Summary
Oxidative stress has been hypothesized to play a role in normal aging. The SKN-1 transcription factor regulates the response to oxidative stress and also is necessary for intestinal development in Caenorhabditis elegans. Using transcriptome analysis, we found that oxidative stress induces almost a thousand genes, including the antioxidant and heat-shock responses, as well as genes responsible for xenobiotic detoxification. There were also 392 down-regulated genes including many involved in metabolic homeostasis, organismal development, and reproduction. Many of these oxidative-stress-induced transcriptional changes are dependent on SKN-1 action; the induction of the heat-shock response is not. When we used RNAi to inhibit genes, we found that most had no effect on either resistance to oxidative stress or longevity; however two SKN-1-dependent genes, nlp-7 and cup-4, that were up-regulated by oxidative stress were found to be required for resistance to oxidative stress and for normal life span. nlp-7 encodes a neuropeptide-like protein, expressed in neurons, while cup-4 encodes a coelomocyte-specific, ligand-gated ion channel. RNAi of nlp-7 or cup-4 increased sensitivity to oxidative stress and reduced lifespan. Among down-regulated genes, only inhibition of ent-1, a nucleoside transporter, led to increased resistance to oxidative stress; inhibition had no effect on lifespan. In contrast, RNAi of nhx-2, a Na+/H+ exchanger, extended lifespan significantly without affecting sensitivity to oxidative stress. These findings show that oxidative stress causes a transcriptional shift from growth and maintenance towards the activation of cellular defense mechanisms; many of these transcriptional alterations are SKN-1-dependent.
Keywords: oxidative stress, C. elegans, microarray, SKN-1, aging, longevity
Introduction
Extensive studies suggest that reactive oxygen species (ROS), byproducts of cellular respiration, play a role in normal aging by causing random deleterious oxidative damage to a variety of tissues (Harman, 1956; Muller et al., 2007). This “oxidative stress theory of aging” explains several aspects of aging, including mitochondrial decline, genomic instability, and telomere shortening (Muller et al., 2007). A considerable body of knowledge in C. elegans suggests that aging may be caused by oxidative stress. Longevity genes also mediate increased resistance to oxidative stress; long-lived dauer mutants show increased resistance to oxidative stress and up-regulation of antioxidant genes (Honda & Honda, 1999; Johnson et al., 2002; Larsen, 1993; Murphy et al., 2003; Yanase et al., 2002). Overexpression of daf-16, a transcription factor mediating most longevity pathways in the worm, also results in increased resistance to oxidative stress in addition to extending lifespan (Henderson & Johnson, 2001). OLD-1, a receptor tyrosine kinase, confers life extension and increased oxidative-stress resistance phenotypes when overexpressed in C. elegans (Murakami & Johnson, 1998; Murakami & Johnson, 2001). Extra copies of hsp-16 also increase both lifespan and oxidative-stress resistance in a daf-16-dependent manner (Walker & Lithgow, 2003). Many C. elegans mutants selected as being resistant to juglone, a free radical generator, show increased mean and maximum lifespan (de Castro et al., 2004). Finally, knockout of sod-1 increased the level of superoxide radicals and shortened the lifespan in C. elegans (Yanase et al., 2008).
However, failing to support the role of oxidative stress in aging in C. elegans are the facts that overexpression of antioxidant enzymes, catalase (CTL) and/or superoxide dismutase (SOD), failed to increase lifespan (Finkel & Holbrook, 2000). Increased expression of gst-4 resulted in increased resistance to various stressors including oxidative stress, but had no effect on lifespan (Leiers et al., 2003). Thus, the role of oxidative stress in causing aging and determining the lifespan in the nematode remains unclear (Muller et al., 2007).
Herein we examine the role of SKN-1 in response to an oxidative stressor, hyperbaric oxygen. SKN-1 is a transcription factor required for response to oxidative stress; SKN-1 activation induces the expression of genes involved in oxidative-stress response, including CTLs, SODs, and several glutathione S-transferases (GSTs) (An et al., 2005). Curiously, skn-1 also is required for intestine development in C. elegans (An & Blackwell, 2003). In adult worms, SKN-1 is mainly expressed in the ASI neurons and in the intestine (Bishop & Guarente, 2007). Oxidative stress stimulates translocation of SKN-1 to the nucleus in a process regulated by several protein kinases, including glycogen synthase kinase-3 (GSK-3), p38 mitogen-activated protein kinase-1 (PMK-1), and four additional kinases required for nuclear localization of SKN-1 in response to oxidative stress: MKK-4, IKKε-1, NEKL-2, and PDHK-2, which were identified through a large scale RNAi screen (Kell et al., 2007). A recent study showed that RNAi knockdown of proteasome core subunits also causes nuclear localization of SKN-1 (Kahn et al., 2007).
SKN-1 also modulates lifespan-extension in addition to stress resistance. skn-1 mutants show decreased resistance to oxidative stress and shortened lifespan, while over-expression of a mutant SKN-1 that constitutively localizes to the nuclei of the intestine leads to increased resistance to oxidative stress and increased longevity (An & Blackwell, 2003; An et al., 2005; Tullet et al., 2008). Reduced insulin/IGF-1 signaling (IIS) also causes nuclear accumulation of SKN-1; the increased stress resistance and lifespan of long-lived daf-2 mutants require nuclear localization of SKN-1 (Tullet et al., 2008). Neuronal expression of SKN-1 is also involved in lifespan-extension in C. elegans by dietary restriction (DR). Recently, Bishop et al. showed that SKN-1 activation in two ASI neurons is required for DR-induced lifespan extension (Bishop & Guarente, 2007); skn-1 mutants failed to show a DR-induced longevity effect and over-expression of SKN-1 in ASI neurons, but not in intestine, rescued the DR-induced longevity in skn-1 mutants (Bishop & Guarente, 2007).
Here, we provide a global gene-expression profile of the response to oxidative stress in adult C. elegans using high-density oligonucleotide microarrays. We also examine the role of SKN-1 in regulating this response and test the involvement of targets of SKN-1 in specifying resistance to oxidative stress and in determining longevity. We find that the expression of nlp-7, a neuropeptide-like protein (NLP) and cup-4, a coelomocyte-specific ion channel, are required for normal lifespan and that knockdown of nhx-2, a Na+/H+ exchanger, can increase the lifespan of C. elegans.
Results
Transcriptional profile of oxidative stress caused by hyperbaric oxygen
We compared transcripts from unstressed hermaphrodites with those of hermaphrodites stressed with hyperbaric oxygen (99% O2, 40 PSI, 6 hours). We used a sterile strain, TJ1060 [fer-15(b26); spe-9(hc88)] to eliminate transcripts from developmental stages. Of 22,548 transcripts surveyed, 13,652 (61%) transcripts were expressed in controls and 14,063 (62%) in oxidative-stressed animals (more than one present or marginal call by Affymetrix algorithm). Of these, 948 transcripts were up-regulated and 392 transcripts were decreased statistically significantly by oxidative stress as assessed by SAM (Significance Analysis of Microarray) analysis (false discovery rate (FDR) = 11%) (Tusher et al., 2001). We then classified the 948 up-regulated genes according to their biological functions using DAVID (Database for Annotation, Visualization and Integrated Discovery) (http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003). Many genes involved in cellular response to stress were induced (Table 1). Seven antioxidant-response genes, including ctl-2, ctl-3, and sod-1, and six heat-shock transcripts were significantly up-regulated in oxidatively-stressed worms. Many genes encoding GST and cytochrome P450 family members showed a significant increase in expression. Induction of genes involved in neuropeptide signaling, such as nlp-6, nlp-7, nlp-14, and nlp-16, was also observed (Table 1 and Table S1). Among the 392 down-regulated genes were genes involved in cellular homeostasis, especially osmoregulation (Table 2 and Table S2). Genes in pathways associated with development, reproduction, and growth were also reduced in expression. However, genes involved in three different biological functions, cellular localization, metabolic transport, and metabolism/catabolism, were included in both up-regulated and down-regulated classes of genes.
Table 1.
Functional classification of genes up-regulated by oxidative stress
| GO Term | Number of Genes† | P‡ |
|---|---|---|
| Antioxidant Response | ||
| antioxidant activity | 7 | 0.004 |
| oxygen and reactive oxygen species metabolism | 5 | 0.017 |
| response to oxidative stress | 4 | 0.044 |
| GSTs | ||
| glutathione transferase activity | 7 | 0.000 |
| glutathione S-transferase, N-terminal | 20 | 0.000 |
| glutathione S-transferase, C-terminal | 20 | 0.000 |
| Heat Shock Response | ||
| heat shock protein Hsp20 | 6 | 0.001 |
| Electron Transport | ||
| generation of precursor metabolites and energy | 34 | 0.000 |
| electron transport | 32 | 0.000 |
| Neropeptide Signaling | ||
| neuropeptide signaling pathway | 6 | 0.017 |
| Localization | ||
| establishment of localization | 104 | 0.000 |
| Transport | ||
| vesicle-mediated transport | 7 | 0.035 |
| metal ion transport | 16 | 0.039 |
| transition metal ion transport | 4 | 0.023 |
| potassium ion transport | 11 | 0.048 |
| copper ion transport | 4 | 0.012 |
| Metabolism/Catabolism | ||
| Amino acid metabolism | 13 | 0.006 |
| Amino acid and derivative metabolism | 13 | 0.010 |
| Amine metabolism | 14 | 0.017 |
| aromatic compound metabolism | 9 | 0.002 |
| aromatic amino acid family metabolism | 3 | 0.042 |
| fatty acid metabolism | 6 | 0.043 |
| lipid metabolism | 18 | 0.008 |
| nitrogen compound metabolism | 15 | 0.008 |
| carboxylic acid metabolism | 22 | 0.000 |
| organic acid metabolism | 22 | 0.000 |
| Catabolism | 15 | 0.040 |
Selected functional classes of genes that are significantly increased in expression under oxidative stress conditions in adult worms. The number of classified genes was determined using DAVID.
Number of genes called.
Modified Fisher Exact as determined by DAVID.
Table 2.
Functional classification of genes down-regulated by oxidative stress
| GO Term | Number of Genes† | P‡ |
|---|---|---|
| Homeostasis | ||
| homeostasis | 12 | 0.018 |
| osmoregulation | 11 | 0.017 |
| Development | ||
| development | 97 | 0.000 |
| embryonic development | 82 | 0.000 |
| morphogenesis | 38 | 0.001 |
| post-embryonic morphogenesis | 17 | 0.003 |
| post-embryonic development | 20 | 0.002 |
| organismal biosynthesis | 5 | 0.010 |
| Reproduction | ||
| reproduction | 51 | 0.022 |
| sexual reproduction | 27 | 0.018 |
| gametogenesis | 26 | 0.017 |
| Growth | ||
| regulation of growth | 57 | 0.000 |
| regulation of growth rate | 50 | 0.001 |
| positive regulation of growth | 53 | 0.002 |
| positive regulation of growth rate | 48 | 0.002 |
| Behavior | ||
| behavior | 38 | 0.029 |
| locomotory behavior | 34 | 0.025 |
| Cuticle Biosynthesis | ||
| cuticle biosynthesis | 5 | 0.010 |
| Localization | ||
| establishment of localization | 65 | 0.002 |
| Transport | ||
| ion transport | 27 | 0.014 |
| Anion transport | 21 | 0.000 |
| inorganic anion transport | 20 | 0.000 |
| phosphate transport | 20 | 0.000 |
| Metabolism/Catabolism | ||
| amino acid metabolism | 11 | 0.002 |
| amino acid and derivative metabolism | 11 | 0.003 |
| amine metabolism | 11 | 0.011 |
| fatty acid oxidation | 3 | 0.029 |
| nitrogen compound metabolism | 11 | 0.013 |
| carboxylic acid metabolism | 14 | 0.002 |
| organic acid metabolism | 14 | 0.002 |
| serine family amino acid metabolism | 5 | 0.001 |
| serine family amino acid catabolism | 3 | 0.010 |
| glycine metabolism | 4 | 0.001 |
| glycine catabolism | 3 | 0.010 |
Selected functional classes of genes that are significantly decreased in expression under oxidative stress conditions in adult worms. The number of classified genes was determined using DAVID.
Number of genes called.
Modified Fisher Exact as determined by DAVID.
To validate the transcriptional alterations observed in microarray analysis, quantitative RT-PCR was performed for some selected genes. Induction of gst-4, hsp-16.2, nlp-7, and cup-4 by oxidative stress was also detected in quantitative RT-PCR. The expression of nhx-2 and ent-1 decreased consistently in both microarray analysis and quantitative RT-PCR (Table S3).
Comparisons with other relevant transcriptional profiles
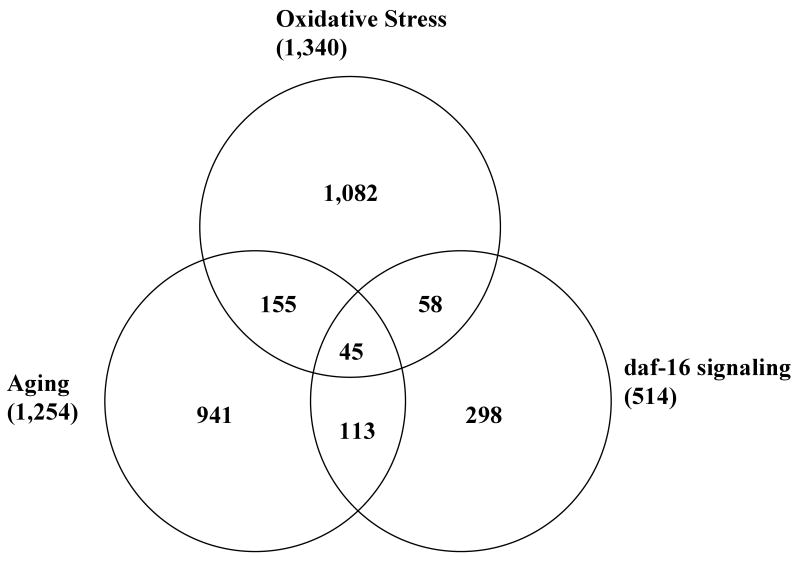
We compared our microarray data with other relevant transcriptional profiles to find overlaps. First, we looked for overlaps between genes regulated by oxidative stress and those that change during aging. A recent transcriptional profile revealed 1,254 genes that change in expression over the C. elegans lifespan (Budovskaya et al., 2008). Among these, 200 genes were also regulated during oxidative stress; 134 genes were up-regulated and 66 genes were down-regulated by oxidative stress (Fig. 1 and Table S4).
Fig. 1.
Venn diagram of differential expression by oxidative stress, aging, and daf-16. Number of overlapping genes in each comparison is greater than expected by chance (P ≪ 0.001 by Fisher's exact test). The representation factors of each comparison are 2.3 for oxidative stress and aging, 2.9 for oxidative stress and daf-16, and 4.8 for aging and daf-16. Lists of overlapping genes are provided as supporting information.
We also looked for overlap with the 514 genes that Murphy et al. (2003) found to be modulated by DAF-16; 89 of the genes that we found to be up-regulated under oxidative-stress conditions overlapped with genes regulated by DAF-16. Most of them, 71 (80%), were genes induced in daf-2 pathway mutants and repressed in daf-16; daf-2 double mutants, while the other 18 genes were repressed in daf-2 mutants. Two catalases, ctl-2 and ctl-3, and two heat-shock proteins, hsp-16.1 and hsp-16.2, were common in both gene-expression profiles (Table S5). In contrast, only 14 genes down-regulated by oxidative stress were targets of DAF-16 and 12 of them (86%) were repressed by DAF-16. Interestingly, the comparison of these transcriptional profiles identified 45 genes that are common in transcriptional profiles of oxidative stress, aging, and targets of daf-16 (Fig. 1 and Table S6). Two well-known biomarkers of oxidative stress, hsp-16.1 and gst-4, were also found in all three profiles.
We also compared transcriptional profiles between C. elegans and D. melanogaster in response to oxidative-stress. Among 13,500 genes screened, 205 genes were up-regulated in D. melanogaster, including genes involved in the heat-shock response, antioxidant response, innate immunity, and purine biosynthesis. Oxidative stress also resulted in down-regulation of 388 genes in flies encoding several proteases, lipases, and transporters (Landis et al., 2004). The comparison between C. elegans and D. melanogaster resulted in only 18 overlapping homologs, including one gene predicted to encode glutathione peroxidase and four cytochrome P450 family genes (Table S7).
SKN-1-dependence of genes involved in antioxidant response and the detoxification pathway
To identify SKN-1-dependent target genes, we compared the transcriptional profiles of oxidative-stressed wild-type worms with that of worms grown on skn-1 RNAi and then treated with the same stressor. Out of 22,548 transcripts, 13,453 (60%) transcripts were expressed in skn-1 RNAi worms and 846 transcripts (4%) are differently expressed in worms grown on skn-1 RNAi. Among 1,340 transcripts altered in expression by oxidative stress 211 transcripts (16%) were SKN-1-dependent, which is more than expected by chance. 119 of the 948 transcripts induced by oxidative stress were significantly reduced in expression by skn-1 RNAi. The induction of many genes involved in both antioxidant response and the detoxification pathway was blocked either completely or in part by skn-1 knockdown (Table 3). For example, the up-regulation of ctl-2 and sod-1 was inhibited 80% and 100% by skn-1 RNAi. The expression of two genes predicted to encode glutathione peroxidase, R03G5.5 and F26E4.12, was also skn-1 dependent (68% and 100%, respectively). SKN-1 regulates the induction of at least nine GSTs. In addition, skn-1 RNAi significantly prevented oxidative-stress-related transcriptional changes of six genes encoding proteins with electron-transfer activity, including cyp-14A2 and glrx-10 (Table 3). (These electron transport genes are not those in the electron transport chain, but are involved in the xenobiotic detoxification.) However, none of the heat-shock proteins that were increased in expression by oxidative stress were affected by skn-1 RNAi (Table S1). Quantitative RT-PCR of selected SKN-1-dependent genes verified the transcriptional changes by skn-1 RNAi observed in microarray data (Table S3). Induction of gst-4, hsp-16.2, nlp-7, and cup-4 by oxidative stress was largely prevented in skn-1 RNAi worms and down-regulation of nhx-2 and ent-1 in oxidatively-stressed worms was also opposed by skn-1 RNAi. We searched for the predicted SKN-binding motif (WWTRTCAT) within a 1 kb upstream region of SKN-1-dependent genes. Of 211 SKN-1-dependent genes, 193 genes (92%) had more than one SKN-1-binding motif (Table 3). The frequency of the SKN-1-binding motif within the 1 kb upstream region of SKN-1-dependent genes was 1 in 395 bp. The SKN-1-binding motif should appear randomly every 2,048 bp (An & Blackwell, 2003). Therefore, genes having a SKN-1-binding motif were over-represented in SKN-1-dependent genes.
Table 3.
The effect of skn-1 RNAi on the expression of genes responding to oxidative stress
| Sequence Name | Gene Name† | Symbol | Fold Δ‡ | P§ | % inhibition by skn-1 RNAi* | Number of SKN-1 binding motif¶ | |
|---|---|---|---|---|---|---|---|
| Antioxidant Response | |||||||
| Y54G11A.5 | catalase 2, peroxisomal | ctl-2 | 2.7 | 0.001 | 80 | 4 | |
| C15F1.7 | superoxide dismutase 1 | sod-1 | 1.4 | 0.007 | 100 | 2 | |
| R03G5.5 | glutathione peroxidase | 3.0 | 0.000 | 68 | 6 | ||
| F26E4.12 | glutathione peroxidase | 1.7 | 0.004 | 100 | 4 | ||
| GSTs | |||||||
| K08F4.7 | glutathione S-transferase 4 | gst-4 | 12.4 | 0.000 | 79 | 3 | |
| Y45G12C.2 | glutathione S-transferase 10 | gst-10 | 3.0 | 0.000 | 100 | 3 | |
| Y45G12C.2 | glutathione S-transferase 10 | gst-10 | 2.9 | 0.000 | 100 | 3 | |
| F37B1.2 | glutathione S-transferase 12 | gst-12 | 9.2 | 0.000 | 79 | 2 | |
| T26C5.1 | glutathione S-transferase 13 | gst-13 | 4.9 | 0.001 | 100 | 4 | |
| F37B1.3 | glutathione S-transferase 14 | gst-14 | 15.9 | 0.000 | 85 | 3 | |
| Y53F4B.37 | glutathione S-transferase 32 | gst-32 | 18.9 | 0.001 | 99 | 0 | |
| F35E8.8 | glutathione S-transferase 38 | gst-38 | 3.9 | 0.000 | 100 | 4 | |
| Y53F4B.33 | glutathione S-transferase 39 | gst-39 | 6.1 | 0.000 | 69 | 2 | |
| Electron Transport | |||||||
| K09A11.3 | cytochrome P450 family 14A2 | cyp-14A2 | 2.4 | 0.007 | 71 | 2 | |
| Y34D9A.6 | glutaredoxin 10 | glrx-10 | 2.2 | 0.001 | 78 | 6 | |
| B0222.9 | xanthine dehydrogenase | 8.0 | 0.001 | 73 | 2 | ||
| F56D5.3 | NADH:flavin oxidoreductase | 15.5 | 0.000 | 93 | 8 | ||
| F17A9.4 | NADH:flavin oxidoreductase | 4.9 | 0.000 | 85 | 5 | ||
| C35B1.5 | thioredoxin, nucleoredoxin and related proteins | 2.4 | 0.000 | 100 | 1 | ||
| Neropeptide Signaling | |||||||
| F18E9.2 | neuropeptide like protein 7 | nlp-7 | 1.2 | 0.025 | 100 | 1 | |
| Homeostasis | |||||||
| B0495.4 | NA(+)/H(+) exchanger 2 | nhx-2 | 0.3 | 0.000 | 31 | 6 | |
| T17E9.2 | N-myristoyl transferase homolog 1 | nmt-1 | 0.7 | 0.016 | 100 | 2 | |
| C54H2.5 | surfeit homolog 4 | sft-4 | 0.6 | 0.016 | 52 | 3 | |
| Y66H1B.4 | sphingosine phosphate lyase 1 | spl-1 | 0.5 | 0.015 | 43 | 3 | |
| F55A11.2 | syntaxin 3 | syn-3 | 0.9 | 0.005 | 100 | 0 | |
| C16A11.4 | PHD Zn-finger proteins | 0.6 | 0.000 | 73 | 2 | ||
| Development | |||||||
| R12C12.2 | associated with RAN function 5 | ran-5 | 0.8 | 0.009 | 94 | 3 | |
| F57B1.2 | SUN domain protein 1 | sun-1 | 0.7 | 0.003 | 72 | 1 | |
| R06C7.5 | adenylosuccinate lyase | 0.6 | 0.002 | 37 | 3 | ||
| F42A8.1 | unknown | 0.4 | 0.000 | 57 | 3 | ||
| Reproduction | |||||||
| T05G5.3 | cyclin-dependent kinase family 1 | cdk-1 | 0.7 | 0.005 | 71 | 5 | |
| ZK809.4 | equilibrative nucleoside transporter 1 | ent-1 | 0.6 | 0.009 | 63 | 0 | |
| C34E10.2 | gro-1 operon gene 2 | gop-2 | 0.8 | 0.009 | 60 | 4 | |
| B0495.4 | NA(+)/H(+) exchanger 2 | nhx-2 | 0.3 | 0.000 | 31 | 6 | |
| T17E9.2 | N-myristoyl transferase homolog 1 | nmt-1 | 0.7 | 0.016 | 100 | 2 | |
| C32E8.8 | patched related family 2 | ptr-2 | 0.7 | 0.004 | 50 | 3 | |
| F23F12.6 | proteasome regulatory particle, ATPase-like 3 | rpt-3 | 0.8 | 0.004 | 62 | 4 | |
| W02B12.3 | SR protein (splicing factor) 1 | rsp-1 | 0.7 | 0.018 | 100 | 2 | |
| F10G7.4 | yeast SCC homolog 1 | scc-1 | 0.8 | 0.015 | 100 | 4 | |
| C54H2.5 | surfeit homolog 4 | sft-4 | 0.6 | 0.016 | 52 | 3 | |
| F55A11.2 | syntaxin 3 | syn-3 | 0.9 | 0.005 | 100 | 0 | |
| C14B1.4 | temporarily assigned gene name 125 | tag-125 | 0.8 | 0.009 | 100 | 7 | |
| F29B9.6 | ubiquitin-conjugating enzyme 9 | ubc-9 | 0.7 | 0.016 | 72 | 1 | |
| R06C7.5 | adenylosuccinate lyase | 0.6 | 0.002 | 37 | 3 | ||
| C50E3.5 | unknown | 0.7 | 0.017 | 96 | 1 | ||
| C09H10.10 | unknown | 0.6 | 0.001 | 93 | 2 | ||
Significant genes and fold change were determined by SAM. Functional classification was determined by DAVID.
The predicted gene titles for unknown genes are shown in Italics.
Fold change under oxidative stress.
Determined by T-test.
% inhibition effect of skn-1 knockdown. All genes are significantly different between control and skn-1 RNAi worms under the same oxidative-stress conditions by SAM (FDR = 22%).
Number of predicted SKN-1-binding motif within 1 kb upstream of the transcription start site.
skn-1 RNAi also inhibits down-regulation of genes involved in homeostasis and reproduction
Of 392 genes down-regulated under oxidative-stress conditions, 92 transcripts (24%) showed significant difference in expression between control and skn-1 RNAi worms. Among 12 down-regulated genes involved in cellular homeostasis, six genes were SKN-1-dependent (Table 3). nhx-2 is known to regulate pH homeostasis and nutrient transport and syn-3 is required for membrane fusion during cytokinesis. RNA levels of many genes involved in reproduction were also affected by skn-1 RNAi. Especially, the reduced expression of six genes by oxidative stress: rsp-1, syn-3, tag-125, scc-1, nmt-1, and C50E3.5, were completely inhibited by skn-1 RNAi. Only 4 of the 20 down-regulated genes involved in post-embryonic development were SKN-1-dependent (Table 3). None of five down-regulated genes involved in cuticle biosynthesis were affected by skn-1 RNAi. For genes involved in ion transport and amino acid/nitrogen compound metabolism, only 2 out of 27 genes and 1 out of 11 genes respectively were SKN-1-dependent (data not shown).
Knockdown of nlp-7 or cup-4 reduces oxidative-stress resistance and shortens the lifespan
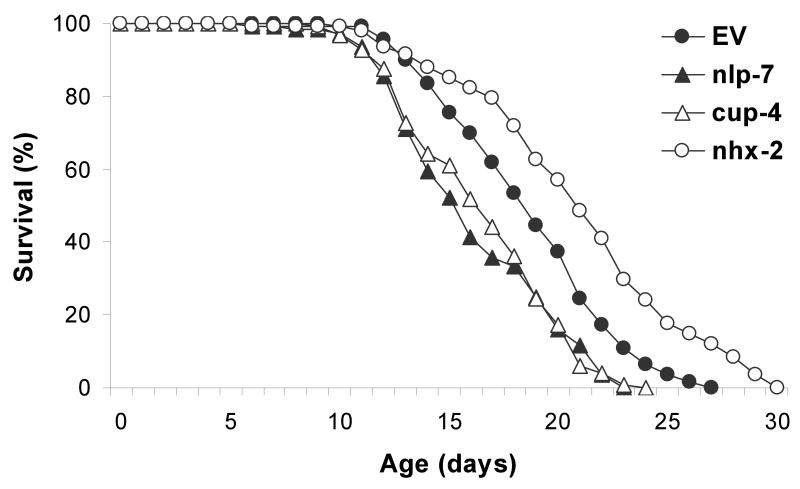
We wanted to further characterize SKN-1-dependent genes using additional types of assays. To determine the role of each SKN-1-dependent gene on oxidative-stress resistance, we tested the effect of gene-specific RNAi on survival under oxidative-stress conditions. Two different generators of reactive oxidants, paraquat and juglone, were used for this experiment. These assays are good for looking at smaller numbers of worms in a very controlled fashion. Among SKN-1-dependent genes that responded to oxidative stress, 24 genes contained in the Ahringer RNAi library (Kamath et al., 2003) were selected for oxidative-stress-resistance assays. The first group of genes includes genes that are induced by oxidative stress. We expected that RNAi of these genes would increase the sensitivity to oxidative stress. Among three antioxidant-response genes tested, ctl-2 and R03G5.5 showed only moderate decreases in resistance to oxidative stress (Table 4). Oxidative stress induced the expression of 20 GSTs and 9 of them were SKN-1-dependent in our microarray analysis (Table S1 and Table 3). The knockdown of gst-4, gst-12, or gst-14 failed to change the oxidative-stress resistance of C. elegans in either paraquat or juglone assays. In contrast, RNAi of genes encoding proteins with electron-transfer activity exhibited significant effects on survival under oxidative-stress conditions (Table 4). We also tested the effect of RNAi of nlp-7 and cup-4, known to encode a NLP (Li, 2005) and a neurotransmitter-gated ion channel (Patton et al., 2005) respectively, on oxidative-stress resistance. Individual knockdown of these two genes significantly reduced oxidative-stress resistance as effectively as skn-1 RNAi (Table 4 and Fig. 2A,C). We then examined the influence of selected genes on longevity and found that the lifespan of N2 worms was not affected by the inhibition of GSTs, or genes involved in the antioxidant response, or electron transport (Table 4). The reduced expression of nlp-7 or cup-4 by RNAi, however, shortened both mean and maximum lifespan, 12-14% (Table 4 and Fig. 3).
Table 4.
Summary of oxidative-stress resistance and longevity using RNAi
| Up-regulated Genes Under Oxidative Stress | |||
|---|---|---|---|
| Gene Symbol† | Paraquat (hours)‡ | Juglone (hours)‡ | Lifespan (days)§ |
| EV | 53.5/40.2 | 1.6/1.8 | 19.3/15.9 |
| skn-1 | 31.2*/22.9* | 0.4*/0.8* | 12.8*/14.5 |
| ctl-2 | 35.8*/40.4 | 0.6*/1.4 | 18.5/15.1 |
| R03G5.5 | 37.5*/45.9 | 0.8/1.1 | 18.8/14.9 |
| gst-4 | 37.7/41.1 | 0.4*/1.8 | 19.2/15.6 |
| fmo-4¶ | 39.7*/30.1* | 0.8*/1.1 | 18.3/16.3 |
| B0222.9 | 30.4*/25.2* | 0.6*/0.7* | 16.2*/15.7 |
| Gene Symbol† | Paraquat (hours)‡ | Juglone (hours)‡ | Lifespan (days)§ |
| EV | 31.3/31.7 | 0.8/1.6 | ND |
| skn-1 | 23.1*/20.9* | 0.2*/*0.6 | ND |
| F26E4.12 | 33.6/32.3 | 0.2/1.7 | ND |
| gst-12 | 30.3/30.0 | 0.3/1.7 | ND |
| gst-14 | 40.9/28.9 | 0.6/1.8 | ND |
| Gene Symbol† | Paraquat (hours)‡ | Juglone (hours)‡ | Lifespan (days)§ |
| EV | 54.7/41.3 | 1.0/2.0 | 18.3/17.2 |
| skn-1 | 37.4*/32.7* | 0.3*/0.7* | 15.6*/17.2 |
| cyp-14A2 | 34.5*/34.1* | 0.3*/0.9* | 16.8/16.2 |
| F17A9.4 | 30.1*/36.5 | 0.4*/1.1* | 16.9/17.1 |
| nlp-7 | 39.2*/33.7* | 0.4*/1.6 | 15.8*/14.6* |
| cup-4¶ | 34.3*/30.0* | 0.4*/1.3 | 16.2*/15.0* |
| Gene Symbol† | Paraquat (hours)‡ | Juglone (hours)‡ | Lifespan (days)§ |
| EV | 31.6/23.4 | 1.2/1.0 | 18.3/17.2 |
| skn-1 | 20.1*/17.1* | 0.2*/0.2* | 15.6*/17.2 |
| spl-1 | 44.0*/35.6 | 0.7/1.1 | ND |
| C16A11.4 | 30.7/31.4 | 0.7/0.7 | ND |
| nhx-2 | 24.9/29.3 | 0.8/0.6 | 19.0*/19.5* |
| syn-3 | 36.5/36.1 | 0.7/1.0 | ND |
| nmt-1 | 34.6/33.4 | 0.7/1.2 | ND |
| tag-125 | 44.1*/33.9 | 0.7/1.2 | ND |
| Gene Symbol† | Paraquat (hours)‡ | Juglone (hours)‡ | Lifespan (days)§ |
| EV | 31.6/23.4 | 1.5/1.1 | 18.3/17.2 |
| skn-1 | 20.1*/17.1* | 0.3*/0.2* | 15.6*/17.2 |
| gop-2 | 38.6/32.4 | 1.3/0.8 | ND |
| scc-1 | 19.3*/24.8 | 1.1/0.8 | ND |
| rsp-1 | 36.7/27.5 | 2.0/1.3 | ND |
| ent-1 | 55.2*/37.9* | 2.5*/1.9 | 17.3/15.9 |
| C50E3.5 | 27.8/32.2 | 1.5/1.2 | ND |
| R06C7.5a | 46.1*/28.2 | 2.4/1.2 | ND |
All assays were replicated twice.
EV: control empty vector, ND: not determined.
Full name and biological function of each gene are provided in Table 3.
Data expressed as mean survival (1st experiment/2nd experiment).
Data expressed as mean lifespan (1st experiment/2nd experiment).
Genes not determined as skn-1-dependent by SAM, but have significant p-value by t-test.
Significantly different from EV control.
Fig. 2.
The effect of RNAi of nlp-7, cup-4, and ent-1 on resistance to oxidative stress. (A and B) Paraquat-resistance assay. Young adult N2 worms were treated with 20 mM of paraquat and survival was scored. skn-1 RNAi showed consistent decreased resistance to paraquat (P < 0.001). Two independent assays showed significant decrease in survival by nlp-7 (P < 0.001, P = 0.027) or cup-4 (P < 0.001, P = 0.007) RNAi, while ent-1 RNAi significantly increased the survival of worms (P < 0.001, P = 0.038). (C and D) Juglone-resistance assay. Adult worms placed on each RNAi plate for 24 hours were transferred to fresh-made juglone plates (30 mg L-1 of juglone) and scored for viability over time. Resistance to juglone was significantly reduced by nlp-7 or cup-4 RNAi (P = 0.041 and P = 0.048, respectively), as observed by skn-1 RNAi (P < 0.01). However, knockdown of ent-1 resulted in significant increase in resistance to juglone (P = 0.045). EV: control empty vector.
Fig. 3.
Lifespan of N2 fed with dsRNA of skn-1-dependent oxidative-stress genes. Animals were scored at 20 °C from egg hatch until all worms were dead. Each longevity assay was replicated and the average of two assays was plotted. Mean lifespan of nlp-7 RNAi (15.2 ± 0.6 SEM) and cup-4 RNAi (15.6 ± 0.6 SEM) was significantly reduced (P = 0.004 and 0.001 for nlp-7 RNAi and P = 0.014 and 0.003 for cup-4 RNAi) compared to contol RNAi (17.7 ± 0.5 SEM), while RNAi of nhx-2 increased (P = 0.002 and 0.005) the mean lifespan (19.2 ± 0.3 SEM). Means of duplicates are shown. EV: control empty vector.
We asked whether transcriptional changes driven by exposure to hyperbaric oxygen are similar to those seen in response to exposure to paraquat, a generator of reactive oxidants used in this study. We assessed the expression level of selected genes in worms treated with 6 hours of 20 mM paraquat (Table S3). The expression of gst-4, hsp-16.2, nlp-7, and cup-4 was increased in worms treated with paraquat, similar to what was observed in worms treated with hyperbaric oxygen; however, the fold increase caused by exposure to paraquat was smaller than that caused by hyperbaric oxygen exposure. Unlike the results observed in worms with hyperbaric oxygen, RNAi of skn-1 showed a significant effect on the induction of gst-4 only in worms treated with paraquat (Table S3).
ent-1 and nhx-2 regulate oxidative-stress resistance and lifespan, respectively
Among genes down-regulated under oxidative-stress conditions, we inhibited the expression of 12 genes that are involved in homeostasis and reproduction by RNAi and found that only one gene, ent-1, showed a consistent increase in oxidative-stress resistance (Table 4 and Fig. 2B,D). ENT-1 is a nucleoside transporter expressed in the adult intestine and pharynx (Appleford et al., 2004). Inactivation of ent-1 by RNAi, however, has no effect on lifespan (Table 4). Previously, it was reported that gene inactivation of nhx-2 by RNAi increases the lifespan of C. elegans up to 40% (Nehrke, 2003). RNAi treatment of nhx-2 increased both average and maximum lifespan significantly in our hands as well (Table 4 and Fig. 3). Our microarray analysis revealed that nhx-2 is a downstream target of SKN-1; however, knockdown of nhx-2 failed to show a significant effect on oxidative-stress resistance (Table 4). Quantitative RT-PCR analysis of ent-1 and nhx-2 in worms treated with paraquat showed consistent results. Similar transcriptional changes of ent-1 were observed in both worms treated with hyperbaric oxygen and paraquat: decrease in expression by oxidative stress and prevention of down-regulation by skn-1 RNAi. Unlike the results observed in worms treated with hyperbaric oxygen, however, the mRNA level of nhx-2 was not altered by either paraquat or skn-1 RNAi (Table S3). Overall, comparison of effects of hyperbaric oxygen and paraquat implies that oxidative stress imposed in different ways induces different yet overlapping adaptive responses. They suggest that regulation by SKN-1 may occur in some genes in response to diverse forms of oxidative stress, but in others be specific to particular forms of oxidative stress.
Discussion
Here we performed gene-expression-profiling in C. elegans under oxidative-stress conditions using high-density oligonucleotide microarrays. As expected, oxidative stress induced the expression of genes involved in antioxidant response, but it also induced several other classes of genes, including heat-shock proteins. Among these was the small heat-shock protein gene hsp-16.2, which is induced by many stressors, including oxidative stress (Link et al., 1999). Up-regulated genes also include many cytochrome P450 family proteins and GSTs. These genes are main components of phase I and II detoxification pathways and their expression is increased by various xenobiotics (Reichert & Menzel, 2005). Oxidative stress induced the expression of six genes involved in neuropeptide signaling, including one FMRF-like peptide (FLP) and three NLPs. To date, 23 flp genes and 32 nlp genes have been predicted from the genome sequence of C. elegans (Husson et al., 2005). Although the biological function of each FLP and NLP is not fully understood, the increased expression of these genes; and presumably proteins, suggests that neuronal signaling involving FLP or NLP plays a role in the response to oxidative stress in C. elegans. Presumably, then, the response to oxidative stress is coordinated by the nervous system.
One class of genes down-regulated by oxidative stress includes those involved in homeostasis. Two transporters known to regulate nutrient absorption in the intestine, nhx-2 and opt-2, were reduced 3-fold and 10-fold, respectively in worms exposed to oxidative stress. NHX-2 is a Na+/H+ exchanger expressed at the apical membrane of intestinal epithelial cells and works together with OPT-2 to regulate nutrient uptake (Walker et al., 2005). The expression of nhx-2 and opt-2 was also reduced in both daf-2 mutants and dauer larvae (McElwee et al., 2004). Oxidative stress also reduces the expression of many genes involved in development and reproduction. A previous study reported that C. elegans grown under hyperoxia shows abnormal gonad appearance and decreased fertility and fecundity (Goldstein & Modric, 1994). Taken together, global transcriptional profiling suggests that oxidative stress causes a transcriptional shift from genes involved in reproduction and homeostasis to the activation of genes encoding proteins for the cellular stress response and detoxification in adult worms.
In C. elegans, lifespan is modulated by an IIS pathway and longevity genes involved in this pathway also mediate increased stress resistance (Honda & Honda, 1999; Johnson et al., 2002). As predicted by the Stress Response Hypothesis (Johnson et al., 1996), comparison of our transcriptional profiles of response to oxidative stress with profiles of genes altered by mutations of the IIS pathway (Murphy et al., 2003) revealed that induction of genes involved in antioxidant response, heat-shock response, and detoxification was common in both. Among 1,340 genes regulated under oxidative-stress conditions, 200 (15%) genes were also altered in expression during aging, including hsp-16.1, gst-4, cup-4, and many collagens. Previously, similar gene-expression patterns were observed between aging and oxidative stress in D. melanogaster (Landis et al., 2004). In D. melanogaster, oxygen exposure was characterized by up-regulation of genes involved in antioxidant response, heat-shock protein, innate immune response, and cytochrome P450 family proteins (Landis et al., 2004). We observed similar functional categories to be induced by oxidative stress in C. elegans. Comparison on an individual gene level, however, revealed a surprisingly small number of overlapping genes, although a similar stress inducer was used in both experiments. Only 18 genes were common in both species, including four cytochrome P450 family proteins and one antioxidant gene. In support of this finding is a recent study by McElwee et al. showing that the longevity assurance mechanisms in different species are also species-specific at the gene level, but conserved at the process level (McElwee et al., 2007). Cross-species comparisons of transcriptional profiles of long-lived IGF-1 mutants in C. elegans, D. melanogaster, and mice showed little evolutionary conservation of the response of individual orthologs, while many functional categories were evolutionarily conserved in long-lived mutants of all three species (McElwee et al., 2007).
One focus of our studies has been the SKN-1 transcription factor which accumulates in the nucleus in response to oxidative stress and regulates oxidative-stress resistance in C. elegans (An & Blackwell, 2003; An et al., 2005). skn-1 mutants have reduced resistance to oxidative stress and shortened lifespan (An & Blackwell, 2003). We asked which oxidative-stress-response genes are under the regulation of SKN-1. Among genes induced by oxidative stress, several genes involved in antioxidant response and many GSTs were affected by skn-1 RNAi. The decreased expression of many genes involved in homeostasis and reproduction by oxidative stress was also significantly opposed by skn-1 RNAi. Interestingly, none of the heat-shock proteins up-regulated under oxidative stress were SKN-1-dependent. Previous study with RNAi of different nuclear respiratory genes showed that mitochondrial dysfunction activates the transcription of a number of heat-shock proteins in C. elegans and the induction is not associated with oxidative stress (Kuzmin et al., 2004). These observations suggest that the transcriptional activation of heat-shock proteins under oxidative-stress conditions may be attributed to mitochondrial impairment caused by increased ROS and may not be related to the activation of SKN-1 by oxidative stress.
Our goal was to initially identify genes involved in the response to oxidative stress using microarrays and then further examine and characterize these genes in various proven ways to look at their role in resistance to oxidative stress and lifespan. We measured the effect of inhibition of SKN-1-dependent oxidative-stress-response genes on survival, under conditions of oxidative stress caused by paraquat or juglone. A significant decrease in survival was observed when expression of several up-regulated genes was inhibited by RNAi. However, RNAi of all three GSTs tested, gst-4, gst-12, and gst-14, failed to show significant effects on resistance to oxidative stress caused by paraquat or juglone. The C. elegans genome contains more than 50 putative GSTs (Leiers et al., 2003). The lack of effect on resistance to oxidative stress by RNAi-induced knockdown of a single GST could be due to their functional redundancy. Other studies show that specific GSTs can modulate the response to oxidative stress in C. elegans; overexpression of gst-4 or gst-10 increased resistance to oxidative stress and RNAi-mediated knockdown led to a significant decrease in resistance to oxidative stress (Ayyadevara et al., 2005; Leiers et al., 2003). Among down-regulated genes, 12 genes involved in homeostasis and reproduction were tested and only one gene, ent-1, significantly increased resistance to oxidative stress. These findings suggest that up-regulated SKN-1-dependent genes play a more critical role in subsequent resistance to oxidative stress than do down-regulated genes.
The free-radical theory of aging proposes that the accumulation of oxidative damage plays a key role in the aging process (Harman, 1956). In C. elegans, life extension in many gerontogene mutants is associated with increased resistance to stress. Long-lived mutants, such as age-1, clk-1, and spe-26, have increased resistance to several stressors, including reactive oxidants (Johnson et al., 2001; Johnson et al., 2002). However, the role of oxidative stress in aging is still controversial. A significant body of evidence is in conflict with a causal role for oxidative stress in aging (Muller et al., 2007). Treatment with SOD mimetics have been reported to increase life span in C. elegans (Melov et al., 2001); however these findings have been questioned in subsequent studies where EUK-8 failed to increase C. elegans lifespan (Keaney et al., 2003). This inconsistency of SOD mimetics might be due to differential efficacy that is specific to culture conditions and possible species specificity (Sampayo et al., 2003). In Drosophila, overexpression of Cu-Zn SOD and CAT has been reported to lengthen lifespan in strains having short lifespan (Orr & Sohal, 1994), but again additional evidence questions this. In relatively long-lived strains, simultaneous overexpression of major antioxidant genes does not extend lifespan (Orr et al., 2003). Transgenic mice overexpressing Cu-Zn SOD in various tissues have a slightly decreased lifespan compared to non-transgenic littermates (Huang et al., 2000).
With selected genes regulated by oxidative stress, we also assessed the effect of RNAi of individual genes on lifespan. Longevity assays identified three SKN-1-dependent genes which modulate the lifespan of C. elegans: nlp-7, cup-4, and nhx-2. NLP-7 is a neuropeptide-like protein as predicted by genome sequence (Li, 2005), but little is known about the biological functions of different neuropeptides. Cellular expression pattern analysis using green fluorescent protein (GFP)-reporter construct shows that NLP-7 is primarily expressed in neurons, including the ASI (Nathoo et al., 2001) and is predicted to be involved in neuronal signaling. cup-4 encodes a ligand-gated ion channel highly homologous to mammalian nicotinic acetylcholine receptors and predicted to regulate endocytosis through modulation of phospholipase C activity (Patton et al., 2005). CUP-4 is solely expressed in coelomocytes, the scavenger cells in the worm body cavity (Patton et al., 2005). RNAi-induced inactivation of nhx-2 led to a 40% increase in C. elegans lifespan (Nehrke, 2003), while knockdown of opt-2, a gene that works with nhx-2 in nutrient uptake in intestine, did not change the lifespan (Meissner et al., 2004). Both nhx-2 and opt-2 were down-regulated by oxidative stress, but only nhx-2 expression was affected significantly by skn-1 RNAi. These observations demonstrate the existence of overlapping pathways activated by SKN-1 in response to oxidative stress that modulate both stress resistance and longevity. Identification of other genes or pathways overlapping between oxidative stress and aging will provide additional evidence to support the Stress Response Hypothesis of aging (Johnson et al., 1996). Further studies regarding nlp-7 and cup-4 function in oxidative stress and aging should help us elucidate the mechanism of SKN-1 action in adult C. elegans.
Experimental procedures
Strains and worm culture
The wild-type N2 CGCb strain was used in all RNAi experiments. TJ1060 [fer-15(b26); spe-9(hc88)] was used for microarray experiments to eliminate progeny production. All strains were grown at 20 °C on NGM plates (1.7% agar, 2.5 mg mL-1 peptone, 25 mM NaCl, 50 mM KH2PO4 pH 6.0, 5 μg mL-1 cholesterol, 1 mM CaCl2, 1 mM MgSO4) with fresh Escherichia coli OP50 as a food source (Sulston, 1988). Synchronous populations of hermaphrodites were established by placing five young adults onto a fresh NGM plate and permitting eggs to be laid for 4 hours at 20 °C.
Oxidative stress and microarray experiments
TJ1060 is temperature sensitive and was grown at 16 °C. Eggs were collected by hypochlorite, hatched overnight at 20 °C (Emmons et al., 1979) and first-stage larvae (L1's) were placed onto 2% peptone plates (1.7% agar, 20 mg mL-1 peptone, 25 mM NaCl, 50 mM KH2PO4 pH 6.0, 5 μg mL-1 cholesterol, 1 mM CaCl2, 1 mM MgSO4) at 25 °C. Sterile 3-day-old young-adult animals on peptone plates with E. coli RW2 were treated with 99% O2 at 40 PSI for 6 hours at 20 °C. We used a hyperbaric chamber (designed by Dr. Chris Link) because it allowed us to sufficiently stress many worms and get good gene expression differences over many tissue types without generating a lot of damage. Half of the worms were not exposed and served as a control population. A small plate of strain CL2166 [dvIs19 pAF15(gst-4∷GFP∷NLS)] (Leiers et al., 2003) was used as an indicator strain and checked for GFP induction in both control and experimental conditions. Worms were harvested by washing them off with S-basal and were then quick frozen in liquid nitrogen. The worms grown on skn-1 RNAi bacteria were prepared exactly as described above. The RNAi bacterial strain was E. coli HT115. The skn-1 RNAi bacterial strain was isolated from the Ahringer RNAi library (Kamath et al., 2003). It is well-known that worm strains grown on different strains of bacteria can show different phenotypes in various assays. Here, to save on arrays, we compared the same strain, TJ1060, grown on either RW2 or on HT115 carrying the skn-1 RNAi sequence. Both are E. coli strains and grow well on the media employed. We assumed that we would be able to identify genes of interest in spite of the difference in bacterial strains. Total RNA was prepared from ∼3,000 worms using the TRIZOL reagent and standard protocols and cleaned up on Qiagen RNeasy columns. The Affymetrix labeling kit was used to prepare labeled cRNA as a target for the Affymetrix GeneChip® C. elegans Genome Array. Four independent total RNA samples were extracted per group and cRNA samples from each total RNA sample were hybridized to independent DNA chips (n = 4). Gene chip hybridization and scanning were done at Genome Explorations (Memphis, MN) and UC-Boulder Gene Chip Core Facility (Boulder, CO).
Analysis of microarray data
The Affymetrix GeneChip® C. elegans Genome Array is designed to monitor gene expression of over 22,500 predicted transcripts. Each gene is represented by ∼11 perfectly matched (PM) and mismatched (MM) control probe pairs. The MM probes act as specificity controls that allow the direct subtraction of both background and cross-hybridization signals. The GeneChip Operating Software (GCOS), version 1.4, was used to analyze the image data. GCOS determines the presence of mRNA in samples and computes the signals of probe sets using the differences and ratios between PM and MM signals. To minimize variability in hybridization, the global scaling method normalizes signals of each chip. The global scaling method is a computational technique in which the average signal of all probe sets in one chip is scaled to a target average intensity, which was 250 in this analysis, by multiplying a scaling factor. This scaling factor is multiplied by each probe set signal to give the raw expression. The microarray gene expression data are available at http://www.ncbi.nlm.nih.gov/geo/ (accession number GSE9301). The fold change (Fold Δ) was calculated using mean signal intensity of each individual gene in each study group.
After array normalization, Significance Analysis of Microarray (SAM) was performed to obtain a list of genes that were significantly altered in expression. SAM assigns each gene a score according to its change in expression relative to the standard deviation of the repeated measurements for that gene and identifies genes whose scores are greater than a threshold as a significant gene. False discovery rate (FDR), the percentage of significant genes that would be identified by chance, is also calculated.
To classify the gene-expression profile according to functional annotation of each gene and test the statistical significance of age-related changes in each category, we used web-based DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/). DAVID allows us to access a relational database of functional annotation and facilitates the biological interpretation of gene lists derived from the results of microarrays and provides statistical methods for discovering enriched biological themes within gene lists.
The mean signal intensities of each gene were used to calculate the effect of skn-1 RNAi on the expression of each gene. Percent inhibition of skn-1 RNAi was calculated as ((O-S)/(O-C))*100, where C is the unstressed control, O is worms with 6 hours of hyperbaric O2 exposure, and S is the average signal intensity obtained from worms treated with skn-1 RNAi under the same oxidative-stress conditions. For genes showing (O-S)/(O-C) > 1, we put the % inhibition effect by skn-1 RNAi as 100.
The predicted SKN-1-binding motif was identified using TFSEARCH (Heinemeyer et al., 1998). 1 kb upstream region of the transcription start site was uploaded and the number of predicted SKN-1-binding motif was counted for each gene.
Quantitative RT-PCR
5 μg of total RNA extracted from worms using RNeasy Mini kit and RNase-free DNase (Qiagen, Valencia, CA, USA) was converted to cDNA by Superscript First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). Quantitative PCR was performed using SYBR Green 2X Mater Mix (Applied Biosystems, Foster City, CA, USA) and the amplification plot was detected with ABI prism 7000 Sequence Detection System (TaqMan) (Applied Biosystems, Foster City, CA, USA). Primer sets used are shown in Table S8. ama-1 was used as control gene for normalization.
Assessment of resistance to oxidative stress
All data were generated with age-synchronized N2 hermaphrodites at 20 °C. 50 young adult worms grown on NGM plates supplemented with 100 μg mL-1 ampicillin, 12.5 μg mL-1 tetracycline, and 0.4 mM IPTG, and spotted with bacteria expressing each dsRNA, were transferred to plates containing 20 mM paraquat and 0.5 mg mL-1 5-fluoro-2′-deoxyuridine or 30 μg mL-1 juglone. For the paraquat-sensitivity assay, worms were transferred to fresh NGM plates with paraquat and dsRNAi-expressing bacteria every 2 days until all worms were dead. All dsRNAi vectors used in this study were verified to include the target genes by DNA sequencing. Agar plates containing 30 μg mL-1 juglone were made as previously described (de Castro et al., 2004). The survival of the worms was monitored every hour after transferring to juglone plates with dsRNAi-expressing bacteria. For quantitative RT-PCR, ∼1000 adult worms treated with 20 mM paraquat for 6 hours were harvested and used to extract total RNA.
Longevity assessment
Longevity assessments were performed on NGM plates with age-synchronized N2 hermaphrodites at 20 °C. For RNAi assessment, 5 L4/young adult worms cultured on NGM plates were transferred to a fresh NGM plate, containing 100 μg mL-1 ampicillin, 12.5 μg mL-1 tetracycline, 0.4 mM IPTG, and 0.5 mg mL-1 5-fluoro-2′-deoxyuridine and spotted with bacteria expressing each dsRNA. After laying eggs for 4 hours, all five adult worms were removed. 60 young adults were picked 3 days after hatching and were transferred to a fresh plate with dsRNA expressing bacteria daily during the egg-laying period. Thereafter, worms were transferred every 2-3 days until all worms were dead. Dead worms were scored daily and removed from the plates immediately. An animal was scored as dead when it did not respond to mechanical stimulation. For the knockout strains, NGM plates containing 0.5 mg mL-1 5-fluoro-2′-deoxyuridine and OP50 as a food source were used. All assessments were duplicated and replicated at least twice. For statistical analysis, the log-rank test was used.
Supplementary Material
Acknowledgments
We wish to thank members of the Johnson and Link labs for helpful advice and critical discussion and Dr. Katerina Kechris for advice on statistical analysis of microarray data. This work was supported by the National Institute on Aging (AG016219).
References
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleford PJ, Griffiths M, Yao SY, Ng AM, Chomey EG, Isaac RE, Coates D, Hope IA, Cass CE, Young JD, Baldwin SA. Functional redundancy of two nucleoside transporters of the ENT family (CeENT1, CeENT2) required for development of Caenorhabditis elegans. Mol Membr Biol. 2004;21:247–259. doi: 10.1080/09687680410001712550. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Dandapat A, Singh SP, Benes H, Zimniak L, Reis RJ, Zimniak P. Lifespan extension in hypomorphic daf-2 mutants of Caenorhabditis elegans is partially mediated by glutathione transferase CeGSTP2-2. Aging Cell. 2005;4:299–307. doi: 10.1111/j.1474-9726.2005.00172.x. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Hegi de Castro S, Johnson TE. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic Biol Med. 2004;37:139–145. doi: 10.1016/j.freeradbiomed.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Emmons SW, Klass MR, Hirsh D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Goldstein P, Modric T. Transgenerational, ultrastructural analysis on the antioxidative effects of tocopherol on early gametogenesis in Caenorhabditis elegans grown in 100% oxygen. Toxicol Appl Pharmacol. 1994;124:212–220. doi: 10.1006/taap.1994.1025. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Huang TT, Carlson EJ, Gillespie AM, Shi Y, Epstein CJ. Ubiquitous overexpression of Cu,Zn superoxide dismutase does not extend life span in mice. J Gerontol A: Biol Sci Med Sci. 2000;55:B5–B9. doi: 10.1093/gerona/55.1.b5. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun. 2005;335:76–86. doi: 10.1016/j.bbrc.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Lithgow GJ, Murakami S. Hypothesis: interventions that increase the response to stress offer the potential for effective life prolongation and increased health. J Gerontol A Biol Sci Med Sci. 1996;51:B392–B395. doi: 10.1093/gerona/51a.6.b392. [DOI] [PubMed] [Google Scholar]
- Johnson TE, de Castro E, Hegi de Castro S, Cypser J, Henderson S, Tedesco P. Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. Exp Gerontol. 2001;36:1609–1617. doi: 10.1016/s0531-5565(01)00144-9. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Henderson S, Murakami S, de Castro E, de Castro SH, Cypser J, Rikke B, Tedesco P, Link C. Longevity genes in the nematode Caenorhabditis elegans also mediate increased resistance to stress and prevent disease. J Inherit Metab Dis. 2002;25:197–206. doi: 10.1023/a:1015677828407. [DOI] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteosomal dysfunction activates SKN-1 and produces a selective oxidative stress response in C. elegans. Biochem J. 2007;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Keaney M, Gems D. No increase in lifespan in Caenorhabditis elegans upon treatment with the superoxide dismutase mimetic EUK-8. Free Radic Biol Med. 2003;34:277–282. doi: 10.1016/s0891-5849(02)01290-x. [DOI] [PubMed] [Google Scholar]
- Kell A, Ventura N, Kahn N, Johnson TE. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic Biol Med. 2007;43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin EV, Karpova OV, Elthon TE, Newton KJ. Mitochondrial respiratory deficiencies signal up-regulation of genes for heat shock proteins. J Biol Chem. 2004;279:20672–20677. doi: 10.1074/jbc.M400640200. [DOI] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, Tavare S, Tower J. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiers B, Kampkotter A, Grevelding CG, Link CD, Johnson TE, Henkle-Duhrsen K. A stress-responsive glutathione S-transferase confers resistance to oxidative stress in Caenorhabditis elegans. Free Radic Biol Med. 2003;34:1405–1415. doi: 10.1016/s0891-5849(03)00102-3. [DOI] [PubMed] [Google Scholar]
- Li C. The ever-expanding neuropeptide gene families in the nematode Caenorhabditis elegans. Parasitology. 2005;131(Suppl):S109–127. doi: 10.1017/S0031182005009376. [DOI] [PubMed] [Google Scholar]
- Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D. Evolutionarily conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Meissner B, Boll M, Daniel H, Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J Biol Chem. 2004;279:36739–36745. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Murakani S, Johnson TE. Life extension and stress resistance in Caenorhabditis elegans modulated by the tkr-1 gene. Curr Biol. 1998;8:1091–1094. doi: 10.1016/s0960-9822(98)70448-8. [DOI] [PubMed] [Google Scholar]
- Murakami S, Johnson TE. The OLD-1 positive regulator of longevity and stress resistance is under DAF-16 regulation in Caenorhabditis elegans. Curr Biol. 2001;11:1517–1523. doi: 10.1016/s0960-9822(01)00453-5. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci U S A. 2001;98:14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrke K. A reduction in intestinal cell pHi due to loss of the Caenorhabditis elegans Na+/H+ exchanger NHX-2 increases life span. J Biol Chem. 2003;278:44657–44666. doi: 10.1074/jbc.M307351200. [DOI] [PubMed] [Google Scholar]
- Orr WC, Mockett RJ, Benes JJ, Sohal RS. Effects of overexpression of copper-zinc and manganese superoxide dismutases, catalase, and thioredoxin reductase genes on longevity in Drosophila melanogaster. J Biol Chem. 2003;278:26418–26422. doi: 10.1074/jbc.M303095200. [DOI] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- Patton A, Knuth S, Schaheen B, Dang H, Greenwald I, Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Reichert K, Menzel R. Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere. 2005;61:229–237. doi: 10.1016/j.chemosphere.2005.01.077. [DOI] [PubMed] [Google Scholar]
- Sampayo JN, Olsen A, Lithgow GJ. Oxidative stress in Caenorhabditis elegans: protective effects of superoxide dismutase/catalase mimetics. Aging Cell. 2003;2:319–326. doi: 10.1046/j.1474-9728.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis Elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Houthoofd K, Vanfleteren JR, Gems D. Dietary restriction in C. elegans: from rate-of-living effects to nutrient sensing pathways. Mech Ageing Dev. 2005;126:929–937. doi: 10.1016/j.mad.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Yanase S, Onodear A, Tedesco P, Johnson TE, Ishii N. SOD-1 deletions in Caenorhabditis elegans alter the localization of intracellular ROS and show molecular compensation. J Gerontol A Biol Sci Med Sci. 2008 doi: 10.1093/gerona/glp020. in press. [DOI] [PubMed] [Google Scholar]
- Yanase S, Yasuda K, Ishii N. Adaptive responses to oxidative damage in three mutants of Caenorhabditis elegans (age-1, mev-1 and daf-16) that affect life span. Mech Ageing Dev. 2002;123:1579–1587. doi: 10.1016/s0047-6374(02)00093-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.