Abstract
DNA damage is known to trigger key cellular defense pathways such as those involved in DNA repair. Here we provide evidence for a previously unrecognized pathway regulating transcription in response to DNA damage and show that this regulation is mediated by the abundant nuclear enzyme poly(ADP-ribose) polymerase. We found that poly(ADP-ribose) polymerase reduced the rate of transcription elongation by RNA polymerase II, suggesting that poly(ADP-ribose) polymerase negatively regulates transcription, possibly through the formation of poly(ADP-ribose) polymerase–RNA complexes. In damaged cells, poly(ADP-ribose) polymerase binds to DNA breaks and automodifies itself in the presence of NAD+, resulting in poly(ADP-ribose) polymerase inactivation. We found that automodification of poly(ADP-ribose) polymerase in response to DNA damage resulted in the up-regulation of transcription, presumably because automodified poly(ADP-ribose) polymerase molecules were released from transcripts, thereby relieving the block on transcription. Because agents that damage DNA damage RNA as well, up-regulation of RNA synthesis in response to DNA damage may provide cells with a mechanism to compensate for the loss of damaged transcripts and may be critical for cell survival after exposure to DNA-damaging agents.
Poly(ADP-ribose) polymerase (PARP) is a highly abundant nuclear protein (for reviews, see refs. 1–6), the physiological role of which is not yet clear. In damaged cells, PARP binds to DNA breaks and becomes enzymatically activated (1, 5). In the presence of its substrate, NAD+, activated PARP automodifies itself through the addition of ADP-ribosyl polymers (1, 5). This automodification inactivates PARP and leads to its dissociation from DNA breaks (7), a prerequisite for DNA repair (8–11). In contrast, in undamaged cells, PARP is found to be associated with regions actively transcribed by RNA polymerase II (POL II) (12), as well as with nucleoli, where ribosomal RNA is transcribed by RNA polymerase I (12). Furthermore, treatment of nuclei with RNase results in the release of PARP (13). In addition, exposure of cells to the transcription inhibitors 5,6-dichloro-1-β-ribofuranosylbenzimidazole or actinomycin D disperses PARP from nucleoli (14). These observations suggest that PARP may play a role in transcription. Here we demonstrate that PARP mediates a link between DNA damage and elongation of transcription by RNA polymerase II.
Materials and Methods
Cell-Free Transcription/DNA Repair Assay.
Lymphoblastoid GMO1953C cells (NIGMS Human Mutant Cell Repository, NJ) were cultured in RPMI medium 1640, and whole cell-free extracts were prepared as described by Manley et al. (15). The reactions were carried out with 50 μg of cell-free extract, 0.125 μg of either pGf1a or pΔGf1a plasmid (16), and 0.125 μg of γ-irradiated pBluescript KS (+) plasmid (pBS; Stratagene), prepared as described previously (10). The transcription–DNA repair assay reaction conditions have been described previously (16). Briefly, the reaction was carried out in a mixture containing Hepes-KOH (pH 7.9), dNTPs, NTPs, [α-32P]CTP, an ATP-regenerating system, and RNase T1 in the presence or absence of 2 mM NAD+ for various lengths of time at 30°C and was terminated by the addition of SDS, EDTA, and proteinase K. After purification of the DNA and RNA by phenol/chloroform extraction and ethanol precipitation, the pellets were resuspended in 10 mM Tris⋅HCl (pH 8.0)/1 mM EDTA. Labeled RNA transcripts were analyzed by either conventional urea gel electrophoresis (6% acrylamide/8 M urea gel), using a Bio-Rad Mini gel apparatus, or by sequencing gel electrophoresis (6% acrylamide/8 M urea gel). Gels were exposed to x-ray film for autoradiography, or radioactive bands were quantified with an InstantImager (Packard).
Depletion of PARP from Whole Cell-Free Extracts.
Depletion of PARP from whole cell-free extracts was carried out essentially as described earlier (10). Briefly, double-stranded DNA-cellulose (0.7-ml bed volume) was packed into a column (1-cm diameter) and washed with 35 ml of 0.3 M NaCl/50 mM Tris⋅HCl (pH 8.0)/10 mM 2-mercaptoethanol/10% glycerol (buffer A). Cell-free extracts (2 mg/ml protein in 1 ml of buffer A) were loaded onto the column and quickly passed through the column by applying high pressure. The column was then washed with 1 ml of buffer A followed by 2 ml of buffer A containing 0.4 M NaCl. Eluted fractions were pooled and concentrated by using Centricon 10 spin columns (Amicon) after dialysis against buffer A. This PARP-depleted fraction was designated Fr 0.4. PARP was then eluted from the column, using buffer A containing 1.0 M NaCl. After dialysis, the eluate was concentrated in a Centricon 10 spin column; this fraction was designated Fr 1.0. Depletion of PARP by this method is about 95% efficient, and recovery of protein in Fr 0.4 is typically 80%.
Preparation of Recombinant PARP.
PARP cDNA was cloned into pET3a (Novagen) and used to transform Escherichia coli HMS174 (DE3) pLys E (Novagen). Expressed PARP was purified by chromatography on a phosphocellulose column, a double-stranded DNA-cellulose column, and a heparin-Sepharose column. The final preparation was free of RNase contamination.
Preparation of Automodified Recombinant PARP.
To prepare automodified PARP, a double-stranded DNA-cellulose column with a 20-μl bed volume was prepared, and 600 ng of human recombinant PARP was applied. After washing with 400 μl of 25 mM Hepes-KOH (pH 7.9)/100 mM KCl/12 mM MgCl2/1 mM EDTA/17% glycerol/2 mM DTT (buffer B), 15 μl of buffer containing 5 mM Tris⋅HCl (pH 8.0)/5 mM MgCl2/250 μM NAD+ was added and incubated for 10 min at 37°C to allow for automodification of PARP. Automodified PARP was then recovered from the column with 15 μl of buffer B, and the protein was quantified.
Plasmid Construction.
A G-less cassette was cloned upstream of the initiation codon for cystic fibrosis transmembrane conductance regulator (CFTR), superoxide dismutase, and firefly luciferase cDNAs. pG-CW5.30 (CFTR), pG-SOD (superoxide dismutase), and pGL3Ad (firefly luciferase) were constructed by replacing the NdeI–PstI sequence of pCW5.30 (17), the EcoRI–SmaI sequence of pT18Kan1 (18), and the KpnI–HindIII sequence of pGL3 (Promega), respectively, with the G-less cassette.
Elongation Assay.
pΔGf1, pG-CW5.30 (CFTR), pG-SOD (superoxide dismutase), and pGL3Ad (firefly luciferase) were digested with PacI (a PacI restriction site is located 18 bases from the +1 position in the G-less cassette), creating a 90-base G-less sequence at one end of the DNA. A C-tail was added by incubating these DNAs with dCTP and terminal transferase. The linearized pG-CW5.30, pG-SOD, and pGL3Ad plasmids were then digested with KpnI, BamHI, and XbaI, respectively, to extract the full-length cDNA with the G-less cassette. pGL3Ad was alternatively digested with ScaI to obtain a 190-base cDNA fragment. The cDNAs were separated from other fragments by ethidium bromide–agarose gel electrophoresis and were used in an elongation assay after extraction from the gel.
Either POL II (40 units/ml) (19) or T7 RNA polymerase (240 units/ml) was allowed to load onto the DNA (15 fmol/ml) from the G-less sequence located at one end of the DNA in the presence of 2 units/μl RNasin/50 μM ATP/50 μM UTP/2 μM CTP/1.3 μCi/μl [α-32P]CTP at 37°C for 30 min as described by Shilatifard et al. (20). A chase was initiated by adding 50 μM GTP and 500 μM CTP in the presence or absence of human recombinant PARP. The reaction was terminated by addition of SDS, EDTA, and proteinase K, and transcripts were analyzed on a sequencing gel (6% acrylamide/8 M urea gel).
Gel Retardation Assay.
The CFTR sequence corresponding to the region of predicted RNA secondary structure (100 bases from the start ATG) was cloned downstream of the T3 RNA polymerase promoter. A 32P-labeled RNA probe was then synthesized using T3 RNA polymerase and used in a gel retardation assay. Probe (2 pmol/ml) was incubated with PARP in a reaction mixture containing 25 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 1 mM MgCl2, 5 mM spermidine, 0.5 mM DTT, 5 mM EDTA and fractionated by native 6% acrylamide gel electrophoresis.
Treatment of HeLa Cells with N-Methyl-N′-nitro-N-nitrosoguanidine (MNNG), Analysis of mRNA, and Western Blotting.
Actively growing HeLa S3 cells were harvested and resuspended in DMEM. Cells (1 × 105) were transferred to 1.5-ml tubes, incubated in the presence or absence of 5 mM 3-aminobenzamide (3AB) for 20 min at 37°C, and treated with 150 μM MNNG for various times. After incubation, cells were spun down by centrifugation at 80 × g for 1 min, and pellets were frozen on dry ice.
Total RNA was extracted with an RNA extraction kit (Promega) and used for mRNA analysis. Total RNA (0.5 μg) was incubated with 100 nmol of oligo(dT) primers (Amersham Pharmacia) in 12 μl of water at 70°C for 10 min, quickly chilled on ice, then combined with 8 μl of a reverse transcriptase reaction mixture containing 2.5× first-strand buffer (GIBCO/BRL), 25 μM DTT, 1,250 μM each dATP, dGTP, and dTTP, 62.5 μM CTP, and 0.1 μCi of [α-32P]CTP. Reactions were initiated by addition of 100 units of Superscript II (GIBCO/BRL) and were incubated for 50 min at 42°C. cDNA was precipitated by ethanol in the presence of ammonium acetate and then electrophoresed on an ethidium bromide/agarose gel (1% agarose). Gels were dried and exposed to x-ray film for autoradiography, or labeled cDNAs were quantified with an InstantImager (Packard).
For Western blotting, frozen cells were lysed with 50 mM Tris⋅HCl (pH 7.5)/500 mM NaCl/10 mM EDTA/10 μM ADP-dihydroxypyrrolidine (21) [an inhibitor of poly(ADP-ribose) glycohydrolase], and samples were used for Western blotting analysis, using the C-II-10 monoclonal antibody against PARP (22).
Results
Promotion of Transcription by Automodification of PARP in a Cell-Free Transcription–Repair Assay.
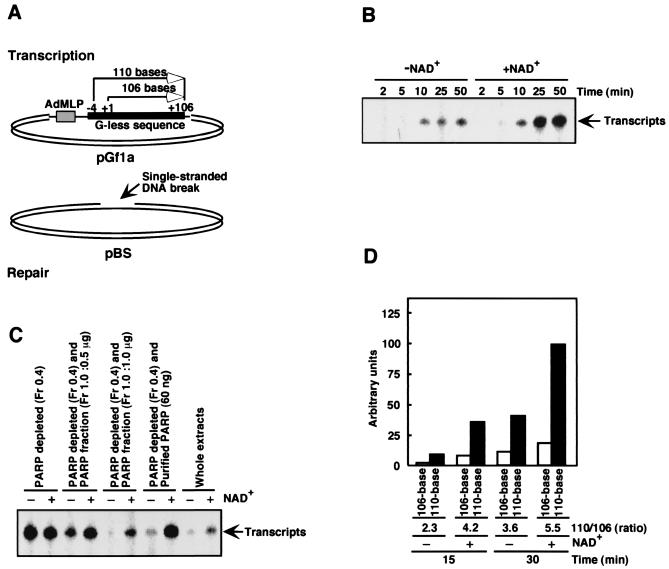
To investigate transcriptional activity (POL II-dependent) in the context of DNA damage and PARP activity, we modified a cell-free transcription–DNA repair assay developed previously (16). This assay relies on the use of two different plasmids (Fig. 1A): undamaged pGf1a containing the adenovirus major late promoter and a G-less cassette for transcription (supercoiled), and damaged pBS containing γ-ray-induced single-stranded DNA breaks (open circular form). These plasmids are incubated with human cell-free extracts in the presence of an ATP-regenerating system, [α-32P]CTP, dNTPs, NTPs, and RNase T1 in the presence or absence of the substrate for PARP, NAD+.
Figure 1.
Cell-free transcription–DNA repair assay. (A) pGf1a plasmid was used as the template for transcription by POL II. pBS plasmid containing γ-ray-induced single-stranded DNA breaks (an average of one per molecule) served as the substrate for DNA repair. (B) Transcription in the presence or absence of 2 mM NAD+. 32P-labeled RNase T1-resistant transcripts generated in the cell-free transcription–DNA repair assay (16) were visualized by autoradiography. (C) Effect on transcription of using PARP-depleted cell-free extracts (4) [0.4 M NaCl eluate (50 μg)] and depleted extracts reconstituted with either partially purified PARP (1.0 M NaCl eluate, 0.5 μg or 1.0 μg) or highly purified PARP from calf thymus [60 ng, obtained from G. G. Poirier (Laval University)] was analyzed. Reactions were carried out for 30 min. (D) The cell-free assay was carried out as described above. Transcripts (110-base and 106-base) were fractionated by sequencing gel electrophoresis and quantified.
After incubation of both plasmids with the cell-free extract, production of RNase T1-resistant transcripts was promoted by 3- to 4-fold by the addition of 0.25 mM (data not shown) or 2 mM (Fig. 1B) NAD+. We also observed NAD+-dependent transcription with the use of a pG-CMV plasmid containing a cytomegalovirus promoter instead of adenovirus major late promoter (data not shown). It is known that POL II can randomly initiate transcription from nicked sites in DNA (23). During the assay, less than 5% of closed circular pGf1 was converted to the open circular form, indicating that negligible amounts of DNA nicking occurred. In addition, when a version of the pGf1a plasmid lacking adenovirus major late promoter (pΔGf1a) was used, only background levels of transcription were observed (data not shown). These results indicate that transcription in our assay is not initiated nonspecifically from DNA nicks.
In the presence of DNA breaks and NAD+, PARP is automodified in this assay (data not shown and refs. 9 and 10). When such automodification was inhibited by 1,5-dihydroxyisoquinoline (24), NAD+ was not sufficient to promote transcription (data not shown). When undamaged pBS plasmid was used instead of damaged pBS, the level of PARP automodification was significantly reduced (about 10% relative to that with damaged pBS), and no apparent promotion of transcription was found with the addition of NAD+ (data not shown). Because automodification inactivates PARP (7), these results suggest that inactivation of PARP is required for transcriptional up-regulation by NAD+.
Reduction of RNA Synthesis by Unmodified PARP.
If inactivation of PARP results in up-regulation of transcription, removal of PARP from cell-free extracts should have a similar effect. We therefore prepared PARP-depleted extracts by double-stranded DNA-cellulose chromatography (10) and found that when these extracts [Fig. 1C, 0.4 M NaCl eluate (Fr 0.4)] were used in the cell-free transcription–DNA repair assay, transcription did in fact increase relative to transcription with whole extracts. The addition of NAD+ had no effect on transcription when the depleted extracts were used (Fig. 1C, Fr 0.4). Adding back crude PARP recovered from double-stranded DNA-cellulose [Fig. 1C, 1.0 M NaCl eluate (Fr 1.0)] or purified PARP (from calf thymus) reduced transcription in the absence of NAD+ and restored the NAD+-dependent promotion of transcription. Therefore, we conclude that unmodified PARP acts to suppress transcription and that inactivation of PARP by automodification in the presence of NAD+ relieves the transcriptional block, resulting in increased transcription.
Reduction of RNA Elongation by Unmodified PARP.
PARP could suppress transcription by POL II either at the level of initiation or at the level of RNA elongation. To distinguish between these two possibilities, we analyzed the ratio of 110-base transcripts to 106-base transcripts produced during the transcription reaction. As illustrated in Fig. 1A, 106-base transcripts (Fig. 1A, +1 to + 106) should be produced when transcription is initiated from adenovirus major late promoter. If unmodified PARP acts to suppress transcription initiation, then the addition of NAD+ should increase the production of 106-base transcripts. In contrast, if PARP suppression acts at the level of elongation, automodification of PARP in the presence of NAD+ should increase the production of 110-base transcripts. In this assay, RNase T1 was typically added to visualize transcripts from the G-less sequence. In the absence of RNase T1, transcripts over 3 kb long were produced (data not shown), suggesting that POL II transcribed the entire circular pGf1 (2.8 kb) and retranscribed the G-less sequence. Because the G-less sequence represents an additional 4 bases (Fig. 1A, −4 to + 106) upstream of the transcription initiation site (Fig. 1A, +1), RNA produced by retranscription of the G-less sequence (110 bases long) in the presence of RNase T1 should represent the effect of PARP on elongation. As seen in Fig. 1D, NAD+ increased production of 110-base transcripts relative to 106-base transcripts, resulting in a ratio of 110-base to 106-base transcripts that was approximately 2-fold higher than the ratio obtained in the absence of NAD+. These results suggest that the suppression by PARP of transcription occurs primarily at the level of transcription elongation. Consistent with our observations, Sawadogo and Roeder (25) reported that PARP has no effect on transcription initiation.
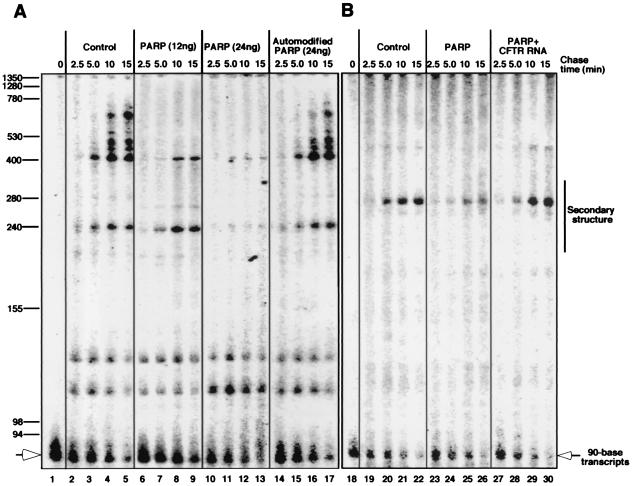
To confirm the results obtained by the assay with cell-free extracts, we performed an elongation pulse-chase experiment, using highly purified POL II (19), recombinant human PARP, and linearized pΔGf1a containing a 90-base G-less cassette at one end. Pulse labeling of transcripts was carried out by loading POL II onto DNA and transcribing the G-less sequence in the presence of ATP, UTP, and [α-32P]CTP, but not GTP, resulting in the production of 90-base transcripts (Fig. 2A, lane 1). A chase was initiated by adding GTP and excess nonradioactive CTP. Discrete bands, formed by pausing of POL II, were visualized (Fig. 2A, lane 2–5). When the chase was carried out in the presence of different amounts of recombinant PARP, synthesis of RNA was reduced in a concentration-dependent manner (lanes 6–9 and 10–13). In addition, similar reductions in transcription were observed when firefly luciferase (data not shown), superoxide dismutase (data not shown), and CFTR cDNAs (Fig. 2B, lanes 19–22 and 23–26) were used. In contrast, as shown in Fig. 2A, addition of purified human recombinant PARP that had been automodified did not reduce elongation (lanes 14–17). Furthermore, the addition of PARP to the reaction mixture had no effect on the elongation of RNA by either T7 RNA polymerase or T3 RNA polymerase (data not shown). Together, these data indicate that PARP can reduce the synthesis of RNA by POL II and that the automodification of PARP relieves this transcriptional inhibition, resulting in increased RNA synthesis.
Figure 2.
Effect of PARP on RNA elongation. (A) Pulse–chase elongation assays using linearized pΔGf1a, highly purified POL II (1.0 unit), and either recombinant PARP (12 or 24 ng) or automodified recombinant PARP (24 ng) were carried out in 25 μl of reaction mixture. During the pulse, 90-base transcripts that were labeled with [α-32P]CMP were generated. Then, the addition of GTP and excess nonradioactive CTP initiated the chase, which was carried out in the presence or absence of PARP. (B) A G-less cassette was cloned upstream of the coding sequence for human CFTR, and the resulting cDNAs were used in a pulse–chase transcription assay. Reactions were also carried out with 40 ng/μl CFTR RNA produced by T3 RNA polymerase.
Interaction of PARP with RNA.
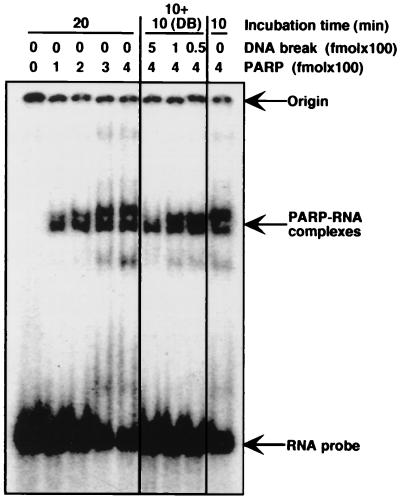
When excess CFTR RNA (synthesized in vitro by T3 RNA polymerase) was added to the elongation–pulse assay, PARP no longer reduced the transcription of CFTR cDNA (Fig. 2B, lanes 27–30), suggesting that the capacity of PARP to block transcription was dependent on an interaction with CFTR RNA. As indicated in Fig. 2B, strong pausing of POL II was observed within a specific region of CFTR RNA. Analysis of CFTR RNA predicts that this region contains a significant secondary structure (see ref. 26 and personal communication with S. Sato and R. R. Kopito). To investigate whether PARP interacts with CFTR RNA secondary structure, gel retardation assays were carried out. As shown in Fig. 3, PARP does in fact appear to interact with the CFTR RNA sequence containing the region of predicted secondary structure. Consistent with the fact that this secondary structure is predicted to contain two stem loops, two retarded bands were observed. When RNA from a TAR sequence (containing one RNA stem loop) from HIV-1 was used, only one discrete retarded band was found (data not shown).
Figure 3.
Analysis of PARP–RNA complexes by gel retardation. 32P-labeled RNA stem loop (20 fmol) synthesized from CFTR cDNA was incubated with various amounts of PARP for the indicated times at 30°C in 10 μl of reaction mixture and was analyzed by 6% acrylamide gel electrophoresis. For analysis in the presence of DNA breaks (DB) (λ-DNA–BstEII digest), PARP–RNA complexes were preformed (10-min incubation) and incubated for a further 10 min after the addition of DNA breaks.
When shorter firefly luciferase cDNA, which contained only minor pause sites, was prepared and used in the pulse–chase elongation assay, over 5 times more PARP was required to reduce transcription compared with the full-length firefly luciferase cDNA (data not shown), further suggesting that pause sites are involved in the capacity of PARP to inhibit transcription. On the other hand, the presence of minor pause sites may give rise to the faint smears seen in Fig. 2 and contribute to transcriptional inhibition by PARP, although transcripts with minor pause sites seem to have less affinity for PARP than those with major pause sites. It has been demonstrated that RNA secondary structure is involved in pausing of POL II on transcripts (for a review, see ref. 27). Thus these data suggest a scenario in which PARP inhibits POL II transcription by binding to RNA stem loops.
After the formation of PARP–RNA complexes, the addition of DNA breaks resulted in the resolution of such complexes (Fig. 3). Because PARP has been demonstrated to have a significantly higher affinity for DNA breaks than for RNA (28), PARP complexed with RNA likely moves to DNA breaks. In the presence of NAD+, the DNA-bound PARP then becomes automodified, resulting in dissociation from DNA breaks and inhibiting rebinding of PARP to RNA. Damage-dependent up-regulation of transcription may therefore occur in two steps, with resolution of PARP–RNA complexes followed by inactivation of PARP through automodification (see Discussion).
Up-Regulation of Transcription by Automodification of PARP in Vivo.
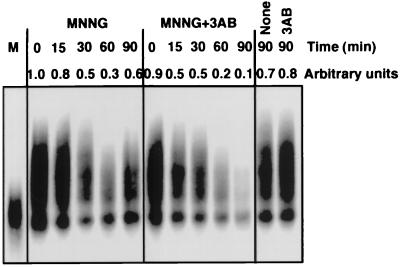
To investigate transcriptional up-regulation by PARP in vivo, we analyzed mRNA synthesis after treatment of cells with the DNA-damaging agent MNNG in the presence or absence of an inhibitor of PARP, 3AB. To avoid any complication of analysis arising from the activation of the apoptotic cell death pathway, we used HeLa cells, which are less sensitive to the induction of apoptosis in response to DNA damage due to the repression of p53 (29). Over 80% of PARP molecules appeared to be automodified after treatment of cells with MNNG when analyzed by Western blotting, and this automodification was effectively inhibited by 3AB (data not shown).
mRNA transcripts from HeLa cells were then analyzed. Total RNA was extracted and cDNA molecules were generated, using reverse transcriptase to initiate synthesis from oligo(dT) primers. cDNAs of a variety of lengths were synthesized from control RNA (Fig. 4, 0 min). When RNA extracted from MNNG-treated HeLa cells was reverse-transcribed, the resulting cDNAs were found to be reduced in length compared with control lengths. Treatment of RNA with MNNG in vitro also reduced the lengths of cDNA observed (Fig. 4, lane M), suggesting that RNA damage was responsible for the inhibition of cDNA synthesis by reverse transcriptase. A reduction in cDNA lengths was also observed under conditions where cells were treated with MNNG and where PARP activity was inhibited by 3AB. However, in the case of cellular exposure to MNNG alone, the cDNA molecules synthesized were measurably longer than those generated from cells exposed to MNNG together with 3AB (Fig. 4). 3AB itself had no effect on the size of cDNA molecules observed (Fig. 4, 3AB, 90 min). Identical results were obtained with 1,5-dihydroxyisoquinoline, another inhibitor of PARP (data not shown; ref. 24). Therefore, we conclude that enzymatically active PARP is responsible for the difference in cDNA lengths observed, presumably by promoting overall levels of mRNA synthesis in MNNG-damaged cells. These results agree well with our in vitro results.
Figure 4.
Effect of DNA damage and PARP inhibition in vivo. Total RNA was extracted from HeLa S3 cells treated with MNNG (150 μM), 3AB (5 mM), or MNNG together with 3AB. 32P-labeled cDNA was synthesized from mRNA transcripts by reverse transcription, using oligo(dT) as a primer. The resulting cDNA molecules were then fractionated on an ethidium bromide/agarose gel (1% agarose), and labeled cDNAs were visualized by autoradiography. As a positive control, mRNA extracted from nontreated HeLa S3 cells (MNNG, 0 min) was exposed to 150 μM MNNG in vitro for 30 min and reverse-transcribed (lane M).
Discussion
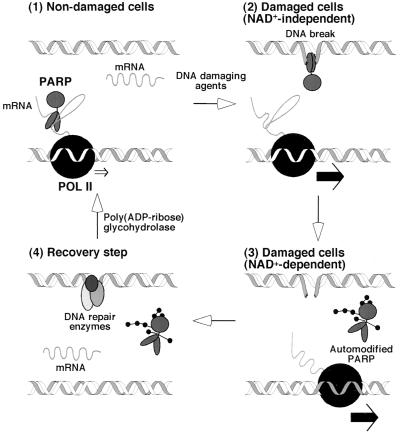
In this report, we have demonstrated that transcription elongation is negatively regulated by PARP in the absence of DNA breaks and is up-regulated in the presence of DNA breaks and NAD+. Previously, a model for the role of PARP in DNA repair was proposed (10); we have modified this model to explain our results as summarized in Fig. 5.
Figure 5.
Model for poly(ADP-ribosyl)ation-mediated up-regulation of transcription. See text for details.
(i) In undamaged cells, mRNA synthesis is negatively regulated by PARP–RNA complexes at the level of elongation. (ii) When DNA breaks are generated, PARP–RNA complexes are resolved as a consequence of the fact that PARP has a significantly higher affinity for DNA breaks than for RNA (27) (NAD+-independent). (iii) If PARP were to persist on DNA breaks, DNA repair would be inhibited (10). However, binding of PARP to DNA breaks activates PARP in the presence of NAD+, promoting extensive automodification (over 200 residues of ADP-ribose conferred on a single PARP molecule) and resulting in the dissociation of PARP from the DNA strand (NAD+-dependent) (1, 7, 10). The release of automodified PARP from DNA breaks allows DNA repair to begin (10), and the automodified PARP is likewise prevented from binding to RNA. Thus mRNA synthesis is up-regulated. (iv) After DNA repair, the ADP-ribose polymers are degraded by poly(ADP-ribose) glycohydrolase, regenerating unmodified PARP (1, 7), which is capable of binding to RNA once more. A significant population of automodified PARP molecules is probably required for PARP to regulate the overall level of transcription by such a process. In fact, PARP is highly abundant in nuclei, and, in response to DNA damage, a significant portion of PARP molecules have been shown to be poly(ADP-ribosyl)ated (see ref. 1 and this work).
In connection to nucleosomes, ADP-ribose polymers are found on core histones in damaged cells (30, 31). In addition, histone H1 (32, 33) and high mobility group proteins (34) are also often poly(ADP-ribosyl)ated. It has been demonstrated that poly(ADP-ribosyl)ation of histone results in opening up of the chromatin structure (35, 36). Acetylation of histones and high mobility group proteins is also known to alter chromatin structure in this way. Deacetylation results in transcriptional repression, so an open chromatin structure is thought to be critical for maintaining transcription (for reviews, see refs. 37–39). Poly(ADP-ribosyl)ation of histones and high mobility group proteins in response to DNA damage may act to regulate transcription in a similar manner.
Because RNA and DNA are chemically similar, RNA is sensitive to many agents known to damage DNA. Thus a mechanism that acts to promote overall levels of transcription may enable cells to compensate for the collateral loss of mRNA after exposure of cells to DNA-damaging agents. The physiological consequences of RNA damage are not well understood, although cytotoxicity is likely to result from the depletion of biologically active mRNA molecules. Thus the pathway described above may serve as part of an overall defense mechanism for cell survival after exposure to DNA-damaging agents. In fact, inhibition of PARP can sensitize cells to DNA-damaging agents (1). More recently, PARP-deficient cells have been shown to be sensitive to exposure to DNA-damaging agents (40, 41), despite the fact that these cells contain normal DNA repair activity (M. D. Vodenicharov, F. R. Sallman, Z.-Q. Wang, M.S.S., and G. G. Poirier, unpublished work; ref. 42). Thus the capacity of cells to compensate for the loss of damaged RNA through PARP activation downstream of DNA damage may help cells counteract the cytotoxic effects of mutagenic agents and anticancer drugs.
Acknowledgments
We thank Philip C. Hanawalt for allowing initiation of this research during the postdoctoral training of M.S.S., Tomas Lindahl for critical reading and various comments, Cristina Ward for editing and scientific comments, Sachiko Sato for discussion, and Ron R. Kopito for providing information related to CFTR RNA secondary structure. This work was supported by the National Cancer Institute of Canada with funds from the Terry Fox Run (for M.S.S.) and in part by the American Cancer Society and the American Heart Association (Heartland Affiliate) (for H.S.). M.S.S. is supported by a salary support award (scholarship) from the Medical Research Council of Canada. The Canada Foundation for Innovation and the Quebec government provided infrastructure support.
Abbreviations
- PARP
poly(ADP-ribose) polymerase
- POL II
RNA polymerase II
- CFTR
cystic fibrosis transmembrane conductance regulator
- 3AB
3-aminobenzamide
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170280397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170280397
References
- 1.Althaus F R, Richter C. ADP-Ribosylation of Proteins: Enzymology and Biological Significance. Berlin: Springer-Verlag; 1987. [PubMed] [Google Scholar]
- 2.de Murcia G, Ménissier de Murcia J. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 3.Jeggo P A. Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 4.Le Rhun Y, Kirkland J B, Shah G M. Biochem Biophys Res Commun. 1998;245:1–10. doi: 10.1006/bbrc.1998.8257. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T, Satoh M S, Poirier G G, Klungland A. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 7.Zahradka P, Ebisuzaki K. Eur J Biochem. 1982;127:579–585. [PubMed] [Google Scholar]
- 8.Smulson M, Istock N, Ding R, Cherney B. Biochemistry. 1994;33:6186–6191. doi: 10.1021/bi00186a018. [DOI] [PubMed] [Google Scholar]
- 9.Satoh M S, Poirier G G, Lindahl T. J Biol Chem. 1993;268:5480–5487. [PubMed] [Google Scholar]
- 10.Satoh M S, Lindahl T. Nature (London) 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 11.Molinete M, Vermeulen W, Burkle A, Ménissier-de Murcia J, Küpper J H, Hoeijmakers J H J, de Murcia G. EMBO J. 1993;12:2109–2118. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakan S, Leduc Y, Lamarre D, Brunet G, Poirier G G. Exp Cell Res. 1988;179:517–526. doi: 10.1016/0014-4827(88)90289-3. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann S H, Brunet G, Talbot B, Lamarr D, Dumas C, Shaper J H, Poirier G G. Exp Cell Res. 1991;192:524–535. doi: 10.1016/0014-4827(91)90072-3. [DOI] [PubMed] [Google Scholar]
- 14.Desnoyers S, Kaufmann S H, Poirier G G. Exp Cell Res. 1996;227:146–153. doi: 10.1006/excr.1996.0259. [DOI] [PubMed] [Google Scholar]
- 15.Manley J L, Fire A, Samuels M, Sharp P A. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 16.Satoh M S, Hanawalt P C. Nucleic Acids Res. 1996;24:3576–3582. doi: 10.1093/nar/24.18.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 18.Renard P, Zachary M D, Bougelet C, Mirault M E, Haegeman G, Remacle J, Raes M. Biochem Pharmacol. 1997;53:149–160. doi: 10.1016/s0006-2952(96)00645-4. [DOI] [PubMed] [Google Scholar]
- 19.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Nature (London) 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 20.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 21.Slama J T, Simmons A M, Hassan M E, Aboul-Ela N, Jacobson M K. In: ADP-Ribosylation Reactions. Poirier G G, Moreau P, editors. Berlin: Springer; 1992. pp. 316–320. [Google Scholar]
- 22.Satoh M S, Poirier G G, Lindahl T. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 23.Slattery E, Dignam J D, Matsui T, Roeder R G. J Biol Chem. 1983;258:5955–5959. [PubMed] [Google Scholar]
- 24.Shah G M, Poirier D, Desnoyers S, Saint-Martin S, Hoflack J C, Rong P, ApSimon M, Kirkland J B, Poirier G G. Biochim Biophys Acta. 1996;1312:1–7. doi: 10.1016/0167-4889(96)00004-3. [DOI] [PubMed] [Google Scholar]
- 25.Sawadogo M, Roeder R D. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato S, Ward C L, Kopito R R. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 27.Uptain S M, Kane C M, Chamberlin M J. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 28.Ohgushi H, Yoshihara K, Kamiya T. J Biol Chem. 1980;255:6205–6211. [PubMed] [Google Scholar]
- 29.Hoppe-Seyler F, Butz K. J Virol. 1993;67:3111–3117. doi: 10.1128/jvi.67.6.3111-3117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krupitza G, Cerutti P. Biochemistry. 1989;28:4054–4060. doi: 10.1021/bi00435a063. [DOI] [PubMed] [Google Scholar]
- 31.Adamietz P, Rudolph A. J Biol Chem. 1984;259:6841–6846. [PubMed] [Google Scholar]
- 32.Aubin R J, Dam V T, Miclette J, Brousseau Y, Huletsky A, Poirier G G. Can J Biochem. 1982;60:1085–1094. doi: 10.1139/o82-139. [DOI] [PubMed] [Google Scholar]
- 33.Adamietz P, Bredehorst R, Hilz H. Eur J Biochem. 1978;91:317–326. doi: 10.1111/j.1432-1033.1978.tb12682.x. [DOI] [PubMed] [Google Scholar]
- 34.Tanuma S, Yagi T, Johnson G S. Arch Biochem Biophys. 1985;237:38–42. doi: 10.1016/0003-9861(85)90251-6. [DOI] [PubMed] [Google Scholar]
- 35.Poirier G G, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Proc Natl Acad Sci USA. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aubin R J, Fréchette A, de Murcia G, Mandel P, Lord A, Grondin G, Poirier G G. EMBO J. 1983;2:1685–1693. doi: 10.1002/j.1460-2075.1983.tb01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howe L, Brown C E, Lechner T, Workman J L. Crit Rev Eukaryotic Gene Expression. 1999;9:231–243. doi: 10.1615/critreveukargeneexpr.v9.i3-4.80. [DOI] [PubMed] [Google Scholar]
- 38.Cheung W L, Briggs S D, Allis C D. Curr Opin Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 39.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Murcia J M, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, et al. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E W. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]