250 μg/ml). However, though an increase in net charge to +9 and +10 further improved antimicrobial activity it had a dramatic affect on increasing unwanted hemolytic activity. These results suggest that there is a critical limit to the net positive charge or positive charge density on a given α-helical antimicrobial peptide. In our peptides, the maximum allowed charge density of +8 per 26-residues (30.8% positively charged residues) provides the best therapeutic index while increasing the charge density to +9 per 26-residues (34.6% positively charged residues) dramatically increases hemolytic activity and thus decrease the therapeutic index. Comparing therapeutic indices to different microorganisms, peptide +8 is the best choice for maintaining antimicrobial activity without affecting hemolytic activity.
The dramatic increase in hemolytic activity on increasing the net charge from +8 to +9 (250 μg/ml to <7.8 μg/ml, respectively) can be explained by the increase in net charge favoring the formation of transmembrane channels/pores in eukaryotic cells. In other words, the placement of a lysine residue in the center of nonpolar face destabilizes transmembrane helix formation in human red blood cells. However, the effect of this specificity determinant can be overridden in two ways: (1) increasing the hydrophobicity on the nonpolar face of our lead compound V13K allows transmembrane penetration of the helix by stabilizing pore/channel formation by increase hydrophobic interactions of the hydrophobic surface of the pore/channel with the hydrophobic lipid (see our previous publication12); (2) increasing the net positive charge beyond +8 could stabilize the pore/channel by increasing hydrophilicity of the pore. It seems reasonable to propose that both surfaces of a transmembrane pore/channel could contribute to formation and stability of the such a pore/channel.
Acknowledgments
This research was supported by the John Stewart Chair in peptide chemistry to R.S.H., and the Biophysics Core Facility at the University of Colorado at Denver and Health Sciences Center.
This report is a tribute to and in memory of Robert Bruce Merrifield, Nobel Laureate, who died in May 2006. Dr. Merrifield was the post-doctoral mentor of R.S.H. from 1971 to 1974 at Rockefeller University, New York, N.Y. and introduced Dr. Hodges to the field of peptide chemistry. One area of Dr. Merrifield's research was in the area of antimicrobial peptides where he made major contributions. His leadership and friendship will be greatly missed.
Contract grant sponsor: NIH Contract grant number: R01GM61855 Contract grant sponsors: NIAID Contract grant number: AI 15940
Footnotes
Publisher's Disclaimer: This article was originally published online as an accepted preprint. The “Published Online” date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com
REFERENCES
- 1.Hancock RE, Lehrer R. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE, Chapple DS. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenssen H, Hamill P, Hancock RE. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall SH, Arenas G. Electron J Biotechnol. 2003;6:271–284. [Google Scholar]
- 5.Ge Y, MacDonald DL, Holroyd KJ, Thornsberry C, Wexler H, Zasloff M. Antimicrob Agents Chemother. 1999;43:782–788. doi: 10.1128/aac.43.4.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, Cheng FC, Loury DJ, Fiddes JC. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Parente J, Harris SM, Woods DE, Hancock RE, Falla TJ. Antimicrob Agents Chemother. 2005;49:2921–2927. doi: 10.1128/AAC.49.7.2921-2927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock RE. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 9.Oren Z, Shai Y. Biochemistry. 2000;39:6103–6114. doi: 10.1021/bi992408i. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. J Biol Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Vasil AI, Rehaume L, Mant CT, Burns JL, Vasil ML, Hancock RE, Hodges RS. Chem Biol Drug Des. 2006;67:162–173. doi: 10.1111/j.1747-0285.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Antimicrob Agents Chemother. 2007;51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DL, Mant CT, Hodges RS. J Biol Chem. 2003;278:22918–22927. doi: 10.1074/jbc.M301777200. [DOI] [PubMed] [Google Scholar]
- 14.Lee DL, Hodges RS. Biopolymers. 2003;71:28–48. doi: 10.1002/bip.10374. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Mant CT, Hodges RS. J Chromatogr A. 2007;1140:112–120. doi: 10.1016/j.chroma.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberg D, Weiss RM, Terwilliger TC. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 17.Carver T, Bleasby A. Bioinformatics. 2003;19:1837–1843. doi: 10.1093/bioinformatics/btg251. [DOI] [PubMed] [Google Scholar]
- 18.Kovacs JM, Mant CT, Hodges RS. Biopolymers. 2006;84:283–297. doi: 10.1002/bip.20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway BW. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 20.Bjorn MJ, Vasil ML, Sadoff JC, Iglewski BH. Infect Immun. 1977;16:362–366. doi: 10.1128/iai.16.1.362-366.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlovskis OR, Pollack M, Callahan LT, III, Iglewski BH. Infect Immun. 1977;18:596–602. doi: 10.1128/iai.18.3.596-602.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frost LS, Paranchych W. J Bacteriol. 1977;131:259–269. doi: 10.1128/jb.131.1.259-269.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watts TH, Kay CM, Paranchych W. Can J Biochem. 1982;60:867–872. doi: 10.1139/o82-110. [DOI] [PubMed] [Google Scholar]
- 24.Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, Lau GW, Mahajan-Miklos S, Plotnikova J, Tan MW, Tsongalis J, Walendziewicz CL, Tompkins RG. Proc Natl Acad Sci USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stieritz DD, Holder IA. J Infect Dis. 1975;131:688–691. doi: 10.1093/infdis/131.6.688. [DOI] [PubMed] [Google Scholar]
- 26.Mant CT, Chen Y, Hodges RS. J Chromatogr A. 2003;1009:29–43. doi: 10.1016/s0021-9673(03)00621-6. [DOI] [PubMed] [Google Scholar]
- 27.Mant CT, Tripet B, Hodges RS. J Chromatogr A. 2003;1009:45–59. doi: 10.1016/s0021-9673(03)00919-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhou NE, Mant CT, Hodges RS. Pept Res. 1990;3:8–20. [PubMed] [Google Scholar]
- 29.Dolan JW. J Chromatogr A. 2002;965:195–205. doi: 10.1016/s0021-9673(01)01321-8. [DOI] [PubMed] [Google Scholar]
- 30.Wieprecht T, Dathe M, Beyermann M, Krause E, Maloy WL, MacDonald DL, Bienert M. Biochemistry. 1997;36:6124–6132. doi: 10.1021/bi9619987. [DOI] [PubMed] [Google Scholar]
- 31.Avrahami D, Shai Y. Biochemistry. 2002;41:2254–2263. doi: 10.1021/bi011549t. [DOI] [PubMed] [Google Scholar]
- 32.Tachi T, Epand RF, Epand RM, Matsuzaki K. Biochemistry. 2002;41:10723–10731. doi: 10.1021/bi0256983. [DOI] [PubMed] [Google Scholar]
- 33.Pierce GE. J Ind Microbiol Biotechnol. 2005;32:309–318. doi: 10.1007/s10295-005-0225-2. [DOI] [PubMed] [Google Scholar]
- 34.Ehrenstein G, Lecar H. Q Rev Biophys. 1977;10:1–34. doi: 10.1017/s0033583500000123. [DOI] [PubMed] [Google Scholar]
- 35.Shai Y. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 36.Sawai MV, Waring AJ, Kearney WR, McCray PB, Jr., Forsyth WR, Lehrer RI, Tack BF. Protein Eng. 2002;15:225–232. doi: 10.1093/protein/15.3.225. [DOI] [PubMed] [Google Scholar]
- 37.Giangaspero A, Sandri L, Tossi A. Eur J Biochem. 2001;268:5589–5600. doi: 10.1046/j.1432-1033.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- 38.Tossi A, Sandri L, Giangaspero A. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 39.Han X, Kang W. Bioinformatics. 2004;20:970–973. doi: 10.1093/bioinformatics/bth027. [DOI] [PubMed] [Google Scholar]
- 40.Dennison SR, Harris F, Phoenix DA. Protein Pept Lett. 2003;10:497–502. doi: 10.2174/0929866033478663. [DOI] [PubMed] [Google Scholar]
- 41.Dathe M, Nikolenko H, Meyer J, Beyermann M, Bienert M. FEBS Lett. 2001;501:146–150. doi: 10.1016/s0014-5793(01)02648-5. [DOI] [PubMed] [Google Scholar]
- 42.Scott MG, Yan H, Hancock RE. Infect Immun. 1999;67:2005–2009. doi: 10.1128/iai.67.4.2005-2009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dennison SR, Wallace J, Harris F, Phoenix DA. Protein Pept Lett. 2005;12:31–39. doi: 10.2174/0929866053406084. [DOI] [PubMed] [Google Scholar]