Abstract
Hepatitis C virus (HCV) is a leading cause of liver disease worldwide. The development of much needed specific antiviral therapies and an effective vaccine has been hampered by the lack of a convenient small animal model. The determinants restricting HCV tropism to human and chimpanzee hosts are unknown. Replication of the viral RNA has been demonstrated in mouse cells1,2, but these cells are not infectable with either lentiviral particles bearing HCV glycoproteins (HCVpp)3 or HCV produced in cell culture (HCVcc)(unpublished data), suggesting a block at the level of entry. Through an iterative cDNA library screening approach we have identified human occludin (OCLN) as an essential HCV cell entry factor that is able to render murine cells infectable with HCVpp. Similarly, OCLN is required for HCV-susceptibility of human cells, since its overexpression in uninfectable cells specifically enhanced HCVpp uptake while its silencing in permissive cells impaired both HCVpp and HCVcc infection. In addition to OCLN, HCVpp infection of murine cells required expression of the previously identified HCV entry factors, CD814, scavenger receptor class B type I (SR-BI)5, and claudin-1 (CLDN1)6. While the mouse versions of SR-BI and CLDN1 function at least as well as the human proteins for promoting HCV entry; both OCLN and CD81, however, must be of human origin to allow efficient infection. The species-specific determinants of OCLN were mapped to its second extracellular loop. The identification of OCLN as a new HCV entry factor further highlights the importance of the tight junction complex in the viral entry process and provides a major advance towards efforts to develop small animal models for HCV.
HCV virions, lipid-enveloped nucleocapsids bearing the viral glycoproteins E1 and E2, appear to enter a host cell in a highly coordinated process involving components of the virus particle and numerous cellular factors7. From the long list of putative HCV entry factors, strong evidence supports specific roles for the tetraspanin CD814, SR-BI5, and the tight junction protein CLDN16. This list appears incomplete, however, as numerous human cell lines and all non-primate cell lines are resistant to HCV entry even when all three human factors are overexpressed3,6 (and data not shown).
To identify additional factors able to render non-human cells susceptible to HCV entry, we performed a cyclic retrovirus-based repackaging screen of a complementary DNA library derived from the highly HCV-permissive human hepatocarcinoma Huh-7.5 cell line8. We screened for genes that rendered a non-permissive mouse embryonic fibroblast cell line, NIH3T3, infectable with HCV pseudoparticles (HCVpp), defective lentiviral particles that display the HCV glycoproteins and measure only viral entry9, 10, 11 (Supplementary Fig. S1a and Supplementary Methods). To maximize the likelihood identifying a novel component that would permit HCV entry into non-human cells, we utilized an NIH3T3 subclone overexpressing human CD81, SR-BI, and CLDN1. This screen identified human occludin (OCLN) as a potential novel HCV entry factor. OCLN is a four transmembrane domain protein present in the tight junction complex of polarized epithelial cells, where it likely functions to regulate paracellular permeability and cell adhesion12,13. Expression of human CD81, SR-BI, CLDN1, and OCLN in NIH3T3 cells enhanced HCVpp infectivity by approximately 120-fold (Fig. 1c and Fig. S1c).
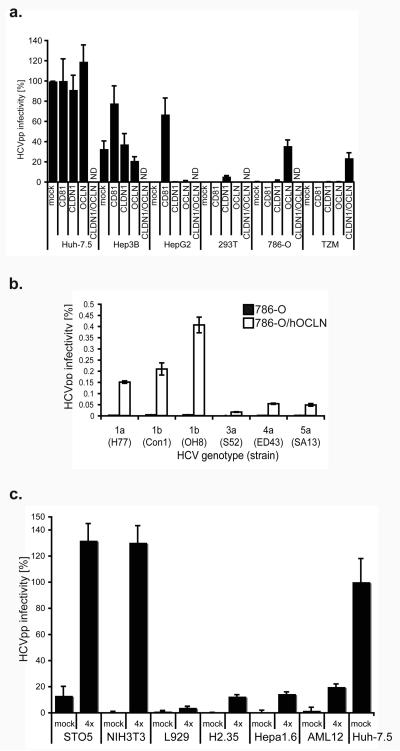
Figure 1. OCLN expression confers susceptibility to HCVpp.
a, Human cells were transduced with human pTRIP-mCherry-CD81 (CD81), pTRIP-Cerulean-CLDN1 (CLDN1), pTRIP-Venus-OCLN (OCLN), both CLDN1 and OCLN, or mock transduced, then challenged in parallel with HCVpp and VSVGpp. HCVpp infectivity is reported as the titer of HCVpp divided by the titer of VSVGpp, after subtraction of the signals from infection with non-enveloped pseudoparticles (Env−pp). Pseudoparticles of different HCV genotypes were used to infect naive (black) or OCLN-expressing (white) 786-O cells (b). c, Mouse cells transduced with human pTRIP-mCherry-CD81 (CD81), pTRIP-SR-BI (SR-BI), pTRIP-Cerulean-CLDN1 (CLDN1) and pTRIP-Venus-OCLN (OCLN) (4×) or mock transduced; challenged and HCVpp infectivity calculated as above. ND, not determined. Means and standard deviation (SD) of at least triplicate experiments are shown.
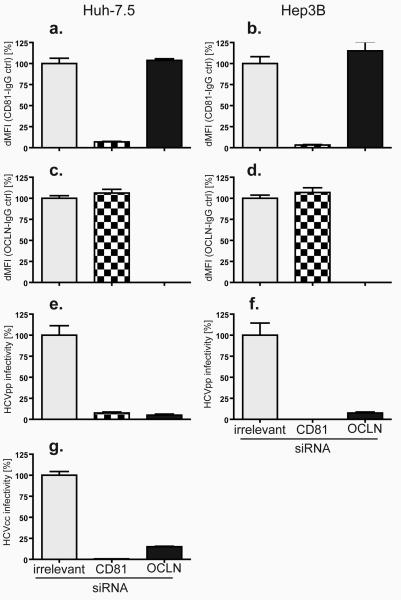
To confirm that OCLN is required for HCV entry, we examined the relationship between its expression and HCV susceptibility in a variety of human cell lines. Naturally HCV-permissive human hepatocyte cell lines (Huh-7.5 and Hep3B) or those previously shown to lack other entry factors (hepatocellular carcinoma HepG2, and embryonic kidney HEK293T) were found to express readily detectable levels of endogenous OCLN (Fig. S1d). In these cells further overexpression of OCLN did not enhance susceptibility to HCVpp (Fig. 1a). Silencing of OCLN, however, inhibited HCVpp infection of Hep3B cells (Fig. 2f), and both HCVpp and HCVcc infection of Huh-7.5 cells (Fig. 2e and g, respectively), indicating that OCLN is essential for HCV infection of naturally permissive cells. This inhibition was HCV-specific, as infection with pseudoparticles bearing the vesicular stomatitis virus glycoprotein (VSVGpp) was not affected (values in Fig 2e-f are corrected for VSV-Gpp infectivity). It should be noted that OCLN knockdown resulted in incomplete inhibition of HCVpp infection for both cell lines. This is likely due to incomplete silencing or stable reservoirs of this protein that remained to contribute some entry functions. In contrast, the naturally HCV-resistant renal carcinoma cell line, 786-O, expresses high levels of the major HCV entry factors CD81, SR-BI, and CLDN1, but approximately 17-fold less OCLN than Huh-7.5 cells (Fig. S1d). OCLN overexpression in 786-O cells specifically enhanced HCVpp infection by over 88-fold (Fig. 1a). Furthermore, this OCLN-dependence of HCVpp infection was observed across a panel of diverse HCV genotypes (Fig. 1b). Another HCVpp-resistant cell line, the HeLa cell-derived cervical carcinoma line TZM, was found to lack both endogenous CLDN1 and OCLN (>100 and 25-fold less mRNA than Huh-7.5 cells, respectively) (Fig. S1d). Overexpression of these factors together increased HCVpp infectivity of TZM cells by 450-fold (Fig. 1a). These results indicate that OCLN is an essential HCV entry factor.
Figure 2. OCLN silencing inhibits HCV entry.
Huh-7.5 (left) and Hep3B (right) cells transfected with irrelevant (grey), CD81 (checkered), or OCLN (black) specific siRNA pools were stained with CD81 (a and b) and OCLN (c and d) antibodies. Expression was analyzed by flow cytometry; the ratio between specific and isotype control staining is shown. Concurrently, siRNA-treated cells were challenged with HCVpp (e and f) and HCVcc (g). HCVpp infectivity was calculated as described in methods; HCVcc infection was quantified by the ratio of viral protein (NS5A) to isotype control staining. All samples are normalized to irrelevant-siRNA treatment. dMFI, difference in mean fluorescence intensity. Means and SD of at least triplicate experiments are shown.
To further examine HCV entry requirements in non-human cells, we transduced murine NIH3T3 cells with all combinations of one, two, three, or all four human CD81, SR-BI, CLDN1, and OCLN. Expression levels were determined by expressing an mCherry/CLDN1 fluorescent protein fusion (Fig. S3a) or by FACS staining for CD81, SR-BI (Fig. S3a) and OCLN (Fig. S3b). No combination of human factors enhanced VSVGpp infectivity (data not shown and results in Fig. 3a are normalized to VSVGpp values. See Fig. S2 for representative raw data). As shown in figure 3a, infectivity was not significantly altered by expression of any of the factors alone. Expression of all four entry factors, however, had a large impact on HCVpp permissiveness, with a 45-fold increase over naïve NIH3T3 cells. Furthermore, expression of all four factors, expressed as fluorescent protein fusions for CD81, CLDN1 and OCLN and un-tagged SR-BI, conferred HCV-susceptibility to mouse embryonic fibroblast cell lines, STO5 and L929, as well as the mouse hepatocyte cell lines, AML12, Hepa1.6, and H2.35, with enhancements of 4 to 85-fold over naïve cells (Fig. 1c). Permissiveness to VSVGpp was not affected by expression of the human proteins. Attempts to infect these murine cells with HCVcc were unsuccessful, likely as a result of inefficient viral RNA replication in these cells, as previously documented2,14. It is interesting that different murine cell lines exhibited varying degrees of HCVpp entry permissiveness after transduction with all four factors. HCV entry factor levels or cell determinants affecting lentivirus reporter expression do not appear to explain the observed differences (since the data were normalized to VSVGpp). Rather, these cell type specific differences may reflect real biological variability in other cellular machinery involved in HCV entry.
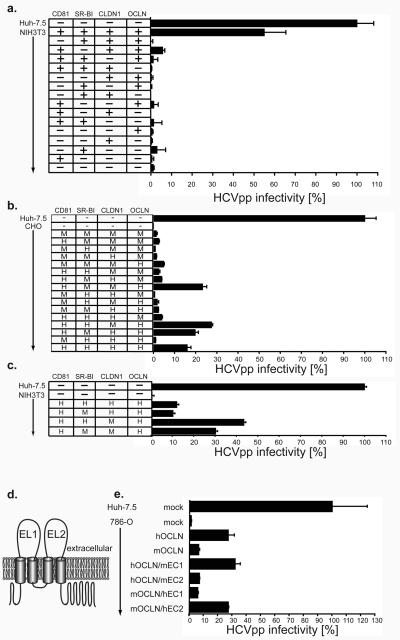
Figure 3. Expression of human OCLN and human CD81 determines HCV species tropism.
a, Mouse NIH3T3 were transduced with combinations of human pTRIP-CD81 (CD81), pTRIP-SR-BI (SR-BI), pTRIP-mCherry-CLDN1 (CLDN1) and pTRIP-OCLN (OCLN). Hamster CHO (b) and NIH3T3 (c) cells were transduced with combinations of human (H) and mouse (M) pTRIP-mCherry-CD81 (CD81), pTRIP-SR-BI (SR-BI), pTRIP-Cerulean-CLDN1 (CLDN1) and pTRIP-Venus-OCLN (OCLN). d, Diagram of OCLN membrane topology12. e, 786-O cells were transduced with mouse/human OCLN chimaeras; all chimaeras were N-terminally tagged with Venus yellow fluorescent protein. Transduced cells (a, b, c and e) were challenged with HCVpp and VSVGpp encoding GFP reporters and HCVpp infectivity calculated and normalized as described in methods. Means and SD of at least duplicate experiments are shown.
To investigate which of the four entry factors are responsible for HCV species-specific tropism, we performed additional experiments in Chinese hamster ovary (CHO) cells, which express low endogenous levels of SR-BI, CLDN1 and OCLN (Fig. S4b). In CHO cells overexpression of all four human factors specifically enhanced HCVpp infection by 60 to 336-fold (Figs. 3b and S4a). While omission of CD81, CLDN1, or OCLN abolished HCVpp entry, a low level of endogenous SR-BI in CHO cells provided some HCVpp entry function when the other three factors were transduced (Fig. S4a). However, since overexpression of SR-BI further enhanced entry by 3-fold in the context of the other human proteins (Fig. S4a), we still considered CHO cells to be an excellent environment in which to study the species-specific functions of each factor. To do so, we transduced CHO cells to express every combination of human and mouse CD81, SR-BI CLDN1, and OCLN. In all combinations, mouse SR-BI functioned equivalently to that of human origin (Fig. 3b). This shows for the first time that mouse and human SR-BI are equally capable of mediating HCV uptake. In CHO cells mouse CLDN1 appears slightly more functional for HCVpp entry than the human protein (Fig. 3b). This is in agreement with a slight preference observed when either of these CLDN1 proteins, which are 90% identical, were expressed in human 293T cells6. These observations were confirmed by transducing mouse NIH3T3 cells with human CD81 and OCLN and all permutations of mouse and human SRBI and CLDN1 (Fig. 3c). While SRBI displayed little to no species specificity, NIH3T3 cells expressing mouse CLDN1, in addition to the other three entry factors, were slightly more permissive to HCVpp than the same cells expressing human CLDN1. While the determinants for this CLDN1 preference have not yet been examined, it remains clear that CLDN1 does not contribute to the restriction to HCVpp entry in mouse cells. It is noteworthy that none of the mouse cells examined above express sufficiently high levels of either murine SR-BI or CLDN1 (Fig. S4d and S4e), thus explaining their dependence on transduction of these proteins for HCVpp permissivity.
In contrast, CD81 and OCLN exhibited human specific HCV entry factor functions in CHO and NIH3T3 cells. CHO cells expressing either of these human proteins in the context of the other three mouse proteins were slightly more permissive to HCVpp than cells expressing only the mouse proteins (Fig. 3b). Furthermore, expression of both human CD81 and OCLN, together with murine SR-BI and CLDN1, rendered CHO cells as infectable with HCVpp as cells expressing all four human factors (Fig. 3b, compare last bar to any with human CD81 and OCLN). These results agree with our previous finding that mouse and hamster CD81, when expressed in the CD81-deficient human hepatocellular carcinoma cell line HepG2, support only low level HCV entry (greater than 10-fold reduced from human CD81)15. Importantly, these data indicate that CD81 and OCLN represent the minimal human-specific entry factors, at least in the context of mouse and hamster cells.
For CD81, the species-specific difference in HCV-entry activity between rat and human proteins maps exclusively to its large extracellular loop15. To determine the regions of OCLN that are responsible for the functional difference observed between mouse and human proteins, which are 91% identical, we expressed chimaeric OCLN molecules in 786-O cells and assayed HCVpp permissivity. In these experiments, a mouse chimaera bearing the second extracellular loop (EC2) of human OCLN was as active as the full-length human protein and at least 4-fold more active than the mouse homologue (Fig 3e, compare mOCLN/hEC2 to hOCLN and mOCLN, respectively). Conversely, a human OCLN mutant with the EC2 of the mouse protein functioned similarly to the full-length mouse factor in HCVpp infectivity (Fig 3e, compare hOCLN/mEC2 to mOCLN). These data suggest that the human-specific determinants of OCLN's HCV entry factor functions are entirely contained within EC2.
This study represents a major step forward in understanding both HCV host cell entry and HCV species tropism. All human, murine, or hamster cell lines that we tested became permissive for HCV entry when engineered to express the molecules CD81, SR-BI, CLDN1, and OCLN. HCV entry into primary mouse hepatocytes might be complicated by expression of additional dominant negative restriction factors absent in cell lines, which is experimentally difficult to address at this point. Our data suggest that OCLN completes the list of cell-type specific HCV entry factors; any other factors required for HCV entry must be ubiquitously expressed, or at least conserved between a wide range of human and mouse cells. The fact the OCLN is a major component of the tight junction complex further highlights the significance of this structure and cell polarity to HCV entry. We previously showed that CLDN1 acts late in the entry process, just prior to virion internalization6. The intimate association of CLDN1 and OCLN at the tight junction suggests that both these factors may function in a similar time frame. The use of multiple uptake factors with distinct cell surface distributions strengthens the hypothesis that HCV follows a coordinated entry pathway similar to that of coxsackievirus B. This virus initially interacts with a primary receptor (decay-accelerating factor) on the luminal cell surface, followed by lateral migration of the virus–receptor complex to the tight junction, where interaction with the coxsackie and adenovirus receptor is immediately followed by uptake into the host cell16. Strikingly, coxsackievirus B entry also requires OCLN17, further suggesting similar entry mechanisms of this virus and HCV. Recent work by Brazzoli et al. supports this step-wise model of HCV entry, demonstrating that initial engagement of CD81 on the cell surface, by either fluorescently labeled CD81 antibodies or soluble forms of the HCV glycoproteins, is followed by GTPase-dependant actin rearrangements that allow lateral movement of the CD81-bound complex into areas of cell-cell contact overlapping with both CLDN1 and OCLN localization18.
The study of HCV pathogenesis and the development of urgently needed effective antivirals and therapeutic and/or preventative vaccines targeting this virus has been severely hampered by the lack of convenient inbred small animal models capable of supporting HCV infection and replication. Numerous blocks will certainly need to be overcome before complete viral replication in a mouse can be achieved. HCV RNA replication in mouse cells is inefficient1,2, and the ability of such cells to support virion assembly is unknown. Our results clearly demonstrate, however, why mouse cells are unable to support HCV entry. This major block to HCV replication in murine cells can now be overcome simply by the expression of human CD81 and OCLN in the context of mouse CLDN1 and SR-BI, providing a clear foundation upon which a mouse model for HCV infection can begin to be constructed.
Methods
For more detailed methods see Supplementary Information
Pseudoparticles
Pseudoparticles were generated by FuGENE6 (Roche Applied Science, Indianapolis) mediated cotransfection of 293T cells with plasmids encoding (1) a minimal HIV (pTRIP, CSGW, CSPW) or MLV (LMN8) provirus encoding green fluorescent protein (GFP) or other transgene, (2) gag-pol (HIV or MLV) and (3) appropriate viral glycoproteins (HCV E1E2 or VSV-G). To generate Env−pp the glycoprotein vector was replaced with empty vector. Pseudoparticle-containing supernatants were harvested at 24, 48 and 72 h, pooled and filtered (0.45 μm pore size). Pseudoparticle infections were performed in the presence of 4 μg/ml polybrene. A minimum of 72 h elapsed between transduction and reporter gene quantification by flowcytometry or subsequent experiments.
Cyclic lentivirus cDNA library repackaging rescue screen
An Huh-7.5-derived cDNA library in the LMN8 retroviral vector (kindly provided by Paul Bieniasz, Aaron Diamond AIDS Research Center, The Rockefeller University, New York, U.S.A.) was packaged into VSVGpp and applied to NIH3T3 cells stably expressing human CD81, SR-BI and CLDN1 (N3xF26), which were then challenged with HCVpp, encoding a puromycin (CSPW) resistance gene, and subjected to antibiotic selection (Fig. S1a). Surviving clones were pooled and a fraction tested for their susceptibility to GFP-encoding HCVpp. The populations were subsequently transfected with MLV gag-pol and VSV-G expression plasmids to package the LMN8 genomes with the cDNA inserts contained in the surviving N3xF26 cells into fresh VSVGpp (cyclic packaging rescue, CPR). These were used to transduce naïve N3xF26 cells for subsequent rounds of screening (see supplementary materials for further details).
Cell culture grown HCV (HCVcc)
A plasmid encoding the chimaeric Jc1FLAG2(p7-nsGluc2A) reporter genome19 was XbaI linearized and transcribed using MEGAscript T7 (Ambion, Austin, Texas). RNA was electroporated into Huh-7.5 cells using a ECM 830 electroporator (BTX Genetronics) and HCVcc was collected from supernatants 48-72 h after transfection20.
Supplementary Figure S1. Identification of OCLN as an HCV entry factor. a, Illustration of the screen for cDNAs conferring HCVpp-susceptibility to NIH3T3 cells expressing human CD81, SR-BI and mCherry/CLDN1 (N3×F26). The primary Huh-7.5-derived library, in a LMN8 murine leukemia virus (MLV)-based retroviral vector, was packaged into VSVGpp by co-transfection of 293T cells with the library DNA and vectors expressing MLV gag-pol and VSV-G. Naïve N3×F26 cells were then transduced with the pseudoparticle library and challenged with HCVpp encoding a puromycin resistance gene (CSPW) followed by antibiotic selection. Surviving clones were pooled, expanded, and tested for susceptibility to pseudoparticles bearing no glycoprotein (Env−pp), HCVpp and VSVGpp encoding a GFP reporter (CSGW). The population was subsequently transfected with MLV gag-pol and VSV-G to produce VSVGpp encoding the LMN8 genomes with the cDNA inserts contained in the surviving N3×F26 cells (cyclic packaging rescue, CPR). These re-packaged pseudoparticles were used to transduce naïve N3×F26 cells for subsequent rounds of screening. CSPW proviruses are HIV-1 based and thus not repackaged during the MLV gag-pol mediated CPR step. In addition, this provirus contains a deletion in the 3'LTR that prevents expression of packagable viral RNA in transduced cells. b, HCVpp permissivity of N3×F26 cells increased during multiple rounds of library transduction and selection. Naïve N3×F26 cells (first column) or aliquots of N3×F26 cells transduced with the parental (2nd column) or repackaged (3rd and 4th column) LMN8-cDNA library were assessed for permissivity to Env−pp, HCVpp, or VSVGpp encoding a GFP reporter (CSGW). GFP expression was measured by flow cytometry 72 h after infection. Actions performed between assays are denoted on the top: library – initial transduction of the cDNA library; puro – transduction with HCVpp encoding a puromycin resistance reporter gene followed by puromycin selection; CPR – transfection of MLV gag-pol and VSV-G to repackage the cDNAs contained in the N3×F26 cell population into pseudoparticles for delivery to naïve N3×F26 cells and further selection. The percentage of GFP positive cells is indicated in the upper right corner of each plot. After two rounds of selection including one repackaging steps the effective HCVpp titer on the selected population increased by about 100-fold (3rd column). After three rounds of selection including two repackaging steps the effective HCVpp titer increased about 20-fold compared to naïve N3×F26 cells (top row) whereas the effective titers of VSVGpp (2nd row) and Env−pp (3rd row) were essentially unchanged or even decreased. Of 24 cell clones isolated by FACS after 3 rounds of selection, eight harbored a cDNA encoding OCLN. No cDNA could was found in the remaining 16 clones, likely representing untransduced cells selected through nonspecific uptake of HCVpp. All of the LMN8-OCLN cDNA clones contained an identical amino-terminally truncated OCLN cDNA fragment that expressed an OCLN protein missing the first 140 amino acids, essentially removing the intracellular N-terminal tail. c, LMN8-VSVGpp encoding the isolated N-terminally truncated OCLN as well as pTRIP-VSVGpp encoding the full-length OCLN were used to transduce naïve N3×F26 cells. These cells were then challenged with GFP encoding pseudoparticles. HCVpp infectivity is reported as the titer of HCVpp divided by the titer of VSVGpp, after subtraction of Env−pp signals, as described in the materials and methods. Values are normalized to parallel infections of highly permissive Huh-7.5 cells. d, Total RNA was isolated from the indicated cell lines and the human CD81, CLDN1 and OCLN mRNA levels were determined by RT-quantitative PCR. Data were normalized to GAPDH expression and plotted as percentage of Huh-7.5 cells. e, Pseudoparticles of different HCV genotypes were used to infect naive Huh7.5 cells and HCVpp infectivity calculated as above. Means and standard deviations (SD) of at least triplicate experiments are shown
Supplementary Figure S2. Selected raw data of HCV entry in human, mouse and hamster cells as analyzed by flow cytometry. Human (Huh7.5, 786-O), mouse (NIH3T3) and hamster (CHO) cells were transduced as indicated with human pTRIP-mCherry-hCD81 (hCD81), pTRIP-Cerulean-hCLDN1 (hCLDN1), pTRIP-Venus-hOCLN (hOCLN) and then challenged in parallel with HCVpp, VSVGpp or Env−pp. Shown are FACS plots representative of those that have been used for the calculations of HCV infectivity in all other analyses.
Supplementary Figure S3. Expression of HCV entry factors in transduced cells. Mouse NIH3T3 were transduced in the indicated combinations with human pTRIP-CD81 (CD81), pTRIP-SR-BI (SR-BI), pTRIP-mCherry-CLDN1 (CLDN1) and pTRIP-OCLN (OCLN). CD81, SR-BI and CLDN1 (a) and OCLN expression (b) were monitored by flow cytometry at the time of infection. dMFI, the difference in the mean fluorescence intensity. Means and SD of at least triplicate experiments are shown.
Supplementary Figure S4. CD81, SR-BI, CLDN1 and OCLN constitute the minimal set for efficient HCVpp entry into nonhuman cells. a, Hamster CHO cells were transduced with the indicated combinations of human pTRIP-mCherry-CD81 (CD81), pTRIP-SR-BI (SR-BI), pTRIP-Cerulean-CLDN1 (CLDN1) and pTRIP-Venus-OCLN (OCLN). Transduced cells were challenged with HCVpp and VSVGpp encoding GFP reporters and HCVpp infectivity calculated and normalized as described in the materials and methods. Total RNA was isolated from CHO (b) and NIH3T3 (d), hamster (c) and mouse (e) liver and the hamster or mouse CD81, CLDN1 and OCLN mRNA levels were determined by RT-quantitative PCR. Data were normalized to GAPDH expression. Means and SD of at least duplicate experiments are shown.
Supplementary Material
Acknowledgements
The authors thank David Bowman, Kathy Mu, Michelle Hunter, Jodie Tassello, and Megan Holz for excellent technical assistance, John L. Law and Nadim Shohdy for advice on RNAi, Thomas von Hahn and Andrew J. Syder for helpful discussions and Catherine Murray for editing the manuscript. Svetlana Mazel, Christopher Bare and Xiaoxuan Fan from the Rockefeller University Flowcytometry Core Facility provided outstanding technical support. This work was supported by the Greenberg Medical Research Institute, the Ellison Medical Foundation, the Starr Foundation, the Ronald A. Shellow Memorial Fund, the Richard Salomon Family Foundation and funded in part by a Grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative (grants to C.M.R.), and the National Institutes of Health (grants to M.J.E. and C.M.R.). C.M.R. is an Ellison Medical Foundation Senior Scholar in Global Infectious Diseases. A.P. and M.J.E. were supported by Kimberly Lawrence-Netter Cancer Research Discovery Fund Award Postdoctoral Fellowships. M.J.E. was also supported by the Charles H. Revson Postdoctoral Fellowship. We would like to thank Paul D. Bieniasz and Theodora Hatziioannou (Aaron Diamond AIDS Research Center, The Rockefeller University, New York, NY) for providing reagents, including the LMN8 retroviral plasmid, and expertise necessary for the cDNA library construction and screening. Roger Tsien (University of California, San Diego, CA) kindly provided the mCherry fluorescent protein encoding plasmid, Atsushi Miyawaki (RIKEN, Saitama, Japan) the Venus/YFP plasmid and David W. Piston (Vanderbilt University, Nashville, TN) for the Cerulean/CFP plasmid. Constructs expressing HCV glycoproteins of diverse genotypes and the JFH-1 subgenomic replicon cDNA were generously provided by Jens Bukh (Copenhagen University Hospital, Hvidovre, Denmark) and Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) respectively.
Footnotes
Competing financial interests The authors declare the following conflicts of interest, which are managed under University policy: C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and the HCVcc cell culture system.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Uprichard SL, Chung J, Chisari FV, Wakita T. Replication of a hepatitis C virus replicon clone in mouse cells. Virol J. 2006;3:89. doi: 10.1186/1743-422X-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Q, Guo JT, Seeger C. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J Virol. 2003;77(17):9204. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197(5):633. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 5.Scarselli E, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO Journal. 2002;21(19):5017. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007 doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 7.von Hahn T, Rice CM. Hepatitis C virus entry. J Biol Chem. 2008;283(7):3689. doi: 10.1074/jbc.R700024200. [DOI] [PubMed] [Google Scholar]
- 8.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 2002;76:13001. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartosch B, et al. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A. 2003;100(24):14199. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummer HE, Maerz A, Poumbourios P. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 2003;546(2-3):385. doi: 10.1016/s0014-5793(03)00635-5. [DOI] [PubMed] [Google Scholar]
- 11.Hsu M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA. 2003;100:7271. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paris L, Tonutti L, Vannini C, Bazzoni G. Structural organization of the tight junctions. Biochim Biophys Acta. 2008;1778(3):646. doi: 10.1016/j.bbamem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Chiba H, et al. Transmembrane proteins of tight junctions. Biochim Biophys Acta. 2008;1778(3):588. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Uprichard S, Chisari F, Wakita T. presented at the 12th International Symposium on Hepatitis C Virus and Related Viruses; Montreal, Canada. 2005. unpublished. [Google Scholar]
- 15.Flint M, et al. Diverse CD81 proteins support hepatitis C virus infection. J Virol. 2006;80(22):11331. doi: 10.1128/JVI.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124(1):119. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 17.Coyne CB, Shen L, Turner JR, Bergelson JM. Coxsackievirus entry across epithelial tight junctions requires occludin and the small GTPases Rab34 and Rab5. Cell Host Microbe. 2007;2(3):181. doi: 10.1016/j.chom.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brazzoli M, et al. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82(17):8316. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marukian S, et al. Cell Culture-Produced Hepatitis C Virus Does Not Infect Peripheral Blood Mononuclear Cells. Hepatology. 2008 doi: 10.1002/hep.22550. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309(5734):623. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.