Summary
The functionalization of alkanes was once thought to lie strictly within the domain of enzymes that activate dioxygen in order to generate an oxidant with suitable potency to cleave inert C–H bonds. The emergence of the radical SAM superfamily of enzymes—those which use S-adenosyl-l-methionine as a precursor to a 5′-deoxyadenosyl 5′-radical—has kindled a renaissance in the study of radical-dependent enzymatic reactions, and is ushering in a wealth of new and intriguing chemistry that remains to be elucidated. This review will focus on a special subclass of radical SAM enzymes that functionalize inert C–H bonds, highlighting the functional groups and the chemistry that leads to their insertion. Within this class are: (1) enzymes that catalyze sulfur insertion, the prototype of which is biotin synthase; (2) enzymes that catalyze P- or C-methylation, such as P-methylase or Fom 3; (3) enzymes that catalyze oxygen insertion, such as the anaerobic magnesium protoporphyrin-IX oxidative cyclase (BchE); (4) and enzymes that functionalize n-hexane or other alkanes as the first step in the metabolism of these inert compounds by certain bacteria. In addition to surveying reactions that have been studied at various levels of detail, this review will speculate on the mechanisms of other types of reactions that this chemistry lends itself to.
The functionalization of unactivated C–H bonds is one of the more demanding tasks with which enzymes are charged. The acid dissociation constants (pKas) of unactivated C–H bonds can approach values of 40 and beyond, rendering these bonds too inert to be acted upon by polar chemical processes—meaning those in which key steps involve proton or hydride transfer. Indeed, these kinds of reactions, almost by definition, involve paramagnetic species or intermediates, and are initiated by removal of a key hydrogen atom (H•). Compounding the dilemma, many unactivated C–H bonds exhibit homolytic bond-dissociation energies (HBDEs) that range upwards of 98 to 104 kcal/mol. Therefore, the challenge is to generate, in a controlled fashion, a species that has suitably elevated oxidizing power to cleave bonds of this nature. In a review article written in 1990, Perry Frey proposed that the enzymatic generation of such a species would necessitate the participation of a metal atom [1]. Now, almost two decades later, this hypothesis still holds true for both aerobic and anaerobic mechanisms of C–H functionalization.
Dioxygen-derived Oxidants
In the aerobic world, nature has responded to the challenge of cleaving inert C–H bonds by evolving several mechanisms for the controlled generation of potent oxidants. Consistent with the argument put forth by Frey, each requires a metal ion—most notably iron—which, in the course of cleaving the O–O bond of dioxygen, accesses high valence states that result in formation of the potent oxidant. Although other metals such as copper—and some organic cofactors such as flavins—participate in the generation of oxidants that can cleave C–H bonds, those C–H bonds tend to be mildly to significantly activated, displaying significantly reduced HBDEs. The most common cofactors employed in the aerobic functionalization of C–H bonds include heme systems, non-heme mononuclear iron (II) systems, and non-heme dinuclear iron (II) systems. The commonality among these systems is that each must reduce molecular oxygen by four electrons to complete its catalytic cycle. Two of these electrons are typically provided by the substrate, while the remaining two either derive from NADPH via the intermediacy of other electron carriers, or are provided by a co-substrate [2]. Other enzymes, such as the non-heme mononuclear iron(II) enzyme isopenicillin N-synthase (IPNS) and the non-heme diiron enzyme myo-inositol oxygenase, oxidize their substrates by four electrons, and thus do not require the input of external reducing equivalents [3-5].
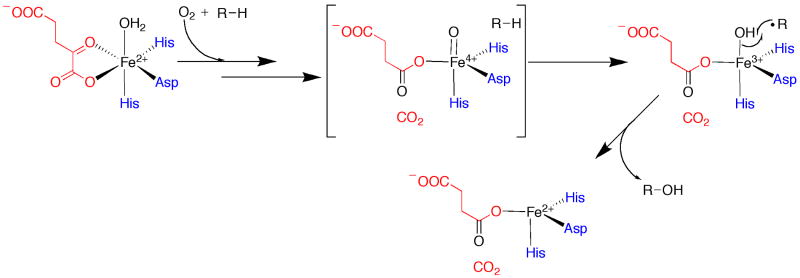
Many, if not most, instances of inert C–H bond functionalization by dioxygen-utilizing enzymes involve the insertion of an oxygen atom into the C–H bond; however, alternative reactions include halogenation, desaturation, cyclization, and epoxidation [4]. For the most common reaction—hydroxylation—abstraction of a hydrogen atom by a high-valent iron–oxo species—most often Fe(IV)–oxo—affords a substrate radical and a reduced iron–hydroxo intermediate. Hydroxylation then arises via a radical-recombination between these two species, which is often described as the radical-rebound step [4]. Figure 1 depicts an abbreviated series of steps for hydroxylation by the α-ketoglutarate (αKG)-dependent oxygenases. In their resting state, these enzymes contain a mononuclear Fe(II) atom that is coordinated by a (His)2-(Asp/Glu) motif—referred to as the 2-His-1-carboxylate facial triad—which is often found in non-heme mononuclear iron(II) enzymes that use dioxygen as a cosubstrate [2]. The octahedral coordination sphere of the iron atom is completed by three water ligands, all of which depart upon binding of α-KG and the appropriate substrate. Upon addition of dioxygen to the pentacoordinated Fe(II) atom, a reductive decarboxylation takes place with formation of a succinate-coordinated Fe(IV)–oxo species (shown in brackets). This potent oxidant abstracts a hydrogen atom from the substrate, resulting in formation of an Fe(III)–hydroxo species. This species then undergoes radical recombination with the substrate radical to afford product and a coordinatively unsaturated Fe(II) species, which can rebind a molecule of water and another molecule of α-KG to return to the resting ternary complex [4]. Amazingly, this class of enzymes also has been shown to catalyze halogenation of unactivated C–H bonds [6]. The crystal structure of the chlorinase SyrB2 showed that the Asp/Glu ligand of the facial triad motif is substituted with an alanyl residue, providing a site for coordination of a chloride ion [7]. Therefore, in chlorination of unactivated C–H bonds, abstraction of a substrate-derived hydrogen atom by an Fe(IV)–oxo intermediate is proposed to be followed by a radical recombination between the substrate radical and the chloride ligand rather than the hydroxide ligand [7,8].
Figure 1.
Abbreviated mechanism for hydroxylation by α-ketoglutarate-dependent enzymes (adapted from reference 4). Shown in red is the α-ketoglutarate co-substrate. Shown in blue are the ligands that compose the 2-His-1-carboxylate facial triad.
5′-Deoxyadenosyl 5′-Cobalamin
The functionalization of inert C–H bonds is not a characteristic that is solely within the purview of aerobic organisms. Many obligate anaerobes also require the ability to cleave strong C–H bonds for a variety of purposes, functionalization of these unactivated positions being only one of them [9]. Moreover, some of these anaerobic strategies for cleaving strong C–H bonds are also found in aerobes and in all three kingdoms of life, suggesting that they are remnants of the anoxic world that were not selected against by evolutionary pressure upon the introduction of dioxygen into the atmosphere.
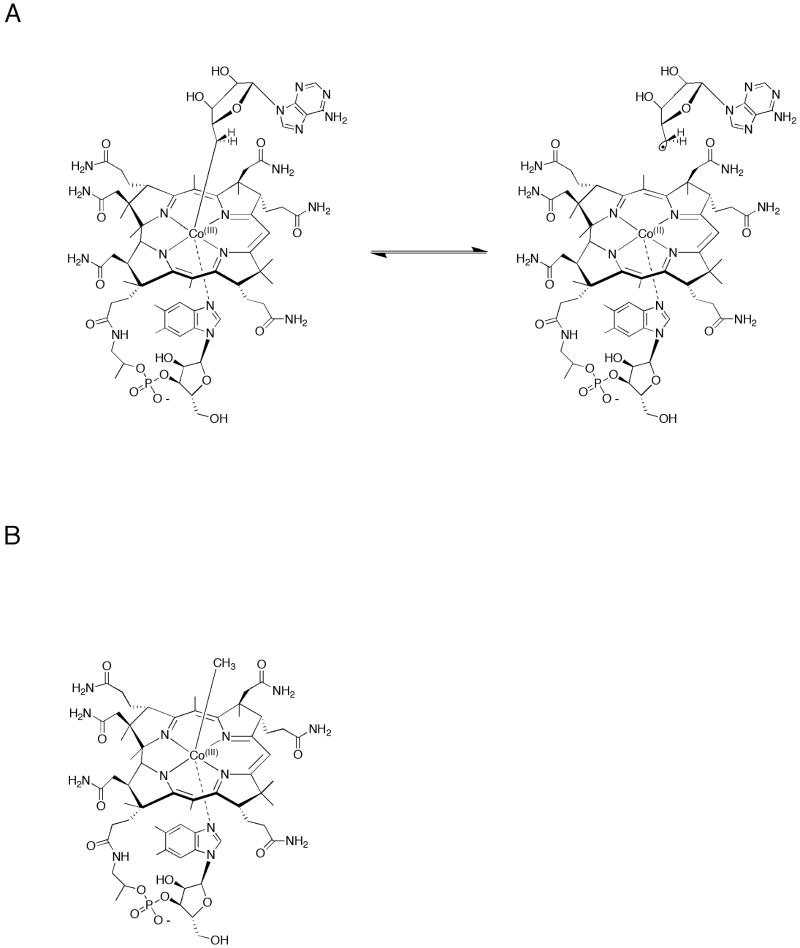
It appears that nature's solution for cleaving strong C–H bonds in the absence of participation by dioxygen is the 5′-deoxyadenosyl 5′-radical (5′-dA•), first suggested in classic and seminal studies of the enzymes dioldehydrase, glyceroldehydrase, and ethanolamine ammonia lyase [1,10]. In these enzymes, the precursor to the 5′-dA• is 5′-deoxyadenosyl 5′-cobalamin (AdoCbl), the coenzyme form of vitamin B12. As shown in Figure 2a, AdoCbl is a large molecule, having several distinct domains. At first glance the core of the molecule, the corrin ring, resembles that of protoporphyrin IX—the organic component of the heme macrocycle—although closer inspection will show that there are differences in saturation, differences in size (the corrin macrocycle is contracted by one carbon), and differences in the substituents that decorate the perimeter of each macrocyle. At the center of the corrin macrocycle is a cobalt atom that is bonded directly to the 5′-carbon of 5′-deoxyadenosine, its distal ligand; a 5,6-dimethylbenzimidazole group that is connected to the corrin macrocyle by a propanolamine-ribofuranosyl phosphate chain serves as the proximal ligand. In some B12-dependent reactions, upon binding of the cofactor to the protein, the 5,6-dimethylbenzimidazole ligand is displaced by the imidazole side chain of a histidinyl residue, which is often found in a conserved DxHxxG motif [11]. These two conformations of the proximal ligand are denoted base-on and base-off, respectively. Methylcoblamin, shown in Figure 2b, is another B12-derived cofactor, and is employed as a methyl donor in a subset of methyltransferase reactions [11]. Its importance in the functionalization of unactived C–H bonds is discussed in more detail below.
Figure 2.
Structures of 5′-adenosyl 5′-cobalamin (AdoCbl) (A); and methylcobalamin (B). As shown, AdoCbl undergoes homolysis to afford a 5′-deoxyadenosyl 5′-radical and cob(II)alamin.
AdoCbl participates in two classes of reactions, none of which involves functionalization of C–H bonds. The first class is the most expansive, and comprises 1,2 cross-migrations, in which a substituent (–NH2, –OH, –CH(NH2)COOH, –CO(SCoA), –C(CH2)COOH) on one carbon atom exchanges positions with a hydrogen on an adjacent carbon atom. The second class consists of a dehydration reaction that is followed by the addition of what is formally a hydride ion. The only established reaction in this class is the reduction of ribonucleotides to deoxyribonucleotides, catalyzed by B12-dependent ribonucleotide reductases (RNRs) [11]. The salient property of AdoCbl is the relative weakness of the Co–C organometallic bond, which exhibits HBDEs ranging from ∼31–34 kcal/mol, depending on whether the cofactor is in the base-on or base-off conformation [11]. In general, binding of substrate (or allosteric effector) to the holoenzyme complex induces homolysis of the Co–C bond, generating a cob(II)alamin species and the 5′-dA• (Figure 2a). Turnover then initiates by abstraction of an appropriate hydrogen atom from the small-molecule substrate in mutase reactions, or abstraction of a hydrogen atom from a protein cysteinyl residue, which in turn abstracts a hydrogen atom from a nucleotide substrate, as is found in RNRs [11]. In all known AdoCbl-dependent reactions, homolysis of the cofactor is a reversible process during normal catalysis; at the end of each turnover the 5′-dA• is regenerated via abstraction of a hydrogen atom from 5′-dA by a product radical, and then recombines with cob(II)alamin to afford the intact cofactor.
Radical SAM
AdoCbl is a structurally complex molecule, and is synthesized de novo by only a select set of microorganisms [12]. About 20 gene products are required for its de novo biosynthesis [12]—an energetically expensive undertaking—begging the question: “Is there an anaerobic counterpart that requires less elaborate cofactors?” The answer is yes! However, it appears that in creating AdoCbl nature has lazily revamped a more primitive design for generating the exact same oxidant so that the cofactor—at the very least—might be more tolerant of the introduction of dioxygen into the atmosphere. Evidence suggests that the primordial radical generator is S-adenosyl-l-methionine (SAM), which, in concert with a [4Fe–4S] cluster, allows for formation of the same 5′-dA• generated by AdoCbl-dependent enzymes. In fact, this radical-generating system has been designated “a poor man's adenosylcobalamin,” because it is not as elaborate and elegant as the more evolved cofactor [13]. As will be described below, the [4Fe–4S]/SAM cofactor participates in a vastly greater variety of reactions than AdoCbl, which has led to its rebranding as “a rich man's adenosylcobalamin” [14].
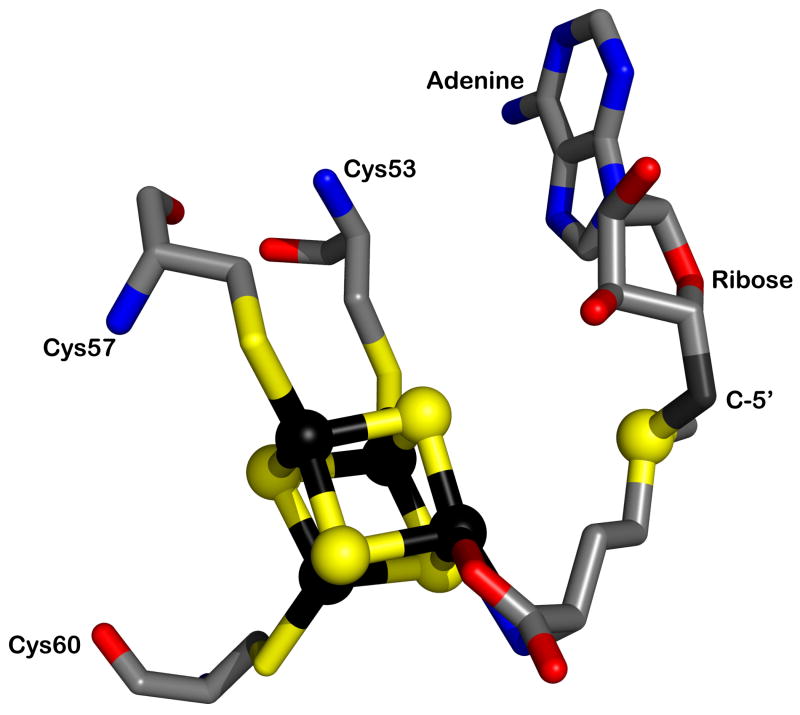
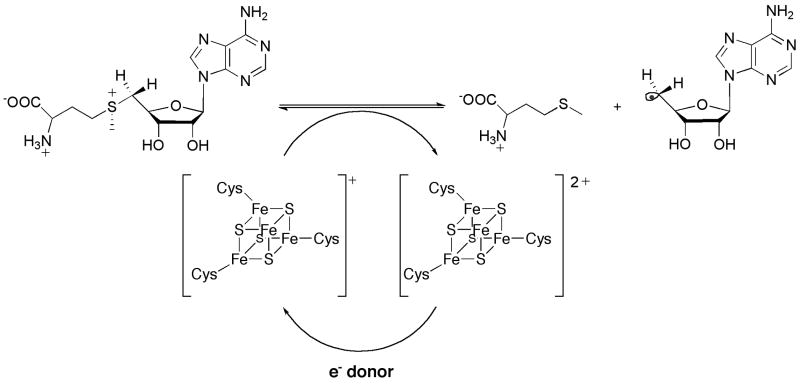
The hallmark of the class of enzymes that use the [4Fe–4S]/SAM cofactor—designated the radical SAM (RS) superfamily—is a CxxxCxxC motif found typically in the N-terminal half of their primary structures [15]. The cysteinyl residues within the motif coordinate a [4Fe–4S]2+/+ cluster that is obligate for turnover, and which functions intimately in the generation of the potent oxidant [15]. One iron atom in the cubane cluster is not ligated by a protein cysteinyl ligand, which presents an open coordination site to which SAM binds via its α-amino and α-carboxylate groups (Figure 3) [16,17]. In instances in which RS proteins are isolated with their iron–sulfur (Fe/S) cluster(s) intact, or in which the clusters are reconstituted by mixing the apo-protein with excess iron and sulfide, the clusters are typically in the diamagnetic +2 oxidation state. The addition of a low potential electron—which is provided by designated proteins in vivo (or in vitro) or by artificial reductants in vitro—affords the catalytically relevant form, in which the Fe/S cluster assumes the +1 oxidation state [18,19]. In contrast to the Co–C bond of AdoCbl, the HBDE of the S–5′-C bond of SAM is too elevated (∼60 kcal/mol) for the 5′-dA• to be generated by simple homolysis. Radical generation requires the injection of an electron from the [4Fe–4S]+ cluster into the sulfonium of SAM, which induces scission of the S–5′-C bond, affording l-methionine as the co-product (Figure 4) [18,19].
Figure 3.
Structure of S-adenosylmethionine bound in contact to the [4Fe–4S] cluster in biotin synthase (PDB 1R30) (adapted from reference 55). The iron atoms of the iron-sulfur cluster are shown in black, while the sulfur atoms are shown in yellow. Oxygen atoms, red; nitrogen atoms, blue; carbon atoms, grey. The structure was prepared using the Pymol Molecular Graphics System (http://www.pymol.org)
Figure 4.
Formation of a 5′-deoxyadenosyl radical via a reductive cleavage of S-adenosylmethionine. The coproduct shown is L-methionine. The iron–sulfur cluster displayed is that which is ligated by cysteines in the CxxxCxxC motif.
In an early review article on RS enzymes, the authors delineated three subclasses, which were distinguished by the fate of the 5′-dA• [20]. This classification is still valid, and serves as a useful means of grouping the reactions for further discussion. Class I enzymes are most analogous to those that require AdoCbl, in that they cleave SAM reversibly. At the end of each catalytic cycle SAM is regenerated, presumably via attack of the 5′-dA• onto l-methionine with concomitant reduction of the [4Fe–4S]2+ cluster to [4Fe–4S]+. In Class II enzymes, the cleavage of SAM is irreversible; however, the 5′-dA• radical abstracts a hydrogen atom from a glycyl residue residing on a cognate protein, creating a glycyl radical cofactor that is stable in the absence of molecular oxygen. The glycyl radical cofactor then initiates catalysis by abstracting a target hydrogen atom, and is itself regenerated after each turnover. In Class III enzymes SAM is cleaved irreversibly for each hydrogen atom abstracted and functions solely as a co-substrate. To date, only Class II and Class III RS enzymes are known to catalyze functionalization of inert C–H bonds.
Class II (Glycyl Radical) RS Enzymes Involved in C–H Functionalization
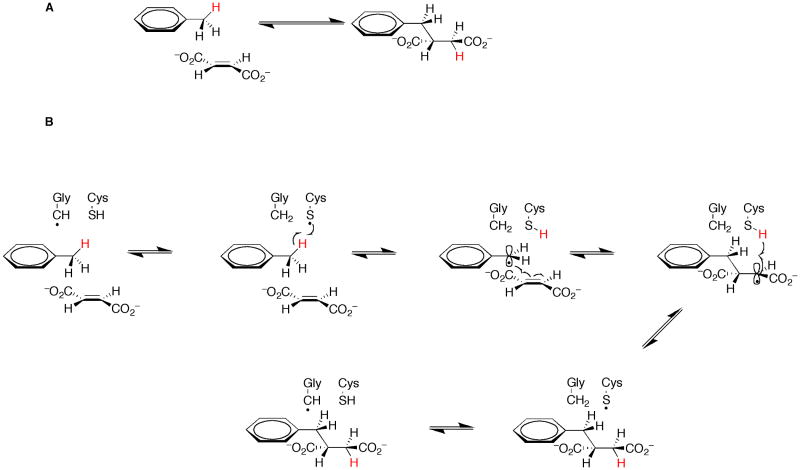
The prototype of glycyl radical enzymes that catalyze C–H functionalization is benzylsuccinate synthase (BSS). This enzyme catalyzes the first step in the anaerobic catabolism of toluene, which is the addition of fumarate to the methyl group of toluene, affording (R)-benzylsuccinate (Figure 5a) [21,22]. Enzymes from Thauera aromatica and Azoarcus sp. strain T have been the subject of most in vitro mechanistic interrogation, and are α2β2γ2 heterohexamers composed of polypeptides of ∼98 kDa, ∼8.7 kDa, and ∼6.6 kDa in molecular mass, respectively [23-25]. The C-terminal half of the α-subunit of BSSs shares strong sequence similarity with enzymes that are known to bear glycyl radicals, such as the anaerobic ribonucleotide reductase (ARR) and pyruvate formate–lyase (PFL). This similarity also extends to an absolutely conserved cysteinyl residue, which functions intimately in their reactions. Substitution of this cysteinyl residue (Cys492) with an alanyl residue by site-directed mutagenesis afforded a protein that was unable to complement a strain carrying a lethal mutation in the gene that encodes the α subunit of BSS [26]. Mechanistic studies indicate that the methyl hydrogen removed from toluene during catalysis is transferred to the C-3 pro-(S) position of benzylsuccinate, indicating a syn addition of fumarate [27]. A mechanism that is consistent with current experimental evidence is shown in Figure 5b. The reductive cleavage of SAM is catalyzed by a separate activating protein [24,26], allowing for formation of a glycyl radical on the α subunit via the intermediacy of a 5′-dA•. The glycyl radical abstracts a hydrogen atom from the conserved cysteine residue to form a thiyl radical, which in turn abstracts a hydrogen atom from the methyl group of toluene to afford a benzyl radical. The benzyl radical adds to the re face of C-2 of fumarate to yield a benzylsuccinyl radical, which is quenched by return of a hydrogen atom from the conserved cysteinyl residue. The resulting thiyl radical is then free to regenerate the glycyl radical cofactor [27-29].
Figure 5.
Reaction catalyzed by benzylsuccinate synthase (A). Proposed mechanism for benzylsuccinate synthase (B) (adapted from reference 27).
The C–H bonds on the methyl substituent of toluene are not completely unactivated; they possess HBDEs of ∼85 kcal/mol [30]. However, there is mounting evidence that a similar system is operative in the anaerobic degradation of n-alkanes! Certain strains or cultures of sulfate- or nitrate–reducing obligate or facultative anaerobes were found to metabolize a variety of straight-chain fatty acids or n-alkanes [31-34]. Metabolite analyses of a sulfate-reducing bacterial culture as well as the nitrate-reducing strain, HxN1, indicated that various n-alkanes were activated by their addition to fumarate to give the corresponding alkylsuccinate. In both instances addition of fumarate took place at one of the subterminal positions of the n-alkane, suggesting that the enzyme system takes advantage of the lower HBDEs of C–H bonds of subterminal carbons [35,36]. Moreover, when perdeuterated n-dodecane was administered to the sulfate-reducing culture under anoxic growth conditions, all deuterium atoms were retained in the product, suggestive of return of the abstracted deuterium atom across the double bond of fumarate, reminiscent of the reaction catalyzed by BSS (Figure 5b) [35].
A recent proteomics analysis of genes that are upregulated in strain HxN1 under conditions of growth on n-hexane led to the identification of an operon that encoded seven proteins, of which several displayed sequence similarity to components of BSS. This operon was given the identifier mas for (1-methylalkyl)succinate synthase-encoding, and the seven open reading frames were labeled masA through masG [37]. MasC, MasD, and MasE are thought to be homologs of the three subunits of BSS. MasD is the largest of the three proteins, and bears the conserved motif (RVxG) that is found in all enzymes known to accommodate glycyl radical cofactors, as well as the conserved cysteine residue that is converted into a thiyl radical during catalysis. In addition, masG encodes a protein that is homologous to glycyl-radical activating enzymes. In support of a BSS-like mechanism for activation of alkanes, a strong EPR signal indicative of a glycyl radical was observed in suspended cells grown on n-hexane but not n-hexanoic acid, which is an already activated compound [36].
Similar findings were obtained with the sulfate-reducing strain AK-01. Two proteins that were homologous to the α subunit of BSS were upregulated when strain AK-01 was grown on n-hexadecane, but not when grown on n-hexadecanoic acid. Sequencing of the genomic DNA of strain AK-01 revealed that these proteins were associated with different operons, which were denoted alkylsuccinate synthase 1 and 2 (ass1 and ass2) [38]. Although in vitro studies of either the MAS or ASS enzyme systems have not been reported, a mechanism very similar to that described for BSS is expected (Figure 5b) [9]. It is interesting that nature would elect to employ a glycyl radical-based mechanism for alkylsuccinate synthases, given the unfavorable equilibrium associated with a glycyl radical ultimately generating a secondary alkyl radical—although via a thiyl radical intermediate. Glycyl and cysteinyl residues possess gas-phase HBDEs of ∼83 (Cα–H) and 90 (S–H) kcal/mol, respectively [39,40], while C–H bonds on secondary carbons possess HBDEs of ∼95 kcal/mol [40]. Therefore, the equilibrium would be expected to lie considerably to the left in the absence of significant influence by the active site of the enzyme.
Class III RS Enzymes Involved in C–H Functionalization by Sulfur Insertion
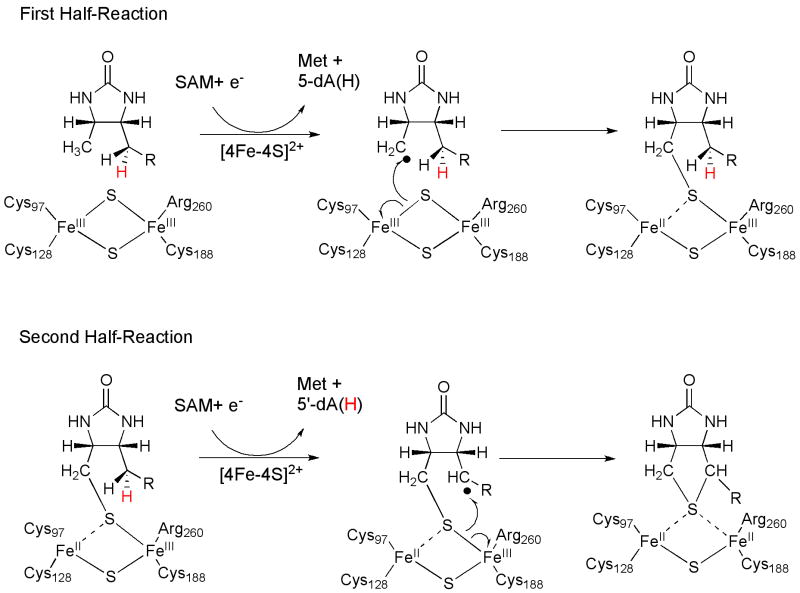
Class III RS enzymes were once defined as those that catalyze the insertion of sulfur atoms into unactivated C–H bonds [20]; however, it is now apparent that enzymes within this class catalyze a far greater diversity of reactions, including sulfur insertion, decarboxylation, methylthiolation, dehydrogenation, C–C bond formation, and various complex transformations involved in the biosynthesis of cofactors and other natural products [41]. Of the enzymes that catalyze sulfur insertion, biotin synthase (BioB) is the best studied, and has served as the prototype. As shown in Figure 6a, it catalyzes C–H bond cleavage with concomitant sulfur insertion between carbons 6 and 9 of dethiobiotin (DTB), resulting in formation of the thiophane ring of biotin [42]. Labeling studies indicated a direct transfer of deuterium to 5′-dA when BioB was incubated with [9-2H3]-, [6-2H2, 9-2H3]-, or [6-2HS, 9-2H1]-DTB under turnover conditions. The near absence of deuterium incorporation when [6-2HR, 9-2H1]-DTB was used as the substrate supports direct hydrogen atom abstraction from C-6 by a 5′-dA•, and indicates that the pro-S hydrogen is the one that is removed [43]. These studies are also consistent with earlier findings that suggested that two equiv of SAM must be expended to form one equiv of biotin [44]. More recent studies have shown that 9-mercaptodethiobiotin is converted into biotin in a kinetically competent fashion, suggesting that it is an intermediate in the reaction [45,46].
Figure 6.
Reactions catalyzed by radical SAM enzymes that are known to catalyze sulfur insertion. Biotin synthase reaction (A); lipoyl synthase reaction (B); MiaB reaction (C); RimO reaction (D). SAM, S-adenosylmethionine; Met, L-methionine; 5′-dA, 5′-deoxyadenosine (figure adapted from reference 56).
The question that has stimulated the greatest interest in the BS reaction pertains to the direct source of the sulfur atom incorporated into biotin and the chemical mechanism for its incorporation. After a sinuous period of investigation by a number of laboratories, two studies provided evidence that the sulfur atom derives from the protein itself, which was consistent with in vitro assays that showed that no more than one equiv of biotin is formed from one equiv of BioB monomer [47,48]. Analysis of the active form of BioB by UV-vis, Mössbauer, and resonance Raman spectroscopies showed that it contains one [2Fe–2S] cluster and one [4Fe–4S] cluster per polypeptide [49-52]. Upon incubation of this form of the protein under turnover conditions, absorption features attributed to the [2Fe–2S] cluster were observed to diminish, while those of the [4Fe–4S] cluster remained relatively constant. The rate of disappearance of the [2Fe–2S] cluster was reported to be similar to that for formation of biotin [53]. A similar observation was made in a separate study in which Mössbauer spectroscopy was used to monitor changes in both cluster forms; however, the [2Fe–2S] cluster was reported to disappear significantly faster than the rate of biotin formation, which is consistent both chemically and kinetically with its involvement in biotin formation [54]. Finally, BioB that was reconstituted and shown spectroscopically to contain a [2Fe–2Se] cluster supported production of selenobiotin [52].
A working model that is consistent with most experimental evidence is shown in Figure 7, and invokes the use of the [2Fe–2S] cluster as the source of the sulfur atom inserted into DTB [53]. Following a reductive cleavage of SAM, the resulting 5′-dA• abstracts a hydrogen atom from C-9 of DTB to afford 5′-dA and a substrate radical. The substrate radical attacks one of the bridging μ-sulfido ligands of the [2Fe–2S] cluster with concomitant inner-sphere electron transfer to one of the ferric irons to afford the mixed-valent Fe3+/Fe2+ pair. Another reductive cleavage of SAM generates a second 5′-dA•, which abstracts the 6-pro-S hydrogen of DTB. The resulting substrate radical attacks the sulfur atom of 9-mercaptodethiobiotin with concomitant inner-sphere electron transfer to the coordinated Fe3+, completing formation of the thiolane ring with retention of configuration at C-6 [53].
Figure 7.
Proposed mechanism for biotin synthase (adapted from reference 53).
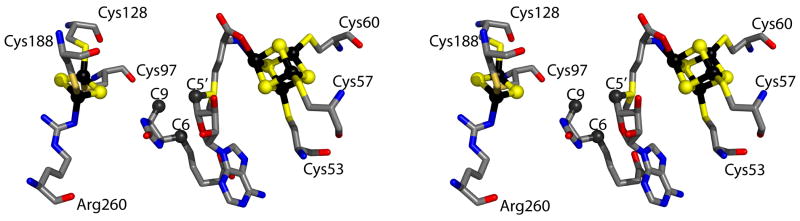
The X-ray crystal structure of BioB from E. coli appears to be generally consistent with the proposed mechanism of catalysis. The active site shows SAM bound in contact with the [4Fe–4S] cluster, and DTB to be in close proximity to the [2Fe–2S] cluster, with both clusters flanking the organic substrates (Figure 8). Of particular note, C-9 of DTB is ∼4.6 Å away from the closest bridging μ-sulfido ligand of the [2Fe–2S] cluster and 3.9 Å away from C-5′ of SAM [55].
Figure 8.
X-ray structure of the biotin synthase active site (PDB 1R30) (Ref 55). The [4Fe–4S] and [2Fe–2S] clusters as well as dethiobiotin and S-adenosylmethionine are shown in stick format. The iron atoms of the iron-sulfur clusters are shown in black, while the sulfur atoms are shown in yellow. Oxygen atoms, red; nitrogen atoms, blue; carbon atoms, grey. The structure was prepared using the Pymol Molecular Graphics System (http://www.pymol.org)
The mechanism for BioB has some precedent in the second half-reaction of the reaction catalyzed by the mononuclear non-heme iron-dependent enzyme isopenicillin N-synthase (IPNS), which catalyzes the four-electron oxidation of δ-(l-α-aminoadipoyl)-l-cysteinyl-d-valine (ACV) tripeptide to isopenicillin N [3]. However, in contrast to BS, both the oxidant—proposed to be an Fe(IV)–oxo species formed via chemical steps that involve dioxygen—and the atom to be inserted are part of the same molecule (Figure 9). The salient feature of the mechanism is the coordination of the sulfur atom to be inserted to an Fe3+ species (Figure 9; shown in brackets), which allows for facile inner-sphere electron transfer concomitant with radical attack and ring closure (Figure 9). Because ring-closure involves insertion of a sulfur atom that derives from the substrate itself, IPNS is readily catalytic. By contrast, BS and other similar RS enzymes that catalyze sulfur insertion are sacrificed after just one turnover by virtue of their roles as both catalysts and substrates [56]. Recent experimental evidence, however, indicates that the BS reaction is catalytic in vivo, suggesting that pathways exist for the facile regeneration of the [2Fe–2S] cluster [57,58].
Figure 9.
Abbreviated mechanism of isopenicillin N-Synthase (adapted from reference 4). The key step, shown in brackets, is the radical recombination with concomitant inner-sphere electron transfer that allows ring formation.
A more detailed analysis of RS enzymes that catalyze sulfur insertion was recently published in this journal, and the reader is invited to scan this review for further insight into this intriguing class of reactions [56]. Other known enzymes within this class include lipoyl synthase (LipA), MiaB protein (MiaB), and RimO. LipA catalyzes the last step in the synthesis of the lipoyl cofactor, which is the insertion of two sulfur atoms into C–H bonds at carbons 6 and 8 of a fatty acyl chain appended in an amide linkage to a lipoyl carrier protein (Figure 6b). MiaB catalyzes the last step in the biosynthesis of the hypermodified tRNA nucleoside 2-methylthio-N6-(isopentenyl)adenosine, which is the insertion of a methylthiol group into the C–H bond at C-2 of a modified adenine ring in certain tRNAs (Figure 6c). One fascinating aspect of MiaB is that a protein composed of a single polypeptide exploits two distinct reactivities of SAM—as a precursor to a 5′-dA• and as a methyl donor [59]. Presently it is not known whether both reactions—sulfur insertion and methylation—take place at the same active site [56]. RimO catalyzes methylthiolation of a conserved aspartate residue (D88 in E. coli) in the ribosomal protein S12 in some organisms, and bears strong sequence similarity to MiaB [60]. No in vitro characterization of this protein has yet been reported. Unlike BS, LS and MiaB do not harbor a [2Fe–2S] cluster, but contain two [4Fe–4S] clusters [56]. It is postulated that the second Fe/S cluster functions in the same capacity as the [2Fe–2S] cluster of BS [56]. The HBDEs of relevant C–H bonds in each of these reactions range as high as ∼98 kcal/mol [40,61], the highest known to date for anaerobic C–H bond cleavage.
Can Functionalization be Cryptic?
One of the exciting aspects of sulfur insertion reactions is the realization that they have the potential to give rise to cryptic outcomes, as is observed for the α-KG-dependent histone trimethyllysyl demethylases, in which oxygen insertion into a C–H bond of a methyl group located on the ε nitrogen of a lysyl residue allows for demethylation via a carbinolamine intermediate [62]. In other words, thiolation or methylthiolation can be the anaerobic counterpart to hydroxylation, opening up possibilities for novel mechanisms of signaling or functionalization/defunctionalization. For example, it has been suggested that the transcription elongator complex protein, Elp3, which harbors N-acetyltransferase activity, might also contain RS-dependent histone demethylase activity, because the protein contains a cysteine rich motif that bears modest resemblance to the CxxxCxxC motif of known RS proteins [63]. Although it is unlikely that Elp3 functions in this capacity, the concept of RS-dependent histone demethylation is an exciting one, especially because, unlike the flavin-dependent histone lysine demethylases, RS chemistry should be amenable to demethylation of trimethyllysyl residues, similarly to the α-KG-dependent histone lysyl demethylases [62]. At least two mechanisms have been proposed for RS-dependent lysine demethylation. One has serious flaws—possibly simple typographical errors [63]—while the other is thermodynamically unlikely [62]. As shown in Figure 10, insertion of a sulfur atom into a methyl group of a trimethyllysyl residue would afford a thiocarbinolamine, which would be expected to fragment to thioformaldehyde and a dimethyllysyl residue. Thioformaldehyde, of course, would degrade further to sulfide as well as formaldehyde, a coproduct of α-KG-dependent histone lysyl demethylases. Although this mechanism is hypothetical—no RS histone demethylase enzymes have yet been discovered—the take-home message is still important, that unknown and/or puzzling reaction mechanisms of RS enzymes might involve cryptic steps.
Figure 10.
Hypothetical mechanism for radical SAM-dependent histone trimethyllysine demethylation.
RS Enzymes that Catalyze Methylation of Unactivated C–H Bonds
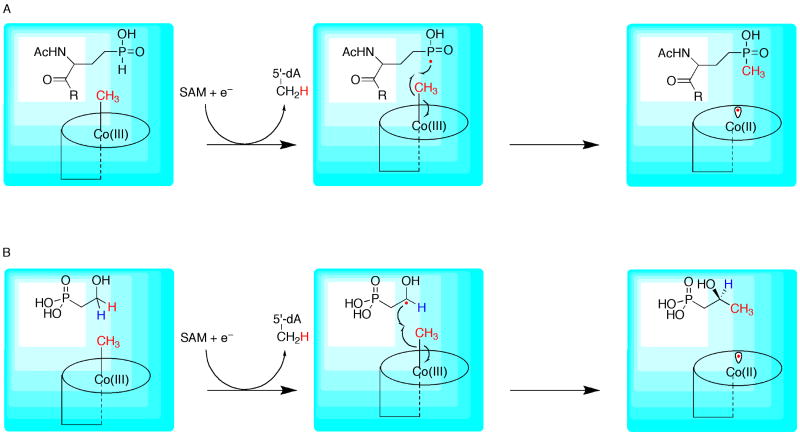
A new and exciting reaction catalyzed by RS proteins is emerging, which involves the methylation of unactivated C–H or P–H bonds. Although others may exist, the most widely found RS methylases appear to be those in the P-methylase family, which are thought to use some form of cobalamin as a cofactor in their reactions [15]. To date, however, none of these enzymes have been isolated and characterized in active forms, presumably because of low solubility and/or labile activity. The canonical P-methylase is involved in the biosynthesis of bialaphos, a tripeptide produced by Streptomyces hygroscopicus SF-1293 that has current commercial use as a herbicide [64]. Seminal genetic and metabolic feeding studies by Seto and coworkers showed that one of the steps in the biosynthesis of bialaphos involves the transfer of a methyl group to a phosphinate phosphorus atom of N-acetyldemethylphosphinothricin (N-AcDMPT) (Figure 11a) [64]. Although this reaction potentially can be mediated via a polar mechanism [64,65], the primary structure of this P-methylase contains the canonical RS signature sequence and shows additional sequence similarities with another methyltransferase (Fom3) that is annotated as an RS protein, but which catalyzes a reaction that cannot be mediated via a polar mechanism [65,66]. This methyltransferase, Fom3, catalyzes the penultimate step in the biosynthesis of fosfomycin—an antibiotic produced by several species of Streptomyces and Pseudomonas—which is the C-2 methylation of hydroxyethyl phosphonate (HEP) (Figure 11b) [64].
Figure 11.
Proposed mechanisms for P-methylase (A) and Fom3 (B). Methylcobalamin is shown in cartoon format (adapted from reference 65).
Several lines of evidence implicate methylcobalamin as the source of the methyl group in P-methylases and other similar transformations. When S. hydroscopicus SF-1293 was cultivated in the absence of cobalt, it was noticed that demethylphosphinothricin (DMPT) and demethylbialaphos accumulated in the fermentation broth. Moreover, P-methylation activity in cell extracts of S. lividans that harbored a plasmid encoding the bialaphos biosynthetic gene cluster was enhanced considerably in the presence of added methylcobalamin, but not SAM, methyltetrahydrofolate, or betaine, other potential methyl donors. Last, incubation of these same cell extracts with [methyl-1-14C]methylcobalamin, but not S-adenosyl-l-[methyl-14C]methionine, resulted in substantial transfer of radioactivity to the expected product [67]. Similar conclusions resulted from analogous genetic and metabolic feeding studies directed at elucidating the pathways for the biosynthesis of fosfomycin [64]. A mechanism for P- or C-methylation has been advanced, and shows a novel role for methylcobalamin in enzymatic catalysis (Figure 11) [65]. With respect to P-methylation a 5′-dA• generated from reductive cleavage of SAM is used to abstract the hydrogen atom from the P–H bond of the substrate, resulting in a phosphinate-centered radical. Subsequent radical recombination between the substrate radical and the methyl group of methylcobalamin affords N-Ac-PTT and cob(II)alamin (Figure 11a). Methylcobalamin could then be regenerated in a manner similar to that observed in methionine synthase, in which cob(II)alamin undergoes a reductive methylation reaction involving reduced flavodoxin and methyltetrahydrofolate [11]. A similar mechanism is displayed in Figure 11b for the C-2 methylation of HEP in the penultimate step in the biosynthesis of fosfomycin. Evidence in support of this role for methylcobalamin is provided by model studies, in which heating of a mixture of 2′-bis(ethoxycarbonyl)propylcobalamin and methylcobalamin resulted in formation of 2-ethyl-2-methylmalonic acid diethyl ester [68]. This result was hypothesized to derive from intermediate formation of a 2′-bis(ethoxycarbonyl)propyl radical via thermolysis of the Co-C bond of 2′-bis(ethoxycarbonyl)propylcobalamin, followed by abstraction of a methyl group from methylcobalamin [68].
Sequence analysis of the primary structures of P-methylase, Fom3, and fortimicin methyltransferase indicates two conserved regions of amino acids that are also found in cobalamin-binding proteins such as methionine synthase, glutamate mutase, 2-methyleneglutarate mutase, and methylmalonyl–CoA mutase. These regions are found in the N-terminal halves of the respective proteins, and contain the sequence DxxGx(S/T) separated from a GG sequence motif by ∼25-30 amino acids [15]. In the X-ray structures of known cobalamin-binding proteins, the conserved amino acids in these two motifs are in the vicinity of the cobalamin cofactor.
RS Enzymes that Catalyze C–O Bond Formation
There are a number of steps in the biosynthesis of various chlorophylls that are catalyzed by enzymes that are annotated as RS superfamily members [69]. For example, two enzymes, BchQ and BchR, catalyze the transfer of methyl groups to the C-82 ethyl and C-121 methyl carbons in the biosynthesis of certain bacteriochlorophylls in Chlorobium tepidum, and have the aforementioned sequence motifs that tentatively assign them to the cobalamin-dependent P-methylase subfamily. One of the most interesting reactions in chlorophyll biosynthesis, however, is formation of the isocyclic ring, which involves oxidation of the C-131 carbon of Mg-protoporphyrin-IX monomethylester (MPE) with concomitant attachment of the propionate methylester side-chain to the macrocycle, affording protochlorophyllide (Figure 12). The entire process is a 6-electron oxidation. Four electrons are removed in the oxidation of C-131 to a carbonyl group, while two additional electrons are removed during oxidative ring cyclization. Intriguingly, this oxidative cyclization can be mediated by two structurally distinct classes of enzymes [69].
Figure 12.
Hypothetical mechanism for the anaerobic Mg-protoporphyrin-IX monomethyl ester cyclase (BchE). Hydroxocobalamin is shown in cartoon format.
Under aerobic conditions the oxidative cyclization of MPE is catalyzed by AcsF—an enzyme purported to contain a nonheme diiron–carboxylate cofactor—with the C-131 carbonyl oxygen deriving from molecular oxygen [70]. The enzyme contains at least two subunits—one of which appears to be membrane associated—and has not been purified to homogeneity. In vitro feeding studies using a partially purified system showed that the C-131-oxo species is converted to product, suggesting that oxidation of C-131 precedes cyclization. In addition, the C-131-hydroxo species was shown to accumulate when the partially purified enzyme was incubated with MPE, the starting substrate [70]. These studies are consistent with a dihydroxylation mechanism involving formation of a geminal-diol intermediate, which collapses to the ketone with concomitant elimination of water. Because of initial installation of the carbonyl functional group, subsequent cyclization could take place via either polar or radical processes, with transfer of electrons back to the enzyme or to an external acceptor.
Certain photosynthetic facultative and obligate anaerobes construct the isocyclic ring of bacteriochlorophyll via an alternative mechanism, in which the C-131 carbonyl oxygen is derived from water. Genetic studies in Rhodobacter capsulatus led to the identification of the bchE gene, which encodes the only known protein to be involved in the anaerobic oxidative cyclization of MPE [70]. R. capsulatus BchE contains 575 amino acids and has a molecular mass of 65.8 kDa. In addition to the canonical CxxxCxxC RS signature sequence, like enzymes within the P-methylase family it also contains the aforementioned motifs that are suggestive of cobalamin binding [15]! There are a number of experimental observations that are consistent with a role for cobalamin in formation of the isocyclic ring of bacteriochlorophyll. Among them, disruption of the bluE and bluB genes in R. capsulatus, which encode proteins involved in the biosynthesis of cobalamin, strongly inhibits formation of bacteriochlorophyll, and results in accumulation of MPE [71]. Similarly, disruption of the bluB gene in Rhodobactor rubrum, which encodes a protein involved in the synthesis of the 5,6-dimethylbenzimidazole ligand of cobalamin, also inhibited the bacterium's ability to convert MPE into protochlorophyllide under anoxic conditions. Accordingly, the bacterium grew poorly under anoxic photoheterotrophic conditions; however, this growth defect could be rescued by introduction of dimethylbenzimidazole to the culture media [72].
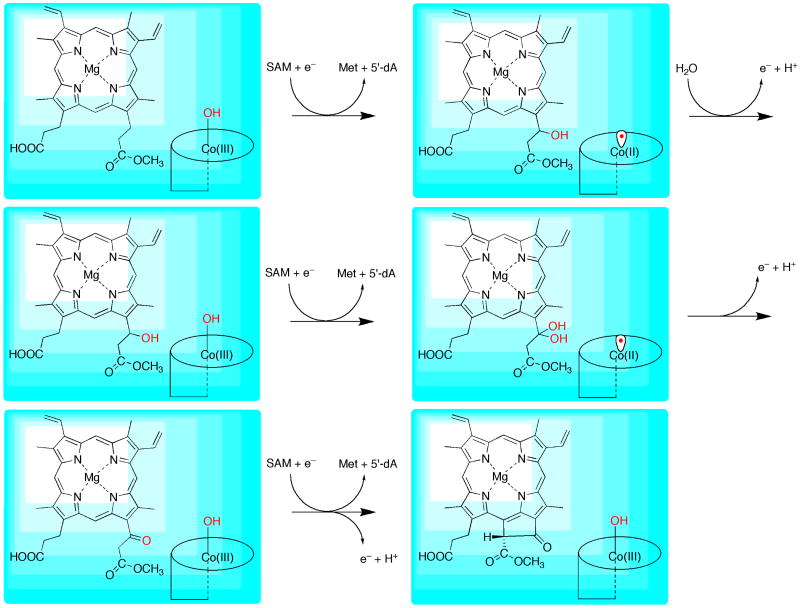
At least one mechanism has been proposed for the BchE-catalyzed conversion of MPE to protochlorophyllide, in which 5′-deoxyadenosyl 5′-cobalamin (AdoCbl) acts as the radical generator [71]. A 5′-dA• radical generated from homolysis of the C–Co bond of AdoCbl is proposed to abstract a hydrogen atom from C-131 to afford a substrate radical. Loss of an electron from the substrate radical intermediate generates a carbocation that is trapped by a molecule of water, affording a C-131-hydroxo intermediate. Abstraction of a second hydrogen atom from C-131—presumably via a second 5′-dA• generated from homolysis of another molecule of AdoCbl, although not explicitly identified in the proposed mechanism—affords a ketyl radical intermediate, which upon loss of a proton and an electron gives rise to the C-131-oxo intermediate. Cyclization to afford protochlorophyllide was proposed to occur via a third hydrogen atom abstraction from the substrate followed by radical attack onto the macrocyle with subsequent loss of a proton and an electron [71].
Although chemically reasonable, the drawbacks of the mechanism proposed by Gough et al are that: (1) it does not make efficient use of the protein's radical SAM machinery—the iron–sulfur cluster was proposed to facilitate proton and electron loss in going from the C-131-hydroxo intermediate to the C-131-oxo intermediate; and (2) at least two—and presumably three—molecules of AdoCbl are irreversibly destroyed during normal turnover, which has no precedent in known AdoCbl–dependent reactions, wherein the cofactor is normally regenerated after each turnover. An alternative mechanism might involve a lipoyl synthase-like reaction [56], in which two sulfur atoms are inserted into C-131 to afford a geminal dithiol, which would collapse to the thioketone concomitant with release of sulfide. The enzyme could then catalyze exchange of the sulfur atom of the thioketone with an oxygen atom from water, affording the expected ketone. Of course, the problem with this cryptic sulfur-insertion mechanism is that it makes no use of the presumed cobalamin cofactor. In addition, a sequence alignment of proposed BchE proteins indicates an insufficient number of conserved cysteine residues to coordinate the required second iron–sulfur cluster.
A third mechanism, shown in Figure 12, is similar to the mechanism proposed for P-methylases, making use of both radical SAM and cobalamin cofactors. However, in contrast to P-methylases, this mechanism proposes that the relevant form of cobalamin in BchE is hydroxocobalamin, a well-known stable variant of vitamin B12 [10]. In this mechanism a 5′-dA• is generated from reductive cleavage of SAM, and abstracts a hydrogen atom from C-131 of MPE, affording a substrate radical. The substrate radical then undergoes radical recombination with hydroxocobalamin, affording a C-131-hydroxo intermediate and cob(II)alamin. Loss of an electron from cob(II)alamin and addition of a water molecule results in reformation of hydroxocobalamin to allow for a second round of the exact same reaction, affording the C-131-geminal diol species, presumed to be an intermediate in the aerobic oxidative cyclase reaction. Collapse of the geminal diol intermediate affords the C-131-keto intermediate. Ring formation can take place upon an additional hydrogen-atom abstraction from C-132 of the substrate followed by attack of the substrate radical onto C-15 of the macrocyle and loss of an electron and a proton. Although this mechanism proposes a novel, though unprecedented, role for cobalamin in enzymatic catalysis, it possesses several attractive features: (1) it makes use of both cofactors (cobalamin and SAM/[4Fe–4S] cluster) that are predicted to bind to the protein; (2) it is similar to the proposed mechanism for P-methylases [65], an enzyme family with which BchE shares sequence similarity; and (3) it is similar to the rebound mechanisms of reactions that use iron-containing cofactors to functionalize unactivated C–H bonds (see Figure 1). The major difference is that the substrate radical is generated by a separate cofactor in BchE, while in iron-containing enzymes that activate dioxygen one cofactor both generates the substrate radical and serves as the source of the activated hydroxo group that is transferred.
Conclusions
The emergence of the radical SAM superfamily of enzymes has greatly contributed to the understanding of anaerobic metabolism and the elucidation of biosynthetic pathways for myriad cofactors and other natural products. Indeed, as shown in this review, RS machinery lends itself to the most kinetically challenging reactions in biology, such as the functionalization of unactivated C–H bonds, which was once thought to be unique to enzymes that activate dioxygen. Moreover, the sheer variety of reactions mediated by RS enzymes is spellbinding, and the pace at which new reactions are discovered suggests that a mother lode of exciting chemistry waits to be unraveled. Despite the progress in elucidating anaerobic mechanisms of C–H bond-functionalization, there is still much to learn, which includes a detailed understanding of the energetics associated with reductive cleavage of SAM, the trapping and characterization of hypothesized intermediates in various reactions, and the elucidation of pathways by which cofactors are reassembled during catalysis, as in RS enzymes that catalyze sulfur insertion.
Perhaps the next big challenge in anaerobic C–H bond functionalization is the unraveling of the mechanism by which methane—containing one of nature's most unactivated C–H bonds—is oxidized to methanol. Although still controversial, evidence suggests that certain consortia of bacteria are capable of carrying out this feat under anoxic conditions [73]. One theory that is gaining momentum suggests that the terminal reaction in methanogenesis, the reduction of methyl–coenzyme M (CoM–S–CH3) with coenzyme B (H–S–CoB) to yield methane and the corresponding heterodisulfide (CoM–S–S–CoB), is simply run in reverse [73-76]. In the forward direction this reaction is catalyzed by methyl-coenzyme M reductase, a 300 kDa enzyme composed of three different subunits in an α2β2γ2 arrangement, and which requires the nickel-containing porphinoid cofactor F430 [75]. Clearly this reaction will involve fascinating chemistry; stay tuned!
Acknowledgments
The author's research on functionalization of unactivated C–H bonds was supported by a grant from the National Institutes of Health (GM-63847)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frey PA. Importance of organic radicals in enzymic cleavage of unactivated carbon-hydrogen bonds. Chem Rev. 1990;90:1343–1357. [Google Scholar]; ••This is a pivotal review article highlighting aerobic and anaerobic mechanisms for cleavage of unactivated C–H bonds. The article advances the thesis that metal ions are required to generate oxidants of sufficient potency to cleave such bonds.
- 2.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin JE, Bradley M. Isopenicillin N synthase: Mechanistic studies. Chem Rev. 1990;90:1079–1088. [Google Scholar]
- 4.Krebs C, Fujimori DG, Walsh CT, Bollinger JM., Jr Non-heme Fe(IV)–oxo intermediates. Acc Chem Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollinger JM, Jr, Diao Y, Matthews MA, Xing G, Krebs C. myo-Inositol oxygenase: A radical new pathway for O2 and C–H activation at a nonheme diiron cluster. Dalton Trans. 2009:905–914. doi: 10.1039/b811885j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaillancourt FH, Yeh E, Vosburg DA, O'Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 7.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 8.Galoníc DP, Barr EW, Walsh CT, Bollinger JM, Jr, Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 9.Buckel W, Golding BT. Radical enzymes in anaerobes. Annu Rev Microbiol. 2006;60:27–49. doi: 10.1146/annurev.micro.60.080805.142216. [DOI] [PubMed] [Google Scholar]
- 10.Babior BM, Krouwer JS. The mechanism of adenosylcobalamin-dependent reactions. Crit Rev Biochem. 1979;6:35–102. doi: 10.3109/10409237909105424. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee R, Ragsdale SW. The many faces of vitamin B12: Catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 12.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 13.Frey PA. Lysine 2,3-aminomutase: Is adenosylmethionine a poor man's adenosylcobalamin? FASEB J. 1993;7:662–670. doi: 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- 14.Frey PA, Magnusson OT. S-Adenosylmethionine: A wolf in sheep's clothing, or a rich man's adenosylcobalamin? Chem Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 15.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper is the first detailed bioinformatics study to recognize that proteins that use S-adenosylmethionine as a precursor to the 5′-deoxyadenosyl 5′-radical constitute a superfamily. It also highlights various subclasses of these enzymes.
- 16.Layer G, Heinz DW, Jahn D, Schubert WD. Structure and function of radical SAM enzymes. Curr Opin Chem Biol. 2004;8:468–476. doi: 10.1016/j.cbpa.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-SAM” protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 18.Cheek J, Broderick JB. Adenosylmethionine-dependent iron-sulfur enzymes: Versatile clusters in a radical new role. J Biol Inorg Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 19.Fontecave M, Mulliez E, Ollagnier-de Choudens S. Adenosylmethionine as a source of 5′-deoxyadenosyl radicals. Curr Opin Chem Biol. 2001;5:506–511. doi: 10.1016/s1367-5931(00)00237-4. [DOI] [PubMed] [Google Scholar]
- 20.Frey PA, Booker S. Radical intermediates in the reaction of lysine 2,3-aminomutase. In: Zard SZ, editor. Advances in Free Radical Chemistry. Vol. 2 JAI Press Inc; 1999. pp. 1–43. [Google Scholar]
- 21.Biegert T, Fuchs G, Heider J. Evidence that oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 22.Rabus R, Heider J. Initial reaction of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch Microbiol. 1998;170:377–384. [Google Scholar]
- 23.Anchong GR, Rodriguez AM, Spormann AM. Benzylsuccinate synthase of Azoarcus sp strain T: Cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol. 2001;183:6763–6770. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leuthner B, Leutwein C, Schulz H, Hoerth P, Haehnel W, Schiltz E, Schaegger H, Heider J. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: A new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 25.Beller HR, Spormann AM. Substrate range of benzylsuccinate synthase from Azoarcus sp strain T. FEMS Microbiol Lett. 1999;178:147–153. doi: 10.1111/j.1574-6968.1999.tb13771.x. [DOI] [PubMed] [Google Scholar]
- 26.Coschigano PW, Wehrman TS, Young LY. Identification and analysis of genes involved in anaerobic toluene metabolism by strain T1: Putative role of a glycine free radical. Appl Environ Microbiol. 1998;64:1650–1656. doi: 10.1128/aem.64.5.1650-1656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper describes the similarity between pyruvate formate–lyase and benzylsuccinate synthase with respect to a conserved glycyl residue and a conserved cysteinyl residue that are both essential for turnover.
- 27.Qiao C, Marsh ENG. Mechanism of benzylsuccinate synthase: Stereochemistry of toluene addition to fumarate and maleate. J Am Chem Soc. 2005;127:8608–8609. doi: 10.1021/ja051972f. [DOI] [PubMed] [Google Scholar]; •This paper describes in vitro mechanistic analysis of the benzylsuccinate synthase reaction and provdes information about the stereochemistry of toluene addition to fumarate.
- 28.Beller HR, Spormann AM. Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J Bacteriol. 1998;62:1188–1196. doi: 10.1128/jb.180.20.5454-5457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spormann AM, Widdel F. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation. 2000;11:85–105. doi: 10.1023/a:1011122631799. [DOI] [PubMed] [Google Scholar]; •This is a resourceful review describing the anaerobic metabolism of a variety of hydrocarbons.
- 30.Lowry TH, Richardson KS. Mechanism and Theory in Organic Chemistry. 3rd. New York: Harper & Row; 1987. [Google Scholar]
- 31.Aeckersberg F, Rainey FA, Widdel F. Growth, natural relationships, cellular fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions. Arch Microbiol. 1998;170:361–369. doi: 10.1007/s002030050654. [DOI] [PubMed] [Google Scholar]
- 32.Reuter P, Rabus R, Wilkes H, Aeckersberg F, Rainey FA, Jannasch HW, Widdel F. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria. Nature. 1994;372:455–458. doi: 10.1038/372455a0. [DOI] [PubMed] [Google Scholar]
- 33.So CM, Young LY. Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl Environ Microbiol. 1999;65:2969–2976. doi: 10.1128/aem.65.7.2969-2976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrenreich P, Behrends A, Harder J, Widdel F. Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria Arch. Microbiol. 2000;173:58–64. doi: 10.1007/s002030050008. [DOI] [PubMed] [Google Scholar]
- 35.Kropp KG, Davidova IA, Suflita JM. Anaerobic oxidation of n-dodecane by an addition reaction in a sulfate-reducing bacterial enrichment culture. Appl Environ Microbiol. 2000;66:5393–5398. doi: 10.1128/aem.66.12.5393-5398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This paper presents evidence that straight-chain alkanes are activated for anaerobic oxidation by addition to fumarate, and hints that the system is similar to benzylsuccinate synthase.
- 36.Rabus R, Wilkes H, Behrends A, Armstroff A, Fischer T, Pierik AJ, Widdel F. Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J Bacteriol. 2001;183:1707–1715. doi: 10.1128/JB.183.5.1707-1715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This paper presents evidence that the first step in the degradation of n-hexane involves a radical addition to fumarate, as is seen in benzylsuccinate synthase.
- 37.Grundmann O, Behrends A, Rabus R, Amann J, Halder T, Heider J, Widdel F. Genes encoding the candidate enzyme for anaerobic activation of n-alkanes in the denitrifying bacterium, strain HxN1. Environ Microbiol. 2008;10:376–385. doi: 10.1111/j.1462-2920.2007.01458.x. [DOI] [PubMed] [Google Scholar]; ••This paper describes the identification of a gene cluster that encodes proteins involved in the anaerobic activation of n-alkanes. The proteins were found to be similar to those in the benzylsuccinate synthase system.
- 38.Callaghan AV, Wawrik B, Chadhain SMN, Young LY, Zylstra GJ. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem Biophys Res Commun. 2008;366:142–148. doi: 10.1016/j.bbrc.2007.11.094. [DOI] [PubMed] [Google Scholar]; •This paper describes the identification of two gene clusters involved in the anaerobic activation of n-hexadecane. The encoded proteins were found to be similar to those in the benzylsuccinate synthase system.
- 39.Armstrong DA, Yu D, Rauk A. Oxidative damage to the glycyl α-carbon site in proteins: An ab initio study of the C–H bond dissociation energy and the reduction potential of the C-centered radical. Can J Chem. 1996;74:1192–1199. [Google Scholar]
- 40.McMillen DF, Golden DM. Hydrocarbon bond dissociation energies. Ann Rev Phys Chem. 1982;33:493–532. [Google Scholar]
- 41.Wang SC, Frey PA. S-adenosylmethionine as an oxidant: The radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Sanyal I, Cohen G, Flint DH. Biotin synthase: Purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- 43.Escalettes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. Biotin synthase mechanism: Evidence for hydrogen transfer from the substrate into deoxyadenosine. J Am Chem Soc. 1999;121:3571–3578. [Google Scholar]
- 44.Guianvarc'h D, Florentin D, Bui BTS, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem Biophys Res Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 45.Taylor AM, Farrar CE, Jarrett JT. 9-Mercaptodethiobiotin is formed as a competent catalytic intermediate by Escherichia coli biotin synthase. Biochemistry. 2008;47:9309–9317. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Further investigation on the turnover of Escherichia coli biotin synthase with dethiobiotin and 9-mercaptodethiobiotin as substrates. Biochemistry. 2004;43:16432–16441. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- 47.Gibson KJ, Pelletier DA, Turner S IM. Transfer of sulfur to biotin from biotin synthase (BioB protein) Biochem Biophys Res Commun. 1999;254:632–635. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- 48.Tse Sum Bui B, Florentin B, Fournier F, Ploux O, Méjean A, Marquet A. Biotin synthase mechanism: on the origin of sulphur. FEBS Lett. 1998;440:226–230. doi: 10.1016/s0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- 49.Cosper MM, Jameson GNL, Hernández HL, Krebs C, Huynh BH, Johnson MK. Characterization of the cofactor composition of Escherichia coli biotin synthase. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- 50.Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron–sulfur binding sites: Chemical and spectroelectrochemical analysis of iron–sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ugulava NB, Surerus KK, Jarrett JT. Evidence from Mössbauer sectroscopy for distinct [2Fe-2S]2+ and [4Fe-4S]2+ cluster binding sites in biotin synthase from Escherichia coli. J Am Chem Soc. 2002;124:9050–9051. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tse Sum Bui B, Mattioli TA, Florentin D, Bolbach G, Marquet A. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe–2S]2+ cluster in the sulfur insertion step. Biochemistry. 2006;45:3824–3834. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]; •This paper describes a study in which a spectroscopically characterized [2Fe–2Se] cluster on biotin synthase was shown to give rise to selenobiotin, supporting the hypothesis that the [2Fe–2S] cluster is the sulfur donor in the biotin synthase reaction.
- 53.Ugulava NB, Sacanell CJ, Jarrett JT. Spectroscopic changes during a single turnover of biotin synthase: Destruction of a [2Fe–2S] cluster accompanies sulfur insertion. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper provides the first evidence that the active form of biotin synthase contains both a [2Fe–2S] cluster and a [4Fe–4S] cluster, and that sulfur in biotin derives from the [2Fe–2S] cluster.
- 54.Jameson GNL, Cosper MM, Hernández HL, Johnson MK, Huynh BH. Role of the [2Fe–2S] cluster in recombinant Escherichia coli biotin synthase. Biochemistry. 2004;43:2022–2031. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]; •This paper describes the use of rapid kinetics methods coupled with Mössbauer, EPR, and UV–vis spectroscopies to show that the [2Fe–2S] cluster decays faster than biotin is produced. It highlights the need to apply these kinds of physical methods to peer deeply into the mechanism of radical SAM proteins.
- 55.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper describes the X-ray structure of biotin synthase. All substrates and cofactors are bound, allowing a glimpse at the arrangement of the active site of an enzyme that catalyzes sulfur insertion.
- 56.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This review article compares and contrasts radical SAM proteins that catalyze sulfur insertion reactions.
- 57.Choi-Rhee E, Cronan JE. Biotin synthase is catalytic in vivo, but catalysis engenders destruction of the protein. Chem Biol. 2005;12:461–468. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Mühlenhoff U, Gerl MJ, Flauger B, Pirner HM, Balser S, Richardt N, Lill R, Stolz J. The iron–sulfur proteins Isa1 and Isa2 are required for the function but not for the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:495–504. doi: 10.1128/EC.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem. 2004;279:47555–47653. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 60.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:1826–1831. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barckholtz C, Barckholtz TA, Hadad CM. C–H and N–H bond dissociation energies of small aromatic hydrocarbons. J Am Chem Soc. 1999;121:491–500. [Google Scholar]
- 62.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem Sci. 2002;27:115–117. doi: 10.1016/s0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- 64.Seto H, Kuzuyama T. Bioactive natural products with carbon–phosphorus bonds and their biosynthesis. Nat Prod Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- 65.van der Donk WA. Rings, radicals, and regeneration: The early years of a bioorganic laboratory. J Org Chem. 2006;71:9561–9571. doi: 10.1021/jo0614240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuzuyama T, Seki T, Dairi T, Hidaka T, Seto H. Nucleotide sequence of fortimicin KL1 methyltransferase gene isolated from Micromonospora olivasterospora, and comparison of its deduced amino acid sequence with those of methyltransferases involved in the biosynthesis of bialaphos and fosfomycin. J Antibiot. 1995;48:1191–1193. doi: 10.7164/antibiotics.48.1191. [DOI] [PubMed] [Google Scholar]; • This paper provides bioinformatics evidence that radical SAM-dependent P-methylation and C-methylation might be catalyzed via similar mechanisms.
- 67.Kamigiri K, Hidaka T, Imai S, Murakami T, Seto H. Studies on the biosynthesis of bialaphos (SF-1293) 12. C–P bond formation mechanism of bialaphos: Discovery of a P-methylation enzyme. J Antibiot. 1992;45:781–787. doi: 10.7164/antibiotics.45.781. [DOI] [PubMed] [Google Scholar]; ••This interesting paper provides compelling evidence that methylcobalamin serves as the methyl donor in P-methylases.
- 68.Mosimann H, Kräutler B. Methylcorrinoids methylate radicals–their second biological mode of action. Angew Chem Int Ed. 2000;39:393–395. doi: 10.1002/(sici)1521-3773(20000117)39:2<393::aid-anie393>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 69.Gomez Macqueo Chew A, Bryant DA. Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Ann Rev Microbiol. 2007;61:113–129. doi: 10.1146/annurev.micro.61.080706.093242. [DOI] [PubMed] [Google Scholar]; •This is an interesting review article on chlorophyll biosynthesis in bacteria. It mentions several steps that potentially involve novel radical SAM chemistry, including C-methylation, oxygenation, and anaerobic oxidative cyclization.
- 70.Bollivar DW. Recent advances in chlorophyll biosynthesis. Photosynth Res. 2006;90:173–194. doi: 10.1007/s11120-006-9076-6. [DOI] [PubMed] [Google Scholar]
- 71.Gough SP, Petersen BO, Duus JØ. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc Natl Acad Sci U S A. 2000;97:6908–6913. doi: 10.1073/pnas.97.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This is a pivotal paper reporting the requirement of cobalamin in the anaerobic oxidative cyclization reaction in chlorophyll biosynthesis.
- 72.Gray MJ, Escalante-Semerena JC. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci U S A. 2007;104:2921–2926. doi: 10.1073/pnas.0609270104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caldwell SL, Laidler JR, Brewer EA, Eberly JO, Sandborgh SC, Colwell FS. Anaerobic oxidation of methane: Mechanisms, bioenergetics, and the ecology of associated microoganisms. Environ Sci Technol. 2008;42:6791–6799. doi: 10.1021/es800120b. [DOI] [PubMed] [Google Scholar]
- 74.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 75.Krüger M, Meyerdierks A, Glöckner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt J, Böcher R, Thauer RK, et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature. 2003;426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]; •This is an interesting paper, which advances the thesis that the oxidation of methane might be catalyzed by a nickel-containing protein in a reaction that is predicted to be the reverse of methyl-coenzyme M reductase.
- 76.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damsté JS, Op den Camp HJ, Jetten MS, et al.: A