Abstract
Rho GTPases regulate the actin cytoskeleton in all eukaryotes. Fission yeast Cdc42 is involved in actin cable assembly and formin For3 regulation. We isolated cdc42-879 as a thermosensitive strain with actin cable and For3 localization defects. In a multicopy suppressor screening, we identified pob1+ as suppressor of cdc42-879 thermosensitivity. Pob1 overexpression also partially restores actin cables and localization of For3 in the mutant strain. Pob1 interacts with Cdc42 and this GTPase regulates Pob1 localization and/or stability. The C-terminal pleckstrin homology (PH) domain of Pob1 is required for Cdc42 binding. Pob1 also binds to For3 through its N-terminal sterile alpha motif (SAM) domain and contributes to the formin localization at the cell tips. The previously described pob1-664 mutant strain (Mol. Biol. Cell. 10, 2745–2757, 1999), which carries a mutation in the PH domain, as well as pob1 mutant strains in which Pob1 lacks the N-terminal region (pob1ΔN) or the SAM domain (pob1ΔSAM), have cytoskeletal defects similar to that of cdc42-879 cells. Expression of constitutively active For3DAD* partially restores actin organization in cdc42-879, pob1-664, pob1ΔN, and pob1ΔSAM. Therefore, we propose that Pob1 is required for For3 localization to the tips and facilitates Cdc42-mediated relief of For3 autoinhibition to stimulate actin cable formation.
INTRODUCTION
The shape and size of cells is achieved through a strict control of polarized cell growth, which requires regulation of actin organization, addition of new membranes to the zones of active growth, and, in fungal and plant cells, biosynthesis of the cell wall. These processes must be tightly coordinated temporally and spatially to permit the correct growth position in each moment of the cell cycle (Hayles and Nurse, 2001).
Rho GTPases are key molecules in polarized growth regulation, which integrate signals from both the cell cycle and the extracellular environment (Hall, 2005; Park and Bi, 2007). These proteins act as molecular switches, inactive when bound to guanosine diphosphate (GDP) and active when bound to guanosine triphosphate (GTP). GTPase activation proteins (GAPs) stimulate the intrinsic GTPase activity of the Rho proteins, leading to their inactivation; by contrast, Guanine nucleotide exchange factor (GEF) proteins promote the GDP-to-GTP exchange, thereby activating Rho proteins. Rho GTPases directly regulate the actin and microtubule cytoskeleton organization (Ridley, 2006), participate in the secretory pathway control (Brennwald and Rossi, 2007), and regulate fungal cell wall biosynthesis (Levin, 2005). To coordinate all these processes, GTP-bound Rho GTPases interact with and stimulate the activity of a variety of effector proteins.
The Rho-family GTPase Cdc42 plays a central role in the establishment of polarized cell growth through interaction with multiple effector proteins (Cerione, 2004; Etienne-Manneville, 2004). Cdc42 interacts with many proteins such as: p21-activated kinases (PAKs), which participate in actin organization (Zhao and Manser, 2005); Wiskott–Aldrich syndrome protein, which stimulates the formation of actin structures through the activation of the Arp2/3 complex (Takenawa and Suetsugu, 2007); the exocyst complex, necessary for tethering secretory vesicles to the plasma membrane (Wu et al., 2008); and formins, which nucleate linear actin filaments (Faix and Grosse, 2006).
Fission yeast Cdc42 is an essential protein that has been implicated in the regulation of the cell shape and growth (Miller and Johnson, 1994). It interacts with the PAKs, Shk1 and Shk2 (Yang et al., 1998; Chang et al., 1999), and it is the main Rho GTPase required for actin cable assembly (Martin et al., 2007). Actin cables are important structures that contribute to polarized cell growth, serving as tracks for delivery of secretory vesicles to cell tips. In Schizosaccharomyces pombe interphase cells the assembly of these structures depends on the formin For3, which localizes to cell tips (Feierbach and Chang, 2001). Surprisingly, For3 is not essential for cell viability, though for3Δ cells lack actin cables detectable by phalloidin staining. Similar to other formins, For3 is controlled by an autoinhibitory interaction between the N-terminal Dia inhibitory domain (DID) with the C-terminal Dia autoregulatory domain (DAD) (Goode and Eck, 2007). The proper localization of For3 depends on relief of this autoinhibition (Martin et al., 2007), and so far the only proteins involved in this process are the actin-interacting protein Bud6 and the Rho GTPase Cdc42 (Martin et al., 2007).
We had previously isolated several cdc42-ts mutant strains; screening them for multicopy suppressors we found Pob1, an essential protein implicated previously in polarized cell growth and cell septation (Toya et al., 1999). Overproduction of Pob1 partially suppresses the actin defects observed in the cdc42-ts mutant cdc42-879. Here, we describe the interactions among Cdc42, For3, and Pob1 and suggest that Pob1 contributes to relief of For3 autoinhibition.
MATERIALS AND METHODS
Strains, Growth Conditions, and Genetic Methods
Standard S. pombe media and genetic manipulations were used (Moreno et al., 1991). All the strains used were isogenic to wild-type strains 972 h− and 975 h+, and they are described in Supplemental Table 1. The strains were constructed by either tetrad dissection or random spore germination method. Cells were usually grown in either rich medium (YES) or minimal medium (EMM) with appropriate supplements. Escherichia coli DH5α was used as host for propagation of plasmids. Cells were grown in Luria-Bertani medium supplemented with 50 μg/ml ampicillin when needed. Solid media contained 2% agar.
Recombinant DNA Methods
All DNA manipulations were carried out by established methods. Enzymes were used according to the recommendations of the suppliers. S. pombe was transformed by the lithium acetate method (Ito et al., 1983). The nmt promoter-containing vectors pREP3X, pREP4X, and pREP1-GST (Forsburg, 1993) were used for the overexpression of cdc42+, gef1+, scd1+, shk1+, shk2+, and pob1+. For the multicopy expression of pob1+ under the control of its own promoter, an EcoRI fragment of 4.1 kb containing pob1+ open reading frame (ORF) and ∼1 kb of its promoter and terminator was subcloned into pAU-KS or pAL-KS vectors. For the two-hybrid analysis, the pob1+ ORF or different pob1+ fragments were cloned into the NdeI-XmaI sites of pAS2. cdc42+ ORF excluding the 3′ region codifying the prenylatable domain of Cdc42 was cloned into the NcoI-XmaI sites of pACT2. For3 two-hybrid plasmids containing for3N (1–702) and for3C (630–1461) were as described previously (Martin et al., 2005). Saccharomyces cerevisiae Y190 (MATa gal4 gal80 his3 trp1-901 ura3-52 leu2-3,-112 URA3::GAL-<lacZ, lys2::GAL (UAS)-<HIS3 cyhr) were transformed and grown on plates without leucine, tryptophan, and histidine and supplemented with 40 mM 3-aminotriazole. β-Galactosidase activity was analyzed in the transformant colonies as described previously (Coll et al., 2003).
Error-prone PCR for obtaining cdc42 ORF mutants was performed as described previously (Martin et al., 2007). cdc42-879 strain identification and isolation were carried out as described previously (Martin et al., 2007). Screening for cdc42-879 multicopy suppressors was performed using a S. pombe genomic library (pURSP1; American Type Culture Collection, Manassas, VA) to transform the mutant strain. Transformant clones were selected at 36°C; the plasmid was recovered, and its DNA insert was sequenced. To substitute the endogenous pob1+ ORF for the region coding for Pob1ΔN (amino acids 314–871), a cassette including 0.7 kb of pob1+ promoter, the hemagglutinin (HA) epitope coding sequence fused in frame to the pob1 fragment, the ura4+ selection marker, and 0.5 kb of pob1+ terminator was constructed in a KS BlueScript vector. Subsequently, this cassette was cut with XhoI-NotI, purified, and used to transform an h+ leu1-32 ura4D-18 (PPG103) strain. Stable haploid transformants were selected and screened by polymerase chain reaction (PCR) for the appropriate gene replacement. A genomic version of pob1+ with the green fluorescent protein (GFP), Cherry, or the HA epitope coding sequences fused to the ORF 5′ end was generated using different cassettes generated as described above. A genomic version of pob1+ with the Myc epitope coding sequence fused to the 3′ end of the ORF was generated as described previously (Bähler et al., 1998).
Glutathione-Sepharose (GS) Pull-Down and Immunoprecipitation
Extracts from 5 × 108 cells expressing the different tagged proteins were obtained as described previously (Arellano et al., 1997) by using 200 μl of lysis buffer. For the analysis of Cdc42–Pob1 interaction the lysis buffer contained 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 137 mM NaCl, 0.5% NP-40, and Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO); and for the analysis of For3–Pob1 interaction, it contained 50 mM HEPES, pH 7.5, 2 mM EDTA, 130 mM NaCl, 20 mM KCl, 1 mM MgCl2, 1% NP-40, and Protease Inhibitor Cocktail. Cell extracts (2–3 mg of total protein) were incubated with GS beads or with the corresponding antibody and protein A-Sepharose beads for 2–4 h at 4°C. The beads were washed four times with lysis buffer and resuspended in sample buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon-P membranes (Millipore, Billerica, MA), and blotted to detect the GFP-, glutathione transferase (GST)-, HA- or Myc-fused epitopes with the corresponding antibodies and the enhanced chemiluminescence detection kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Western-blot analysis of cell extracts (50 μg total protein) was performed to determine the total amount of tagged protein.
In Vivo Analysis of Cdc42 Activity
The expression vector pGEX-Cdc42/Rac interactive binding (CRIB) (Manser et al., 1998) was used to transform E. coli and produce GST fused to the mammalian Pak2 binding domain for Cdc42. The fusion protein was produced according to the manufacturer's instructions and immobilized on GS beads. The amount of GTP-bound Cdc42 was determined using a pull-down assay as described previously (Coll et al., 2003). In brief, extracts from wild-type, cdc42-879, pob1-664, or wild-type overexpressing pob1+ strains carrying integrated HA-cdc42+ were obtained as described previously (Arellano et al., 1997), by using 500 μl of lysis buffer (50 mM Tris, pH 7.5, 20 mM NaCl, 0.5% NP-40, 10% glycerol, 0.1 mM dithiothreitol, 1 mM NaF, and 2 mM Cl2Mg, containing 100 μM p-aminophenyl methanesulfonyl fluoride, leupeptin, and aprotinin). Cell extracts (2 mg of total protein) were incubated with 10 μg of GST–CRIB protein coupled to GS beads for 2 h, washed four times, and blotted with anti-HA monoclonal antibody (mAb). Total HA-Cdc42 levels in whole-cell extracts (30 μg of total protein) were monitored by Western blot.
Microscopy Techniques
For calcofluor white staining of S. pombe cell wall and septum, exponentially growing cells were harvested, washed, and resuspended in a calcofluor solution (0.1 mg/ml) for 5 min at room temperature. After washing with water, cells were observed in a microscope with the corresponding UV filter. Actin staining was performed by using AlexaFluor 488-phalloidin. HA-Cdc42 immunolocalization was performed in exponentially growing cells fixed in 16% ultrapure formaldehyde and stained using anti-HA mAb as described previously (Martin et al., 2007). Cell samples were observed using a DMXRA microscope (Leica Microsystems, Deerfield, IL) equipped for Nomarski optics and epifluorescence, and photographed with an Orca-ER C4742-80 camera (Hamamatsu, Bridgewater, NJ). Actin staining was analyzed using an IX71 microscope (Olympus, Tokyo, Japan) equipped with a personal Delta Vision system and a CoolSNAP HQ2 monochrome camera (Photometrics, Tucson, AZ). Stacks of six z-series sections were acquired at 0.4-μm intervals. Images were analyzed using deconvolution and maximum two-dimensional (2D) projections of z-series. For experiments involving for3DAD*, actin stainings were performed as described previously (Martin et al., 2007), and both actin and For3 localization were imaged on a DMI4000B inverted microscope (Leica Microsystems) equipped with a HCX PL APO 100×/1.46 numerical aperture oil objective and an UltraView spinning disk confocal system (PerkinElmer Life and Analytical Sciences, Boston, MA) (including a Yokagawa CSU22 real-time confocal scanning head, an argon/krypton laser, and a cooled 14-bit frame transfer EMCCD C9100-50 camera) (Martin and Berthelot-Grosjean, 2009). Stacks of z-series confocal sections were acquired at 0.3-μm intervals with the UltraView software, and images were rendered by 2D maximal-intensity projection.
RESULTS
cdc42-879 Is a Thermosensitive Strain That Shows Actin Cable Defects
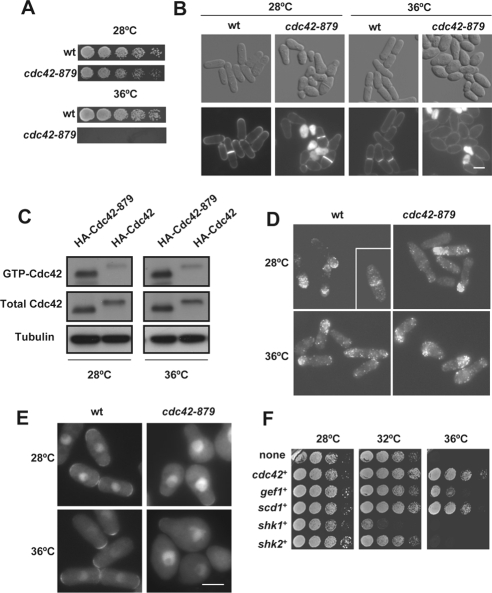
To find new Cdc42 effectors in S. pombe, we searched for suppressors of a cdc42 thermosensitive mutant strain, termed cdc42-879, which was obtained by degenerate PCR, as described previously (Martin et al., 2007). This strain showed a severe growth defect at 36°C (Figure 1A). In addition, it displayed aberrant morphology even at permissive temperature with a variety of cell shapes, namely, pear-like cells, lemon-shaped cells, round cells, and very small cells due to asymmetric septum positioning (Figure 1B). Approximately 15% of the cells grown at 28°C showed cell lysis and >50% displayed monopolar growth (compared with 22% of wild-type cells). At septation, cdc42-879 cells were shorter (10.81 ± 1.64 μm, n = 85) and wider (4.64 ± 0.83 μm, n = 85) than wild-type cells (13.56 ± 0.91 μm, n = 68; 3.64 ± 0.21 μm, n = 51) grown in the same conditions. When mutant cells were transferred to medium at 36°C, they stopped dividing after 3 h and became ovoid (Figure 1B).
Figure 1.
Morphological defects of S. pombe cdc42-879 strain. (A) Growth of wild-type (wt) and cdc42-879 cells spotted at one-fourth dilution in YES medium and incubated at 28 or 36°C for 3 d. (B) Differential interference contrast (DIC) and fluorescence images of wt and cdc42-879 cells stained with calcofluor. Cells were grown at 28 or 36°C for 5 h. Bar, 5 μm. (C) Levels of GTP-bound Cdc42 in cdc42-879 cells. Extracts from wild-type and cdc42-879 cells grown at 28°C and transferred at 28 or 36°C for 3 h were precipitated with GST-CRIB and blotted with anti-HA antibodies (top). Total HA-Cdc42 in cell lysates was visualized by Western blot. Tubulin was used as loading control. (D) Immunofluorescence of HA-Cdc42 (wt) and HA-Cdc42–879 with anti-HA antibody. Both wild-type and mutant cdc42-879 cells were grown at 28°C and transferred at 36°C for 3 h. (E) Localization of GTP-bound Cdc42 by using wt and mutant cdc42-879 cells carrying integrated Gic2 CRIB-GFP. Cells were grown at 28°C and transferred at 36°C for 3 h. (F) Rescue of cdc42-879 thermosensitive growth defect by expression of Cdc42 pathway proteins. cdc42-879 cells were transformed with the expression plasmid pREP81X carrying either no insert, cdc42+, the coding genes for the GEFs gef1+ and scd1+, and the coding genes for the PAKs shk1+ and shk2+. Cells were grown on minimal media plates for 3 d at the indicated temperatures.
To understand the reasons behind the phenotype of cdc42-879 cells, we analyzed whether this mutation caused changes in Cdc42 levels, localization, or activity. Total levels of Cdc42, as detected by Western blot, were similar in wild-type strain and cdc42-879 strains, ruling out the possibility that this strain might have a defect in Cdc42 stability at 36°C (Figure 1C).
Cdc42 localizes to the growing poles and to the division area (Merla and Johnson 2000). HA-Cdc42-879 localization was similar to that of wild-type Cdc42 as observed by immunofluorescence of fixed cells grown at either 28 or 36°C (Figure 1D). We could not observe Cdc42-879 in live cells because we were unable to tag the protein with enhanced GFP in the N-terminal region, probably because the resulting protein is nonfunctional.
Analysis of the total levels of active GTP-Cdc42 in the cdc42-879 mutant strain by pull-down using GST-CRIB from mammalian Pak2 (Manser et al., 1998) showed no decrease with respect to the levels of wild-type cells. On the contrary, a larger proportion of Cdc42-879 seemed to be GTP associated (Figure 1C). However, when we investigated the localization of active Cdc42 by imaging the Gic2 CRIB domain tagged with GFP, which binds to GTP-bound Cdc42 in live cells (Tatebe et al., 2008), we discovered that the intensity of the CRIB-GFP signal was much weaker at the growing poles and the septum region of cdc42-879 cells than in wild-type cells (Figure 1E). Together, these results suggest that cdc42-879 aberrant cell shape and thermosensitivity are not due to mislocalization of Cdc42 or to a global decrease in active Cdc42. Instead, they might be caused by a decrease in the level of GTP-Cdc42 in the areas of growth, as shown in Figure 1E, or by the lack of signaling to a specific effector protein.
Overexpression of cdc42+ or of its GEFs, in particular scd1+, which localizes to the growth areas (Hirota et al., 2003), restored the ability of cdc42-879 cells to grow at 36°C (Figure 1F), suggesting that a lack of localized GTP-Cdc42 might cause the cdc42-879 phenotype. In contrast, overexpression of shk1+ or shk2+ genes, coding for the S. pombe PAK kinases, which are the two main Cdc42 effector proteins in S. pombe, did not suppress cdc42-879 thermosensitivity. Indeed shk1+ overexpression was rather toxic in cdc42-879 cells, which barely grew at 32°C (Figure 1F). These results suggest that the thermosensitivity of this strain is not due to a defect in the PAK signaling pathways.
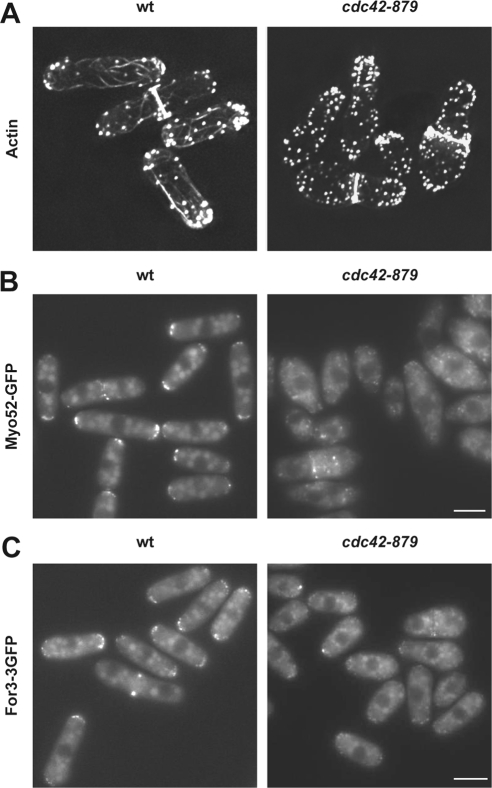
Because Cdc42 has been implicated in the organization of the actin cytoskeleton, we performed actin staining in cdc42-879 strain. The mutant cells have very weak and short actin cables as well as depolarized actin patches (Figure 2A). To corroborate the actin cable defects, localization of Myo52, a type V myosin, and the formin For3 were examined. In wild-type cells, Myo52, which carries secretory vesicles along the actin cables, localizes to the growing poles and to the division area during cytokinesis (Motegi et al., 2001). However, in cdc42-879 cells, Myo52-GFP was barely detectable at the cell poles, displaying a punctuate localization in the cytoplasm (Figure 2B). Similarly, For3-3GFP localization was reduced in cdc42-879 mutant cells compared with wild-type cells even at permissive temperature (Figure 2C). These results show that cdc42-879 cells have a defect in actin cable organization similar to that observed in cdc42-1625 mutant strain (Martin et al., 2007).
Figure 2.
Actin defects of cdc42-879 strain. (A) Actin staining of wild-type (wt) and cdc42-879 strains grown at 28°C by using AlexaFluor 488-phalloidin. Fluorescence images of Myo52-GFP (B) and For3-3GFP (C) in wt and cdc42-879 cells grown at 28°C.
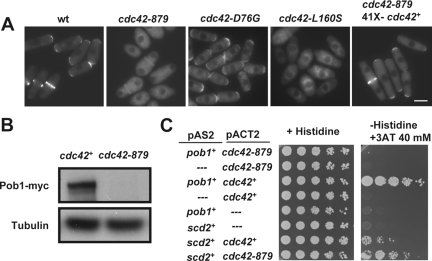
Sequence analysis of the cdc42+ ORF in the cdc42-879 mutant strain showed that it contains two point mutations that generate two amino acid changes, D76G and L160S, in the protein. It is noteworthy that the latter mutation is very close to that harbored by Cdc42-1625 protein A159V (Martin et al., 2007). To know the contribution of the two mutations to the phenotype of the cdc42-879 cells, we generated a cdc42D76G allele and a cdc42L160S allele and used them to replace the endogenous wild-type cdc42+. Although cdc42D76G cells grew at a rate similar to that of a wild-type strain at 36°C, cdc42L160S cells were unable to grow at this temperature (Supplemental Figure 1A). In addition, cdc42D76G cells were shorter and wider than wild-type cells, whereas cdc42L160S cells had aberrant morphology similar to cdc42-879 cells (Supplemental Figure 1B). cdc42L160S cells showed defects in actin cable organization, Myo52-GFP, and For3-3GFP localization similar to those in cdc42-879 cells (Supplemental Figure 1, C–E). In contrast, cdc42D76G cells had no major actin defects. Together, these results indicate that L160S mutation is largely responsible for the cdc42-879 thermosensitivity and actin cable defects.
Pob1 Is a Multicopy Suppressor of cdc42-879 Thermosensitivity That Restores Actin Cable Formation and For3 Localization
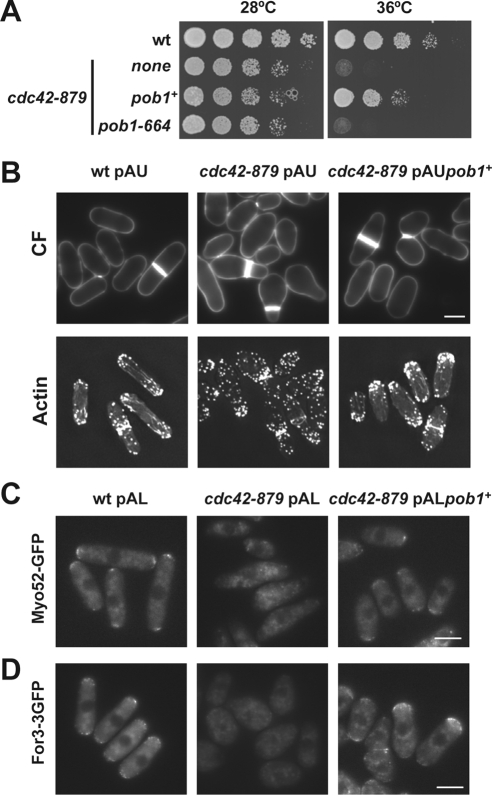
The mutant strains cdc42-879 and cdc42L160S showed similar defects in actin organization and morphology. Moreover, the two mutant strains were thermosensitive and were not suppressed by overproduction of either Shk1 or Shk2. To identify new effectors of Cdc42 we searched for multicopy suppressors of cdc42-879. We used the cdc42-879 allele because it has a more severe thermosensitive phenotype in minimal medium than cdc42L160S cells. S. pombe genomic DNA library pURSP1 was used to transform cdc42-879 cells and transformant clones were selected at 36°C. One of these clones contained a plasmid that included over 9 kb of the cosmid SPBC1289 (nucleotides 4120–13622). This fragment contained several ORFs including pob1+, coding for an essential protein involved in cell morphogenesis. Pob1 is homologous to Boi1 and Boi2 from S. cerevisiae (Toya et al., 1999) and has 871 amino acids, including a Scr homology (SH)3 domain (residues 9–60), a sterile alpha motif (SAM) domain (residues 248–312), and a pleckstrin homology (PH) domain (residues 703–806). pob1-664, a thermosensitive mutant strain, becomes misshapen at 36°C (Toya et al., 1999), displaying lemon-shaped cells similar to cdc42-879 cells. Overexpression of pob1+ partially suppressed cdc42-879 thermosensitivity (Figure 3A). In contrast, overexpression of pob1-664 did not restore the ability of cdc42-879 cells to grow at 36°C (Figure 3A). Multicopy expression of pob1+ under its endogenous promoter also partially suppressed cdc42-879 thermosensitivity. The morphology of cdc42-879 cells overexpressing pob1+ was similar to that of wild-type cells, although they were more irregular, slightly shorter, and wider (Figure 3B). These results suggest that cdc42-879 mutant might have other defects besides those suppressed by pob1+ overexpression.
Figure 3.
Pob1 is a cdc42-879 suppressor. (A) Growth of wild-type (wt) and cdc42-879 transformed with the plasmid pREP42HA empty or carrying pob1+ or pob1-664 as inserts. Cells were spotted at one-fourth dilution in minimal medium and incubated at 28 or 36°C during 3 d. (B) Fluorescence images of wt and cdc42-879 cells transformed with the plasmid pAU or pAU-pob1+. Cells grown at 28°C and shifted to 36°C for 5 h were stained with calcofluor (CF) (top) or AlexaFluor 488-phalloidin (bottom). Fluorescence images of Myo52-GFP (C) or For3-3GFP (D) in the same strains grown at 28°C. Bar, 5 μm.
Because the multicopy expression of pob1+ had an important effect on the thermosensitivity and the morphology of cdc42-879 cells, we analyzed the actin cytoskeleton of these cells by phalloidin staining. Multicopy expression of pob1+ in cdc42-879 cells restored polarized actin patches and longer and stronger actin cables as compared with cdc42-879 cells, although they seemed to be slightly more disorganized than in wild-type cells (Figure 3B). In agreement with the observation that actin cables were restored in cdc42-879 cells overexpressing pob1+, Myo52-GFP and For3-3GFP localized properly to the poles of these cells (Figure 3, C and D, respectively). Together, these results suggest that Pob1 may participate in Cdc42 signaling to activate actin cable formation and to localize For3.
Pob1 Physically Interacts with Cdc42 in a GTP-dependent Manner
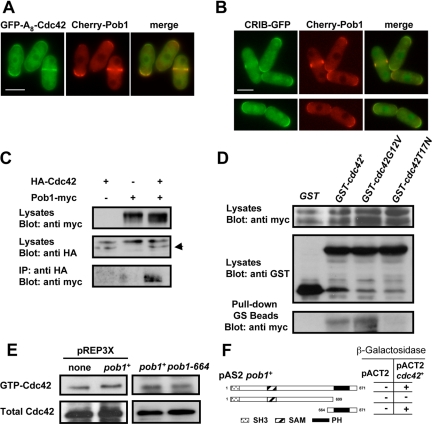
To address the relationship between Cdc42 and Pob1, we first analyzed the localization of both proteins. It had been described previously that both Cdc42 and Pob1 localized to the growing poles and to the division area (Toya et al., 1999; Merla and Johnson, 2000). We found that GFP-A8-Cdc42 partially colocalized with Pob1 (Figure 4A); moreover, CRIB-GFP also colocalized with Cherry-Pob1 (Figure 4B), suggesting that Pob1 and active Cdc42 interact. To test for physical interaction, we performed coimmunoprecipitation experiments. Using strains expressing HA-Cdc42 and Pob1-Myc under the control of their endogenous promoters, we found that anti-Myc antibodies immunoprecipitated HA-Cdc42 together with Pob1-Myc (Figure 4C). To determine whether this interaction was dependent on the activation state of Cdc42, the strain expressing Pob1-Myc was transformed with pREP1-GST plasmids containing either the wild-type cdc42+ allele, the hyperactive allele (cdc42G12V), or the dominant negative allele (cdc42T17N). Pull-down of GST using GS beads detected the highest levels of Pob1-Myc in the pull-down of extracts containing constitutively active Cdc42 (GST-Cdc42G12V). We also detected Pob1-Myc in the pull-down of GST-Cdc42 containing extracts but Pob1-Myc was hardly detected in pull-down experiments with extracts expressing the dominant negative GST-cdc42T17N allele (Figure 4D). These results indicate that Pob1 interacts with GTP-bound Cdc42.
Figure 4.
Pob1 physically interacts with Cdc42 in a GTP-dependent manner. (A) Cells containing GFP-A8-Cdc42 and Cherry-Pob1 were grown to log phase and examined by fluorescence microscopy. (B) Cells containing CRIB-GFP and Cherry-Pob1 were grown to log phase and examined by fluorescence microscopy. (C) Extracts from cells expressing HA-Cdc42 and Pob1-Myc under their endogenous promoters were immunoprecipitated using anti-HA antibody and probed with anti-Myc antibody. Extracts were assayed for the level of HA-Cdc42 and Pob1-Myc by Western blot. Arrow points to HA-Cdc42 band. (D) Cells expressing endogenous Pob1-Myc were transformed with pREP1-GST, pREP1-GSTcdc42+, pREP1-GSTcdc42G12V, or pREP1-GSTcdc42T17N and grown in the absence of thiamine for 14 h. Cell extracts were pulled down with GS beads, and the precipitates were probed with anti-Myc antibody. Expression of Pob1-Myc and GST-Cdc42 alleles was analyzed by Western blot. (E) Pob1 does not regulate the amount of GTP-bound Cdc42. Extracts from wild-type cells, cells overexpressing pob1+, or pob1-664 cells carrying HA-cdc42+ incubated at 36°C for 5 h were precipitated with GST-CRIB and blotted with anti-HA antibodies (top). Total HA-Cdc42 in cell lysates was visualized by Western blot (bottom). (F) β-Galactosidase analysis of the two-hybrid interaction between Cdc42 and different fragments of Pob1. Several regions of the pob1+ ORF were cloned into pAS2 and used as bait for cdc42+ cloned into pACT2.
There are two kinds of proteins that interact with active GTPases: effector proteins and GAPs, which negatively regulate Rho proteins. To determine the role of Pob1 regarding Cdc42, we analyzed the global levels of Cdc42-GTP in a strain overexpressing pob1+ and in a strain containing the pob1-664 allele incubated 5 h at 36°C (Figure 4E). There was no significant difference in the levels of Cdc42-GTP. Therefore, we conclude that Pob1 is not a GAP for Cdc42.
The region of Pob1 that interacts with Cdc42 was mapped using the two-hybrid system by cloning several regions of the pob1+ ORF as bait in pAS2 and cdc42+ in pACT2 (Durfee et al., 1993). Cdc42 interacted with the C-terminal PH-containing region of Pob1 and this interaction was abolished when the PH domain was absent (Figure 4F). These results indicate that Cdc42 interacts with the PH domain of Pob1 that has been described as essential for its function (Toya et al., 1999). Interestingly, in the two-hybrid assay pob1+ not only interacted with cdc42G12V and cdc42+ but also interacted with the dominant-negative cdc42T17N allele (data not shown). Therefore, it might not be Pob1 but other molecule sensing the activation state of Cdc42 in the complex detected by coimmunoprecipitation.
Cdc42 Regulates Pob1 Localization and/or Stability
Because pob1+ overexpression suppressed the phenotype of cdc42-879 and could be a target protein of Cdc42 signaling, we analyzed the localization of GFP-Pob1 to see whether this protein was altered in cdc42-879 cells. We could only detect a faint signal of Pob1 localized to the division area of cdc42-879 cells, whereas polar localization of Pob1 was completely abolished even at permissive temperature (Figure 5A). Overexpression of wild-type cdc42+ in cdc42-879 cells restored Pob1 localization to the growing areas (Figure 5A). We also analyzed the localization of Pob1 in the cdc42D76G and cdc42L160S mutant strains and found that Pob1 was localized in cdc42D76G as in wild-type cells, but the localization at the tips was abolished in cdc42L160S cells (Figure 5A). To see whether the lack of GFP-Pob1 signal was caused by Pob1 dispersion into the cytoplasm or by Pob1 degradation in cdc42-879 cells, we analyzed Pob1-myc levels by Western blotting. Pob1-myc was barely detectable, suggesting that Pob1 may be unstable in cdc42-879 (Figure 5B).
Figure 5.
Cdc42 regulates Pob1 localization. (A) Localization of GFP-Pob1 in wild-type (wt), cdc42-879, cdc42D76G, cdc42L160S, and cdc42-879 cells transformed with the overexpression plasmid pREP41Xcdc42+ grown at 28°C. (B) Total Pob1-myc in lysates from wild-type and cdc42-879 cells grown at 28°C visualized by Western blot by using anti-myc antibody (top). Tubulin was visualized in the same extracts as loading control (bottom). (C) Two-hybrid analysis of the interaction between Pob1 or Scd2 (pAS2) with Cdc42 or Cdc42–879 (pACT2). Interaction was assessed by growth on YNB plates without histidine and supplemented with 40 mM 3-amino triazol (3-AT).
The observed Pob1 localization and/or stability defect in cdc42-879 cells could be caused by a defective interaction of Pob1 with the Cdc42-879–mutated protein. Indeed, the mutated Cdc42-879 protein was unable to interact with Pob1 by two-hybrid analysis (Figure 5C). As a positive control, we used Scd2, a Cdc42 scaffold protein (Chang et al., 1994; Endo et al., 2003), that interacted with both wild-type Cdc42 and mutated Cdc42–879. These results suggest that Pob1 interaction with Cdc42 is important for its localization and/or stability.
Pob1 Binds to For3 and Might Facilitate the Cdc42-mediated Relief of Autoinhibition
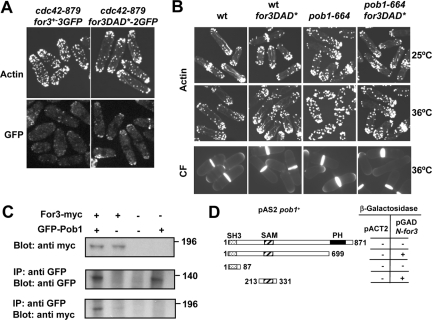
Overexpression of pob1+ partially suppresses the thermosensitivity and restores For3 localization in cdc42-879. These results led us to consider the hypothesis that Pob1 could participate in Cdc42 activation of For3. We first tested whether the cdc42-879 actin defects may be due to a failure to activate For3. We found that expression of the constitutively active for3DAD* allele, which blocks the autoinhibitory intramolecular interaction between For3 DID and DAD domains, improved cell shape and actin patch localization in cdc42-879 as it did in cdc42-1625 mutant strain (Martin et al., 2007). In addition, For3DAD*-2GFP localized more efficiently than wild-type For3-3GFP in cdc42-879 cells (Figure 6A). This suggests that, as is the case for Cdc42-1625, Cdc42-879 fails to efficiently relieve For3 autoinhibition.
Figure 6.
Pob1 binds to For3 and might facilitate the Cdc42-mediated relief of autoinhibition. (A) Actin staining of cdc42-879 endogenously expressing for3+-3GFP or for3DAD*-2GFP alleles (top). Localization of For3-3GFP or For3DAD*-2GFP in cdc42–879 cells (bottom). (B) Actin (top and middle) and calcofluor (CF) staining (bottom) of wild-type (wt), wild-type cells expressing for3DAD*, pob1-664, and pob1-664 cells expressing for3DAD* grown at 25 or 36°C. (C) Extracts from cells expressing GFP-Pob1 and For3-Myc under their endogenous promoters were immunoprecipitated using anti-GFP antibody and probed with anti-Myc antibody. Extracts were assayed for the level of GFP-Pob1 and For3-Myc by Western blot. (D) Two-hybrid analysis of the interaction between different fragments of Pob1 and For3. Several regions of the pob1+ ORF were cloned into pAS2 and the for3 sequence coding for the N-terminal half of For3 (amino acids 1–702), cloned into pGAD was used as bait.
pob1-664 mutant cells grown at restrictive temperature (36°C) display pear-shaped and lemon-shaped morphologies similar to cdc42-879 cells (Toya et al., 1999). They also showed actin cytoskeleton defects similar to cdc42-879 cells (Figure 6B). Although at the permissive temperature of 28°C, GFP-tagged Pob1-664 localized correctly to cell ends and septum and bound Cdc42 in a two-hybrid assay (data not shown), GFP-Pob1-664 seemed to be unstable at 36°C because it could hardly be detected by fluorescence or in cell extracts (Supplemental Figure 2, B and C). To test whether pob1-664 actin defects at 36°C were also due to defective For3 activation, we analyzed pob1-664 cells expressing for3DAD*. We found that these cells were longer (14.69 ± 0.90 μm) than pob1-664 single mutant cells incubated at 36°C (13.13 ± 1.06 μm) (Figure 6B). Moreover, actin patches of pob1-664 for3DAD* cells were better polarized than those of pob1-664 cells and they had slightly more cables (Figure 6B). We tried to analyze For3DAD*-2GFP localization in pob1-664 cells, but it was impossible to reach a conclusion due to imaging problems of For3-GFP at 36°C. Together, these results suggest that pob1-664 defects are due in part to a failure to activate For3.
We next checked whether Pob1 interacts with For3 by using a strain expressing For3-myc and GFP-Pob1 from their own promoters. For3-myc was detected in anti-GFP immunoprecipitates of cell extracts containing GFP-Pob1 (Figure 6C). The regions of For3–Pob1 interaction were mapped using the two-hybrid assay. We could detect interaction of the For3 N-terminal part with the Pob1 SAM domain, and this interaction was abolished when we used as bait the whole pob1+ ORF, including the PH domain (Figure 6D).
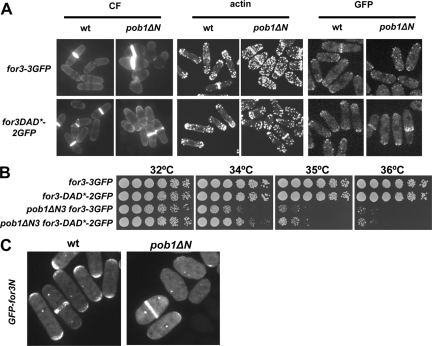
To test the significance of the Pob1–For3 interaction in vivo, we exchanged endogenous pob1+ for pob1ΔN, which lacks the N-terminal region coding for the SH3 and SAM domains (amino acids 1–314). These cells were viable but showed slow growth phenotype that is more pronounced at high temperatures and were pear or lemon shaped. Actin cables were barely visible in these mutant cells and For3-3GFP localization was severely compromised (Figure 7A). Similar to what happens with the pob1-664 mutant, expression of for3DAD* improved cell morphology and actin cytoskeleton organization in pob1ΔN cells. In addition, For3DAD* localized better than wild-type For3 in pob1ΔN (Figure 7A).
Figure 7.
The N-terminal region of Pob1 is required for For3 localization. (A) Wild-type and pob1ΔN strains grown at 30°C in YES expressing either for3-3GFP or for3DAD*-2GFP stained with calcofluor (left) and with AlexaFluor 488-phalloidin (middle) to see the actin. Fluorescence images of For3-3GFP or For3-DAD*-2GFP in the same strains grown at 30°C in YES (right). (B) for3DAD* expression improves the thermosensitive slow growth of pob1ΔN cells. Cells spotted at 1/4 dilution in YES medium and incubated at 32, 34, 35, and 36°C for 3 d. (C) Localization of GFP-For3N (including For3 amino acids 1–702) expressed from a plasmid under control of the medial-strength nmt1 promoter in wild-type and pob1ΔN cells.
We also analyzed whether the expression of for3DAD* was able to improve the slow growth of pob1ΔN cells. As shown in Figure 7B, for3DAD* partially suppresses the slow growth and thermosensitivity of pob1ΔN cells. To test whether activated For3DAD* improves any mutant with defective actin polarity, we analyzed its effect in orb3-167 cells that carry a mutant allele of the kinase nak1+ (Leonhard and Nurse, 2005). However, as shown in Supplemental Figure 3, for3DAD* expression did not suppress the round thermosensitive phenotype of orb3-167, suggesting that its rescuing effects are specific to cdc42 and pob1 mutants.
The above-mentioned results suggest that Pob1 participates in the relief of For3 autoinhibition, which is necessary for For3 localization to cell tips.
For3 anchoring at the tips is mediated by two independent localization domains—one domain in the N terminus and one domain in the C terminus (Nakano et al., 2002; Martin et al., 2005, 2007). Although the C terminus is anchored in a Bud6-dependent manner, the anchor for the N terminus is unknown (Martin et al., 2007). Because Pob1 binds the N terminus of For3 (Figure 6D), we analyzed the localization of a GFP-for3N fragment (including For3 amino acids 1–702) expressed from a plasmid under control of the medial-strength nmt1 promoter in wild-type and pob1ΔN cells. As shown in Figure 7C, GFP-for3N localization at cell tips is very reduced in pob1ΔN cells but still localizes well to the septum (and to the spindle pole body [SPB], which may be an artifact). Thus, these results indicate that the N-terminal region of Pob1 is required for the localization of For3 at cell tips. We were unable to study the localization of GFP-for3N in pob1-664 or cdc42-879 mutants, as GFP-for3N localization to cell tips is unstable at 36°C even in wild-type cells.
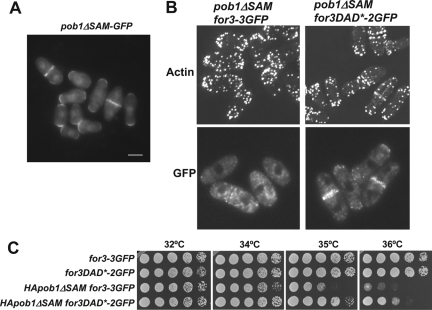
The N-terminal truncation of Pob1 is likely to abolish binding to other proteins in addition to For3. To characterize more closely the function of the domain involved in Pob1-For3 interaction, we made two additional deletions: pob1ΔSH3 (lacking amino acids 1–63) and pob1ΔSAM (lacking amino acids 197–327). Cells carrying pob1ΔSH3 did not have morphology defects and the actin cytoskeleton was similar to wild type (data not shown). GFP-Pob1ΔSAM was properly localized (Figure 8A). However, cells carrying this pob1 mutant allele were pear or lemon shaped and actin cables were difficult to detect (Figure 8B). Expression of for3DAD* improved cell morphology and actin cytoskeleton organization in these cells. Moreover, pob1ΔSAM cells have a growth thermosensitivity defect that was partially suppressed by for3DAD* (Figure 8C). Both the morphology and thermosensitivity defects of pob1ΔSAM cells are not as pronounced as those of pob1ΔN cells, suggesting that the SH3 domain might collaborate in the For3 binding or might bind to other proteins that participate in Pob1 function.
Figure 8.
The SAM domain of Pob1 is required for For3 localization. (A) Fluorescent microscopy of GFP-pob1ΔSAM–expressing cells. Bar, 5 μm. (B) Actin staining of pob1ΔSAM endogenously expressing for3+-3GFP or for3DAD*-2GFP alleles (top). Localization of For3-3GFP or For3DAD*-2GFP in pob1ΔSAM (bottom). (C) for3DAD* expression improves the thermosensitive slow growth of HA-pob1ΔSAM cells. Cells spotted at one-fourth dilution in YES medium and incubated at 32, 34, 35, and 36°C for 3 d.
DISCUSSION
Cdc42 is an essential protein controlling actin cytoskeleton organization and cell morphogenesis. Here, we describe that Pob1, the only protein of the Boi family in S. pombe, is a multicopy suppressor of cdc42-879 thermosensitivity and cytoskeletal defects. We propose that Pob1 is a novel Cdc42 target that facilitates the interaction of GTP-Cdc42 with downstream effectors, including the formin For3.
Regulation of Pob1 by Cdc42
Boi proteins regulate cell growth through their interaction with Rho GTPases in several organisms. Ashbya gossypii Boi1/2 interacts with Rho3 to regulate polarisome localization, including the formin AgBni1 (Knechtle et al., 2006). In S. cerevisiae, it has been shown that Boi proteins also participate in bud emergence (Bender et al., 1996; Matsui et al., 1996) and are required for cell growth (McCusker et al., 2007). Furthermore, Boi1 has been shown to bind activated, but not dominant-negative, alleles of Cdc42 (Bender et al., 1996). We have shown that Pob1 preferentially binds to active Cdc42 in coimmunoprecipitation experiments, although it does not have a CRIB domain. This suggests that the Boi family of proteins may be regulated by Rho-family proteins through similar mechanisms in various species.
We have ruled out the possibility that Pob1 is regulating Cdc42 because neither the overexpression nor the mutation of pob1+ causes a detectable change in the levels of active Cdc42. Localization studies indicate that Pob1 functions downstream of Cdc42, because Cdc42 regulates Pob1 localization and/or stability and not the opposite: in cdc42-879 cells, in which Cdc42 is unable to bind to Pob1, this protein failed to localize to the cell tips and was largely degraded. When Cdc42 was overproduced in cdc42-879 cells, the location of Pob1 to the poles was restored. We note however that Pob1 is delocalized from cell tips in cdc42-879 or cdc42L160S strains even at permissive temperature, suggesting that Pob1 delocalization may not be the only reason for the thermosensitivity of these strains. By contrast, GFP-A8-Cdc42 and GTP-Cdc42 remained localized to the tips of pob1-664 mutant cells at 36°C (Supplemental Figure 2A), a temperature at which Pob1-664–mutated protein is unstable. Together, these results suggest that Pob1 is a novel Cdc42 target. Other examples of Rho GTPases controlling effector localization and stability have been proposed, such as Rho1 and Rho2 regulating Pck1 and Pck2 stability (Arellano et al., 1999; Sayers et al., 2000; Calonge et al., 2003; Villar-Tajadura et al., 2008).
We mapped the region of Pob1 that interacts with Cdc42 to the C-terminal PH-containing region that is essential for the function of Boi proteins (Bender et al., 1996; Toya et al., 1999). In budding yeast, mutant versions of Boi1 with changes in the PH domain still localize to mother/bud necks but not to buds (Hallett et al., 2002). Similarly, we have found that in cdc42-879 cells, Pob1 localized to the division area but not to the poles, suggesting that Cdc42 binding is necessary for the cell tip location of Pob1. It is interesting that pob1-664 thermosensitive allele showed a point mutation that modifies an amino acid (Y719C) located at the beginning of the PH domain (our unpublished data).
Pob1 Regulation of the Formin For3 Localization and Autoinhibition
One important previously described effector of Cdc42 is the formin For3, necessary for actin cable formation (Martin et al., 2007). cdc42-879 mutant cells show For3 localization and actin cable defects, which are partially restored upon Pob1 overproduction. No enzymatic activity has been described for the Boi proteins. Instead they contain several interaction domains with other proteins or with membranes (Bender et al., 1996; Hallett et al., 2002). We also showed that Pob1 binds For3. Therefore, we suggest that Pob1 might function facilitating the interaction of GTP-Cdc42 with other effectors like For3. According to this model (Figure 9), Pob1 would localize to the cell tips by binding to GTP-Cdc42, and in turn would help For3 to localize and facilitates the Cdc42-mediated For3 activation. Thus, the inability of the Cdc42-879–mutated protein to effectively activate For3 and promote actin cable formation may be due to its failure to bind Pob1. In agreement with this model, we found that For3 likely functions downstream of Pob1 as it is dispensable for Pob1 localization (data not shown). It had also been shown that Pob1 localizes correctly in the presence of latrunculin A (Toya et al., 1999). Our model is also supported by the fact that mutations in Pob1 that abolish either Cdc42 or For3 binding (pob1-664, pob1ΔN, and pob1ΔSAM) show similar defective actin cables and delocalized For3 phenotypes, suggesting that Pob1 forms a functional link between Cdc42 and For3.
Figure 9.
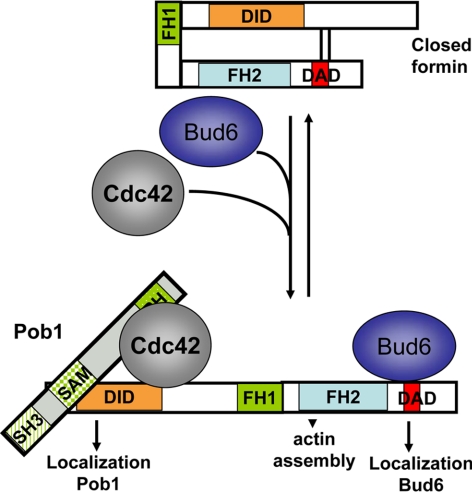
Model for For3 localization and activation at the cell tips. Pob1 localizes to cell tips by binding to GTP-Cdc42, and in turn serves as a tether for For3 N terminus and facilitates the Cdc42-mediated relief of For3 autoinhibition. Bud6 binding at the C terminus of For3 similarly serves as both anchor and activator of the formin (Martin et al., 2007).
Pob1 may regulate For3 in multiple ways. Like many formins, For3 is regulated by an autoinhibitory intramolecular interaction between its N-terminal DID domain and a C-terminal DAD sequence. Relief of this autoinhibition is regulated by Cdc42 binding at the N terminus and Bud6 binding at the C terminus (Martin et al., 2007). Constitutively active For3DAD* improved cell shape and partially restored actin cables, actin patch, and formin localization in pob1 mutant strains, an effect similar to that observed in cdc42 and bud6Δ mutants. This thus suggests that Pob1 contributes to relief of For3 autoinhibition. Because both Cdc42 and Pob1 bind to the N-terminal DID domain-containing fragment of For3, one possibility is that GTP-Cdc42 recruits Pob1 to compete with For3DAD binding. Alternatively, Pob1 may facilitate GTP-Cdc42 binding and competition with the DAD. Future biochemical experiment should clarify the mechanistic details of For3 relief of autoinhibition.
Pob1 also seems to function as one of the cortical tethers that localize For3 to the cell tips. For3 docking at cell tips relies on two independent localization domains—at the N and C termini. For3 C-terminal localization depends on Bud6 (Martin et al., 2007). We now show that localization of an N-terminal fragment of For3 is largely disrupted in pob1ΔN mutants, suggesting that Pob1 serves as a tether for For3 N terminus. Therefore, localization of full-length constitutively active For3 in pob1 mutants may largely depend on the second Bud6-dependent localization domain at the C terminus. In support of this idea, we found that, in the absence of bud6, the for3DAD* mutation largely fails to rescue the poor growth phenotype of pob1ΔN mutants (our unpublished data). Pob1 and Bud6 may thus play largely symmetric roles as both anchors and activators of For3, binding the N and C terminus of the formin, respectively.
Other Potential Roles for Pob1
It is important to note that Pob1, like Cdc42, probably plays additional roles to that of For3 regulation, because Pob1 is an essential protein and For3 is not. One tempting possibility is that Pob1 may function as an effector in other Cdc42-dependent processes essential for cell growth. For example, we observed an apparent increase in the number of patches in cdc42-879 and pob1 mutant strains. Although it is possible that this increase represents a side effect of formin defects, tipping the balance of F-actin toward arp2/3-dependent structures (Gao and Bretscher, 2008), it could also be possible that Pob1–Cdc42 complex regulates directly actin patch biogenesis. Pob1 may also be involved in Cdc42-dependent regulation of exocytosis, as described in S. cerevisiae (Wu et al., 2008). This would provide an interesting point of coordination between actin cable assembly and polarized vesicle delivery. Additional studies are necessary for elucidating new effectors of the Cdc42–Pob1 complex.
ACKNOWLEDGMENTS
We thank H. Valdivieso and J. C. Ribas for useful comments, M. Yamamoto for generous gifts of pob1-664 mutant strain and pob1+ plasmids; C. Castro for technical help with the microscopy, and D. Posner for language revision. This work was supported by grant BFU2007-60675 from the Dirección General de Investigación, MICINN, Spain, and grants SA078A07 and GR231 from Junta de Castilla y León. Work in the laboratory of S.G.M. is supported by a Swiss National Science Foundation Professorship grant (PP00A-114936) and a Human Science Program Career Development Award (CDA0016/2008).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0207) on August 26, 2009.
REFERENCES
- Arellano M., Durán A., Pérez P. Localization of the Schizosaccharomyces pombe rho1 GTPase and its involvement in the organization of the actin cytoskeleton. J. Cell Sci. 1997;110:2547–2555. doi: 10.1242/jcs.110.20.2547. [DOI] [PubMed] [Google Scholar]
- Arellano M., Valdivieso M. H., Calonge T. M., Coll P. M., Durán A., Pérez P. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J. Cell Sci. 1999;112:3569–3578. doi: 10.1242/jcs.112.20.3569. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M. S., Shah N. G., McKenzie A., III, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bender L., Lo H. S., Kokojan V., Peterson J., Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J. Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Rossi G. Spatial regulation of exocytosis and cell polarity: yeast as a model for animal cells. FEBS Lett. 2007;581:2119–2124. doi: 10.1016/j.febslet.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge T. M., Arellano M., Coll P. M., Pérez P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 2003;47:507–518. doi: 10.1046/j.1365-2958.2003.03312.x. [DOI] [PubMed] [Google Scholar]
- Cerione R. A. Cdc42, new roads to travel. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Chang E. C., Barr M., Wang Y., Jung V., Xu H. P., Wigler M. H. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- Chang E., Bartholomeusz G., Pimental R., Che J., Lai H., Wang L., Yang P., Marcus S. Direct binding and In vivo regulation of the fission yeast p21-activated kinase shk1 by the SH3 domain protein scd2. Mol. Cell. Biol. 1999;19:8066–8074. doi: 10.1128/mcb.19.12.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll P. M., Trillo Y., Ametzazurra A., Pérez P. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol. Biol. Cell. 2003;14:313–323. doi: 10.1091/mbc.E02-07-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P.-L., Yeh S.-H., Yang Y., Kilburn A. E., Lee W.-H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Endo M., Shirouzu M., Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42—the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Faix J., Grosse R. Staying in shape with formins. Dev. Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 2001:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Bretscher A. Analysis of unregulated formin activity reveals how yeast can balance F-actin assembly between different microfilament-based organizations. Mol. Biol. Cell. 2008;19:1474–1484. doi: 10.1091/mbc.E07-05-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hallett M. A., Lo H. S., Bender A. Probing the importance and potential roles of the binding of the PH-domain protein Boi1 to acidic phospholipids. BMC Cell Biol. 2002;3:16. doi: 10.1186/1471-2121-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Nurse P. A journey into space. Nat. Rev. Mol. Cell. Biol. 2001;2:647–656. doi: 10.1038/35089520. [DOI] [PubMed] [Google Scholar]
- Hirota K., Tanaka K., Ohta K., Yamamoto M. Gef1p and Scd1p, the Two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 2003;14:3617–3627. doi: 10.1091/mbc.E02-10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knechtle P., Wendland J., Philippsen P. The SH3/PH domain protein AgBoi1/2 collaborates with the Rho-type GTPase AgRho3 to prevent nonpolar growth at hyphal tips of Ashbya gossypii. Eukaryot. Cell. 2006;5:1635–1647. doi: 10.1128/EC.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard K., Nurse P. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J. Cell Sci. 2005;118:1033–1044. doi: 10.1242/jcs.01690. [DOI] [PubMed] [Google Scholar]
- Levin D. E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E., Loo T. H., Koh C. G., Zhao Z. S., Chen X. Q., Tan L., Tan I., Leung T., Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Martin S. G., McDonald W. H., Yates J. R., 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Martin S. G., Rincon S. A., Basu R., Pérez P., Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. G., Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Matsui R., R., A., Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T. A., Yates J. R., 3rd, Gygi S. P., Kellogg D. R. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 2007;9:506–515. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- Merla A., Johnson D. I. The Cdc42p GTPase is targeted to the site of cell division in the fission yeast Schizosaccharomyces pombe. Int. J. Cell Biol. 2000;79:469–477. doi: 10.1078/0171-9335-00073. [DOI] [PubMed] [Google Scholar]
- Miller P. J., Johnson D. I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizasaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Motegi F., Arai R., Mabuchi I. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol. Biol. Cell, 2001;12:1367–1380. doi: 10.1091/mbc.12.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Imai J., Arai R., Toh E. A., Matsui Y., Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Sayers L. G., Katayama S., Nakano K., Mellor H., Mabuchi I., Toda T., Parker P. J. Rho-dependence of Schizosaccharomyces pombe Pck2. Genes Cells. 2000;5:17–27. doi: 10.1046/j.1365-2443.2000.00301.x. [DOI] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tatebe H., Nakano K., Maximo R., Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 2008;18:322–330. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M., Iino Y., Yamamoto M. Fission yeast Pob1p, which is homologous to budding yeast Boi proteins and exhibit subcellular localization close to actin patches, is essential for cell elongation and separation. Mol. Biol. Cell. 1999;10:2745–2757. doi: 10.1091/mbc.10.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Tajadura M. A., Coll P. M., Madrid M., Cansado J., Santos B., Pérez P. Rga2 is a Rho2 GAP that regulates morphogenesis and cell integrity in S. pombe. Mol. Microbiol. 2008;70:867–881. doi: 10.1111/j.1365-2958.2008.06447.x. [DOI] [PubMed] [Google Scholar]
- Wu H., Rossi G., Brennwald P. The ghost in the machine: small GTPases as spatial regulators of exocytosis. Trends Cell Biol. 2008;18:397–404. doi: 10.1016/j.tcb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Kansra S., Pimental R. A., Gilbreth M., Marcus S. Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J. Biol. Chem. 1998;273:18481–18489. doi: 10.1074/jbc.273.29.18481. [DOI] [PubMed] [Google Scholar]
- Zhao Z. S., Manser E. PAK and other Rho-associated kinases effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 2005;386:201–214. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]