Abstract
Although tyrosyl-DNA phosphodiesterase (TDP1) is capable of removing blocked 3′ termini from DNA double-strand break ends, it is uncertain whether this activity plays a role in double-strand break repair. To address this question, affinity-tagged TDP1 was overexpressed in human cells and purified, and its interactions with end joining proteins were assessed. Ku and DNA-PKcs inhibited TDP1-mediated processing of 3′-phosphoglycolate double-strand break termini, and in the absence of ATP, ends sequestered by Ku plus DNA-PKcs were completely refractory to TDP1. Addition of ATP restored TDP1-mediated end processing, presumably due to DNA-PK-catalyzed phosphorylation. Mutations in the 2609–2647 Ser/Thr phosphorylation cluster of DNA-PKcs only modestly affected such processing, suggesting that phosphorylation at other sites was important for rendering DNA ends accessible to TDP1. In human nuclear extracts, about 30% of PG termini were removed within a few hours despite very high concentrations of Ku and DNA-PKcs. Most such removal was blocked by the DNA-PK inhibitor KU-57788, but ~5% of PG termini were removed in the first few minutes of incubation even in extracts preincubated with inhibitor. The results suggest that despite an apparent lack of specific recruitment of TDP1 by DNA-PK, TDP1 can gain access to and can process blocked 3′ termini of double-strand breaks before ends are fully sequestered by DNA-PK, as well as at a later stage after DNA-PK autophosphorylation. Following cell treatment with calicheamicin, which specifically induces double-strand breaks with protruding 3′-PG termini, TDP1-mutant SCAN1 (spinocerebellar ataxia with axonal neuropathy) cells exhibited a much higher incidence of dicentric chromosomes, as well as higher incidence of chromosome breaks and micronuclei, than normal cells. This chromosomal hypersensitivity, as well as a small but reproducible enhancement of calicheamicin cytotoxicity following siRNA-mediated TDP1 knockdown, suggests a role for TDP1 in repair of 3′-PG double-strand breaks in vivo.
1. INTRODUCTION
DNA topoisomerase I relaxes DNA supercoiling by forming a transient single-strand break (SSB) in which Tyr723 of the topoisomerase is covalently linked to the 3′ end of DNA [1]. Tyrosyl-DNA phosphodiesterase (TDP1) is a DNA repair enzyme that cleaves these tyrosyl-phosphate linkages in the rare instances where the topoisomerase fails to religate the break and dissociate [2]. Polynucleotide kinase/phosphatase (PNKP) then removes the resulting 3′-phosphate and phosphorylates the 5′ terminus, and the nick is finally sealed by DNA ligase III [3]. TDP1 can also process the 3′-phosphoglycolate (3′-PG) moieties often formed during free radical-mediated DNA cleavage [4], as well as various other 3′ modifications [5], although much less efficiently than phosphotyrosyl (pTyr) termini. However, at SSBs, 3′-PG moieties can also be efficiently removed by the apurinic/apyrimidinic endonuclease (APE1) [6], and thus it is unlikely that PG processing at SSBs would be an essential function of TDP1. In humans, a missense mutation in TDP1 confers spinocerebellar ataxia with axonal neuropathy (SCAN1) [7], but it remains to be determined how this specific repair defect leads to SCAN1 symptoms.
Despite its demonstrated interactions with SSB repair proteins [8], TDP1 itself is more active toward DSB ends than SSB ends [9], and unlike APE1 [10], it can act on protruding 3′ termini [4]. Moreover, when model DSB substrates bearing protruding 3′-PG termini are incubated in human whole-cell extracts, a substantial fraction of the PG termini are removed, but there is no detectable processing of such termini in SCAN1 extracts, suggesting that resolution of protruding 3′-PG termini is entirely attributable to TDP1, at least in cell extracts [11]. However, SCAN1 cells are at most only marginally sensitive to ionizing radiation [11], which is expected to generate many 3′-PG-terminated DSBs [12]. Moreover, direct measurements of DSBs, by neutral elution and repair focus assays, have failed to detect any significant deficits in repair of radiation-induced DSBS, in either human SCAN1 cells or Tdp1-/- mouse cells [13,14]. Nevertheless, Tdp-/- mice are sensitive to bleomycin, which induces predominantly blunt-ended 3′-PG DSBs [15].
To better understand the possible role of TDP1 in end joining, we examined the interplay between TDP1 and core end joining factors, both as purified proteins and in cell extracts. In addition, chromosome aberrations were examined following treatment of normal and TDP1-mutant cells, following treatment with calicheamicin, which specifically induces 3′-PG DSBs.
2. MATERIALS AND METHODS
2.1 Materials
Native DNA-PKcs and Ku were prepared from HeLa cells [16], and recombinant Ku and XRCC4 / DNA ligase IV complex (X4L4) were isolated from baculovirus-infected insect cells, as described previously [17]. Mutant DNA-PKcs proteins were purified from DNA-PKcs-deficient Chinese hamster ovary V3 cells complemented with human DNA-PKcs cDNA [16].
To obtain TDP1 protein, an insert encoding FLAG-tagged TDP1 was isolated from the vector pFLAG-TDP1 and cloned into the adenoviral vector pZERO-TGCMV. The resulting plasmid was transfected into Ad293 cells to produce a TDP1-expressing Ad-5(delE1,E3) adenovirus [18]. The supernatant from these cultures was used to infect subconfluent Ad293 cells (twenty subconfluent 15-cm dishes) at a multiplicity of 3. After three days, TDP1 protein was isolated from NP-40 lysates and affinity purified on FLAG beads, essentially as described previously [11] except that all volumes were increased 4-fold. Because the FLAG peptide used to elute the tagged protein interfered with chromogenic protein assays, TDP1 concentration was estimated by comparison with known quantities of BSA on Coomassie-stained polyacrylamide gels.
A 3′-pTyr 18-mer with sequence TCCGTTGAAGCCTGCTTT was purchased from Midland Certified Reagents (Midland, TX). Oligomers bearing 3′-PG termini were prepared by bleomycin-induced cleavage of partial duplexes, as described [19]. These oligomers were 5′-32P-end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (Perkin-Elmer). To generate an internally labeled substrate with partially complementary (-ACG/-ACG) 3′-PG overhangs, one labeled and one unlabeled oligomer were ligated into 10- or 11-base 5′ overhangs of 3′-resected plasmid pRZ56, as described previously [20–22].
The DNA-PK inhibitor KU-57788 (aka NU7441, hereafter designated DNA-PKi) and the ATM inhibitor KU-55933 (ATMi) were generously provided by Dr. Graeme Smith, KUDOS Pharmaceuticals. KU-55933 inhibits ATM with an IC50 of 13 nM, vs. 1.8 µM for DNA-PK [23]. KU-57788 inhibits DNA-PK with an IC50 of 14 nM, vs. >100 µM for ATM [24]. Both are at least 100-fold more potent in inhibiting their target kinase than any of 60 other kinases tested [23,24].
2.2 DNA end processing and end-joining reactions
Assays for TDP1 activity toward single-strand 3′-PG and 3′-pTyr oligomers were performed in 60 mM potassium acetate, 50 mM triethanolammonium acetate pH 7.5, 5 mM EDTA, 1 mM dithiothreitol and 50 µg/ml BSA. TDP1 was diluted immediately before use in 0.1 M NaCl, 50 mM Tris pH 8, 5 mM EDTA, 5 mM dithiothreitol, 500 µg/ml BSA and 10% glycerol, and the reactions were incubated for 2 hr at 37°C. In experiments with 3′-PG oligomers, samples were denatured for 2 min at 90°C and then magnesium acetate (15 mM) and PNKP (40 µg/ml, gift of Michael Weinfeld, Cross Cancer Institute, Edmonton, Alberta, Canada) were added and the samples incubated for 1 hr to convert 3′-phosphates generated by TDP1 to 3′-hydroxyls and thereby increase their electrophoretic separation from 3′-PG substrate [4]. Formamide (1–2 volumes) was added and the samples were denatured at 90°C for 2 min and subjected to electrophoresis on 20% polyacrylamide sequencing gels (24% gels for blunt-ended substrates).
End processing reactions with plasmids and purified proteins contained 60 mM potassium acetate, 50 mM triethanolammonium acetate pH 7.5, 1 mM ATP, 1 mM dithiothreitol, 50 µg/ml BSA and 5 mM magnesium acetate. Reactions were stopped by dilution into at least 2 volumes of ice-cold 0.45 M sodium acetate / 10 mM EDTA / 100 µg/ml tRNA, followed immediately by extraction with phenol and then with chloroform at 22°C. Samples were ethanol-precipitated and treated with AvaI (400 units/ml) for 4 hr at 37°C in 25–50 µl of the buffer provided by the vendor (New England Biolabs). Calf intestinal phosphatase (120 units/ml) was added and the samples incubated for an additional 1 hr to remove 3′-phosphates. Phosphatase also removes the 5′-phosphate but the 32P label is 1 base from the AvaI cleavage site and is retained. Alternatively, the DNA was treated with phosphatase first and denatured at 90°C for 5 min, then cut with 400 units/ml TaqαI (New England Biolabs, 65°C, 6 hr), which cleaves single- as well as double-stranded DNA.
For comparison of reaction rates under various conditions, the extent of reaction was calculated as -In(fraction of remaining PG termini). Titrations with TDP1 protein and oligomeric substrates indicated that the extent of reaction was accurately proportional to TDP1 concentration (data not shown).
Reactions with HeLa nuclear extracts contained 3.2 mg/ml extract (Promega in vitro transcription grade), 50 mM triethanolamine-NaOH pH 7.5, 1 mM Mg(OAc)2, 40 mM KOAc, 0.5 mM dithiothreitol, 1 mM ATP, 50 µM of each dNTP or dideoxynucleoside 5′-triphosphate (ddNTP), 50 µg/ml BSA, and 1 µg/ml DNA substrate. In some cases X4L4 (5 µg/ml) DNA-PKi (1 µM) or ATMi (1 µM) were added, as indicated. Following substrate addition, samples were incubated for the indicated times at 37°C, and deproteinized by treatment with proteinase K followed by phenol/chloroform extraction. DNA was digested with TaqI (and in some cases BstXI to detect end joining), precipitated, and analyzed on polyacrylamide sequencing gels [21].
2.3 DNase footprinting
To assess accessibility of DNA ends, the same internally labeled 3′-PG substrates were incubated with Ku, DNA-PKcs and/or ATP as above, and then 1.5 µg/ml DNase I (Sigma) was added and the samples incubated at 37°C for an additional 10 min. Samples were diluted with 1 volume formamide and denatured for 2 min at 90°C prior to analysis on sequencing gels.
2.4 Cytogenetics
Epstein-Barr virus-transformed lymphoblastoid cells from SCAN1 patients [7] were obtained from Dr. James Lupski, Baylor College of Medicine, and maintained in RPMI1640 medium with 10% fetal bovine serum. Calicheamicin (gift of Wyeth Pharmaceuticals) was dissolved in ethanol and stored at −80°C. It was diluted to 20 µM in DMSO and then diluted in complete medium just prior to use. Cells were treated with 0–30 pM calicheamicin for 24 hr and then 1 µg/ml colchicine was added 1 hr before harvest. Metaphase spreads were prepared by standard procedures. For centromere labeling, slides were denatured at 72°C in 70% formamide / 2×SSC for 2 min and then hybridized to a Cy3-labeled pancentromeric probe (Open Biosystems #SFP3336) for 16 hr at 37°C, and extensively washed. The highest-stringency wash was with 0.5×SSC at 37°C. Chromosomes were counterstained with DAPI, slides were coded and metaphases were photographed on a Zeiss LSM510 confocal microscope, using a 405-nm diode laser with 405-nm blocking and 420-nm long pass emission filters for DAPI, and a 543-nm helium laser with 543-nm blocking and 560–615-nm bandpass emission filters for Cy3. Spreads were then scored for acentric fragments and dicentric chromosomes. (See Supplemental Fig. 4 for further details).
2.5 TDP1 knockdown and clonogenic survival assays
HeLa cells were grown in DMEM plus 10% fetal bovine serum and antibiotics (GIBCO). For each experiment, 12-well plates were inoculated with 8 × 104 HeLa cells per well and 24 hr later were transfected with 100 nM TDP1 siRNA (DharmaFECT Smart-Pool) or control siRNA-A (Santa Cruz biotechnology) using DharmaFECT general transfection protocol (Dharmacon) with 2.4 µl DharmaFECT Reagent 1 in 0.8 ml medium per well. After 24 hr, the transfection medium was replaced with complete medium and the incubation was continued for another 48 hr. The cells were then treated with 6, 20 or 60 pM calicheamicin for 4 hr, plated at 500 – 20,000 cells per 100-mm dish and incubated at 37°C. After 12 days, cells were fixed in methanol, stained with crystal violet and colonies were counted. Residual cells not plated were pooled for each siRNA condition and lysed for determination of TDP1 knockdown. The cells were pelleted and washed twice with PBS, then lysed on ice with 50 µl of 10 mM HEPES-KOH pH 7.8, 60 mM KCl, 1 mM EDTA, 0.5% NP-40 plus protease inhibitors (2 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 mM NaVO4, 0.5 mM PMSF). Lysates were cleared by centrifugation (500 × G, 5 min) and TDP1 activity in serial 5:1 dilutions was then determined from the conversion of the 3′-pTyr oligomer to a 3′-phosphate as described above, with incubation at 37°C for 2 hr.
3. RESULTS
3.1 Properties of recombinant TDP1 isolated from human cells
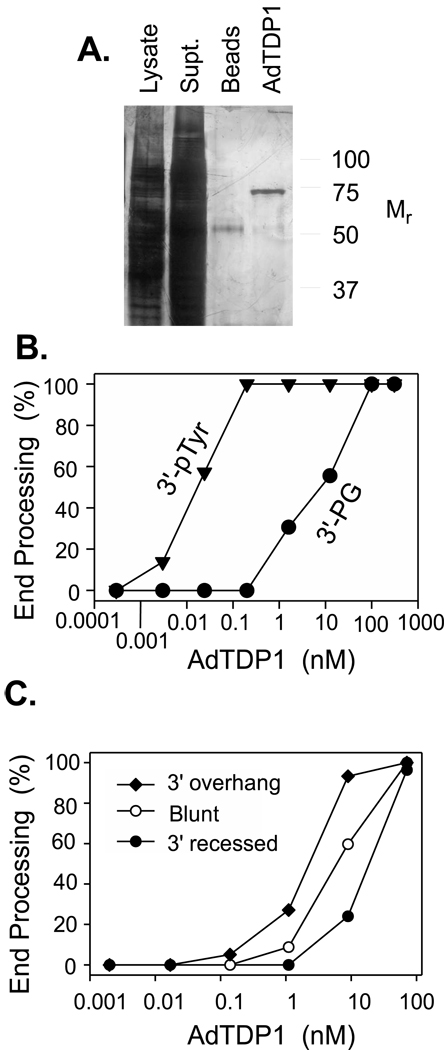
Previously, we showed that when a substrate bearing a 3′-PG terminus on a 3′ overhang was incubated with native Ku purified from human cells, there was significant conversion of the 3′-PG to a 3′-phosphate [4]. This persistent contamination of Ku with TDP1 (which was verified by western blot) after purification through five columns, raised the possibility that Ku might recruit TDP1 to DNA ends and thereby enhance PG processing. However, when recombinant human Ku purified from baculovirus-infected insect cells was added to reactions containing recombinant human TDP1 purified from E. coli, the effect of Ku was always inhibitory [4]. Since it is possible that the bacterially produced TDP1 might lack proper folding or post-translational modifications characteristic of human cells, TDP1 was cloned into an adenovirus, and FLAG-tagged TDP1 (hereafter designated AdTDP1) was affinity-purified from infected HEK293 cells using FLAG antibody-conjugated beads (Fig. 1A).
Figure 1.
Characterization of recombinant AdTDP1 isolated from human cells. A. Silver-stained SDS / 10% polyacrylamide gel showing affinity purification of AdTDP1 on FLAG beads. Samples include the initial lysate (5 µl), the supernatant following pelleting of the beads (10 µl), the beads pelleted after elution of protein with FLAG peptide (5 µl), and the eluted protein (10 µl). B. A mixture of a 3′-pTyr 18-mer (▼) and a 3′-PG 14-mer (●) (20 nM each) was titrated with various dilutions of AdTDP1, and processing was assessed by gel electrophoresis (Supplemental Fig. 1), as described in the Methods. C. A mixture of plasmid substrates bearing 3′-PG termini on a 5-base 3′ overhang (◆), a blunt end (○) or a 2-base 3′-recessed end (●) was titrated with AdTDP1. See Supplemental Fig. 1 and ref. [35] for a detailed description of these substrates. Incubations were for 2 hr at 37°C, and all samples were subsequently treated with PNKP to convert TDP1-induced 3′-phosphate to 3′-hydroxyl termini.
The catalytic properties of recombinant AdTDP1 were similar to those of TDP1 expressed in E. coli, and in particular the ratio of activity toward 3′-pTyr vs. 3′-PG substrates was about 200:1 (Fig. 1B), similar to the ratio of 80:1 seen with the bacterially produced protein [4]. To examine the effect of secondary structure on TDP1-mediated end processing, a mixture of substrates bearing 3′-PG termini on a 6-base 3′ overhang, a blunt end, and a 2-base-recessed end, was titrated with purified TDP1. The 3′ overhang was processed 3 times more efficiently than the blunt end and 10 times more efficiently than the recessed end (Fig. 1C), presumably reflecting differences in accessibility of the termini to TDP1.
3.2 Ku and DNA-PKcs inhibit DNA end processing by TDP1
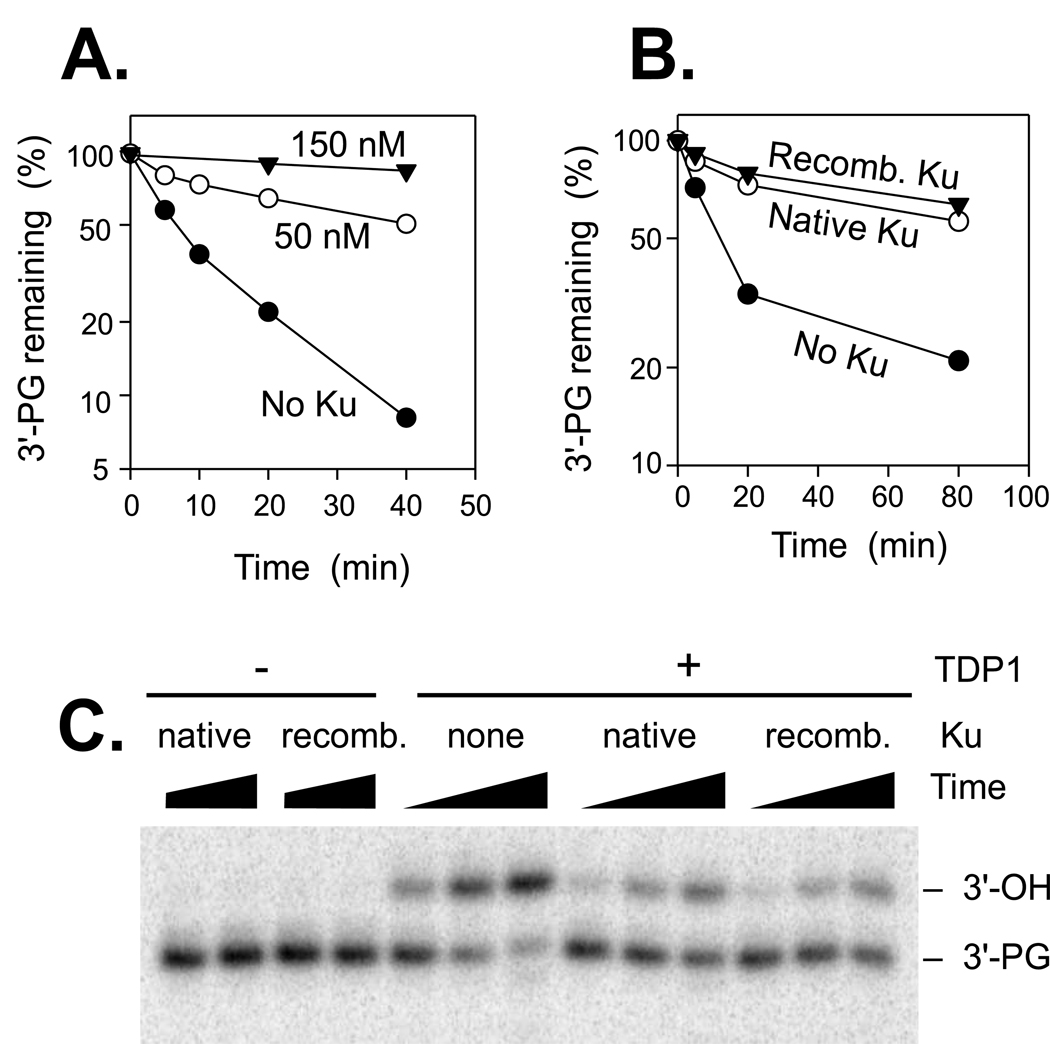
To assess possible recruitment of AdTDP1 by native Ku, processing of a blunt 3′-PG terminus was examined. Because this substrate is inherently less susceptible to TDP1 than the 3′ overhang substrate, there was no detectable processing by the small amount of contaminating TDP1 in native Ku, and so the effect of Ku on PG processing by the recombinant TDP1 could be accurately assessed. Again, however, the addition of Ku was inhibitory, and the recombinant and native forms of Ku were equally effective in inhibition (Fig. 2). Thus, despite presence of TDP1 in preparations of highly purified Ku, it appears unlikely that Ku recruits TDP1 or stimulates its activity at DSB ends. Addition of recombinant X4L4 complex had either no effect or a slightly inhibitory effect on TDP1 activity, in either the presence or absence of Ku (data not shown).
Figure 2.
Inhibition of TDP1-mediated 3′-PG processing by Ku. A. A plasmid bearing a 3′-PG on a 3-base 3′ overhang (see Fig. 3A) was treated with 30 nM AdTDP1 for the indicated times at 37°C in the presence of 0 (●), 50 (○) or 150 nM (▼) native human Ku, and glycolate removal was assessed as described in the METHODS. B. TDP1-mediated glycolate removal was assessed following treatment for the indicated times without Ku (●) or in the presence of 50 nM recombinant Ku (▼) or native Ku (○). C. Gel from which the data in (B.) were derived. Following deproteinization, the samples were treated with AvaI to release a labeled 15-mer from the end of the plasmid, and then with calf intestinal phosphatase to remove the 3′-phosphate.
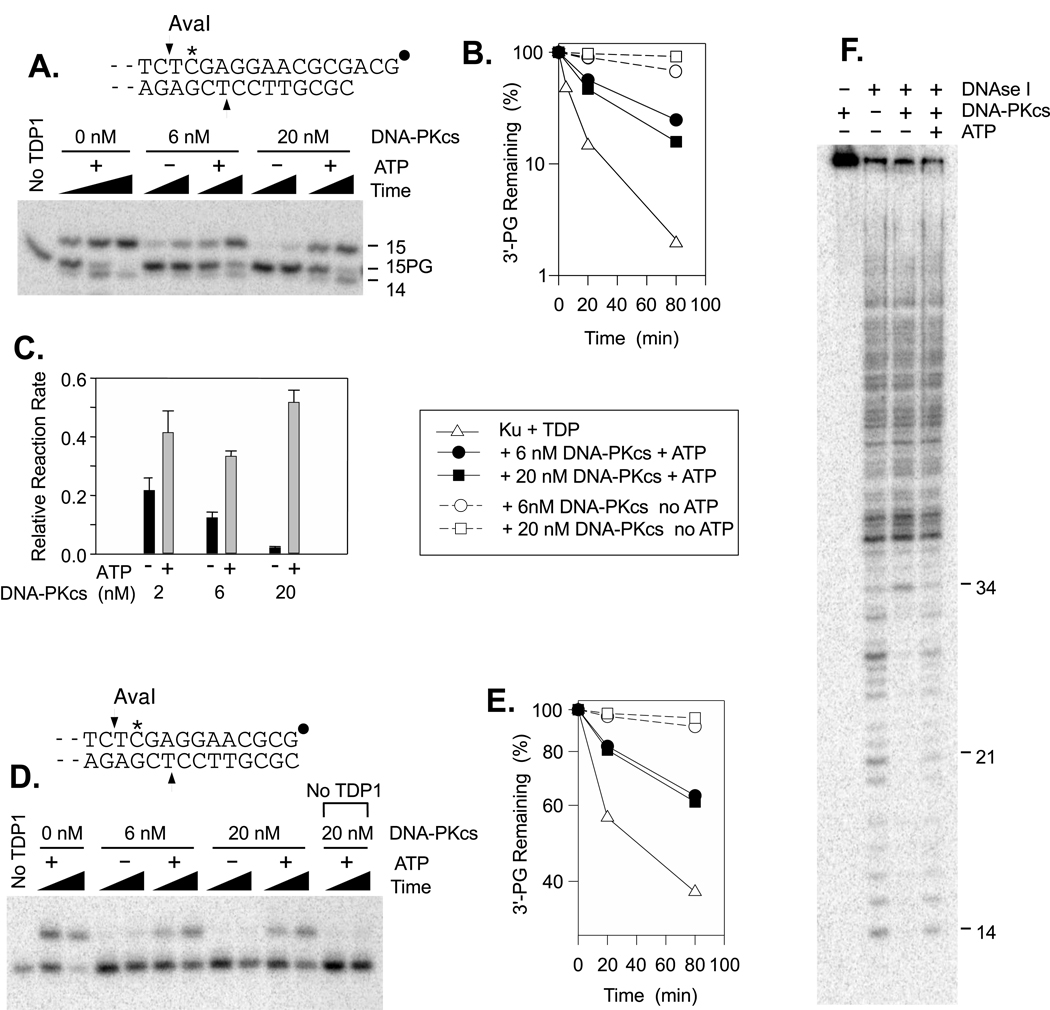
In intact cells, it is likely that free DNA ends would be rapidly sequestered by Ku and DNA-PKcs, given the abundance of these proteins [25,26]. TDP1-mediated PG processing activity was therefore examined in the presence or absence of DNA-PKcs, and a substrate bearing a 3′-PG on either a 3-base 3′ overhang (Fig. 3A; quantitated in Fig. 3B–C) or a blunt end (Fig. 3D; quantitated in Fig. 3E). For comparison, binding of DNA ends by Ku and DNA-PKcs under the same conditions was independently determined by DNase footprinting (Fig. 3F). In the presence of 50 nM Ku, a concentration that by itself reduced the rate of processing about twofold (data not shown), DNA-PKcs further inhibited TDP1 activity at DNA ends; in the absence of ATP, 20 nM DNA-PKcs blocked 3′-PG processing almost completely (Fig. 3C). Similarly, the same combination of Ku and DNA-PKcs completely protected ~30 bp at the DNA end from DNase digestion (Fig. 3F). Thus, in the absence of protein phosphorylation, ends sequestered by Ku and DNA-PKcs are highly resistant if not wholly refractory to TDP1.
Figure 3.
Inhibition of TDP1-mediated 3′-PG processing by DNA-PK in the presence or absence of ATP. All samples also contained 50 nM recombinant Ku and incubation was for 20 or 80 min at 37°C in the presence (closed symbols) or absence (open symbols) of ATP. DNA-PKcs concentration in (B.) and (D.) was 0 (▼), 6 (●,○) or 20 nM (■,□). Substrates were a plasmid bearing 3′-PG termini on either a 3-base 3′ overhang as shown (A., quantitated in B. and C., treatment with 8 nM TDP1) or an otherwise identical substrate with a 3′-PG blunt end (D., quantitated in E., treatment with 37 nM TDP1). Panel (C.) shows pooled data (mean ± SEM) from 3 experiments with the 3′ overhang, with incubation in the presence or in the absence of ATP; the extent of processing for each sample after 80 min incubation was determined and then normalized to that for the sample containing Ku but no DNA-PKcs. In parallel, the 3′-overhang substrate was incubated for 20 min in the presence of Ku, with or without 20 nM DNA-PKcs and/or 1 mM ATP as indicated, and then subjected to DNase footprinting (F.). A replicate footprinting experiment (not shown) gave essentially the same result.
3.3 ATP abrogates DNA-PK-mediated inhibition
Biochemical and cellular studies suggest that access to DNA ends is modulated by DNA-PKcs autophosphorylation, with phosphorylation of the 2609–2647 serine/threonine cluster being particularly important in promoting accessibility [27]. As expected, addition of ATP to reactions containing TDP1, Ku and DNA-PKcs partially restored PG processing (closed symbols in Fig. 3A–E). To replicate conditions used in end joining experiments, 1 mM ATP was added to these reactions; however, consistent with previous studies of DNA-PKcs release from DNA ends [28], 10 µM ATP was sufficient to promote end processing. Furthermore, 1 mM ATP did not stimulate TDP1 in the absence of DNA-PK, but instead had a slight inhibitory effect (Supplemental Fig. 2). Unexpectedly, processing in the presence of ATP was slightly greater at 20 nM DNA-PKcs than at 6 nM (Fig. 3E). Because DNA-PK activation by autophosphorylation requires synapsis of two DNA-PK complexes [29,30], increasing the DNA-PK concentration could increase the rate of autophosphorylation and thereby promote greater access to DNA ends. Nevertheless, processing in the presence of Ku plus DNA-PKcs was always less than in the presence of Ku alone.
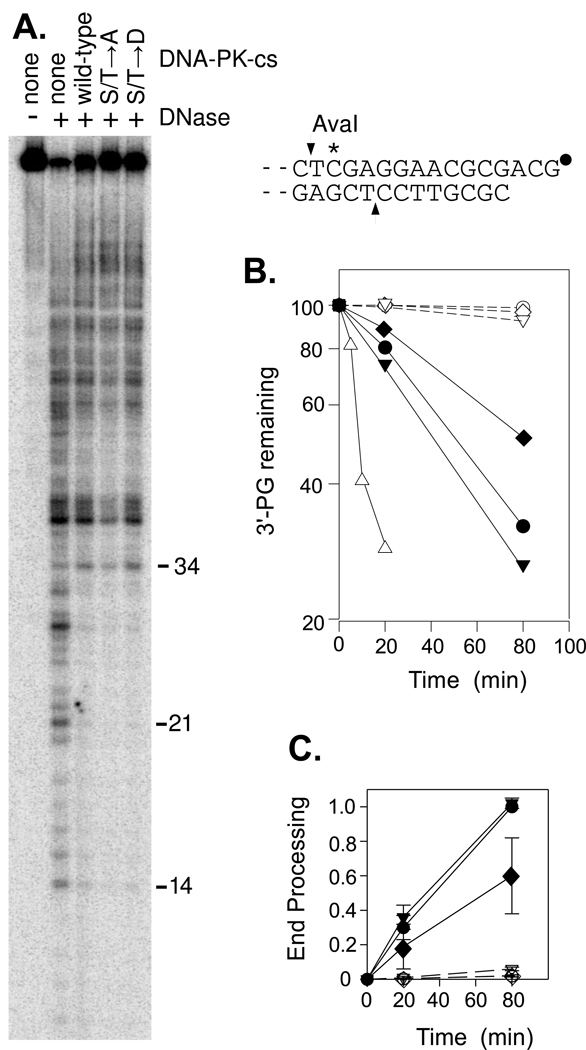
It has been proposed that 2609–2647 cluster phosphorylation can render DNA ends accessible to processing without causing complete DNA-PKcs dissociation [31]. However, in the case of TDP1, conditions that promoted significant PG processing (i.e., addition of ATP) also resulted in a measurable increase in susceptibility to DNase I (Fig. 3F). Thus, these data do not distinguish whether TDP1 could act while phosphorylated DNA-PKcs was still bound to the DSB ends or could only gain access to 3′ termini when DNA-PKcs dissociated. To more directly assess the importance of 2609–2647 cluster phosphorylation in promoting TDP1-mediated end processing, the experiments were repeated with mutant alleles of DNA-PKcs harboring either S/T→A (unphosphorylatable) or S/T→D (mimicking constitutive phosphorylation) mutations at all six target sites in the cluster. These mutant proteins retain full kinase activity [31] and they produced similar footprints upon binding to DNA (Fig. 4A). In the presence of ATP, reactions with the S/T→A mutant showed a modest reduction in processing (closed diamonds in Fig. 4B–C), while the S/T→D mutant gave results similar to wild-type. In the absence of ATP, the S/T→D mutant showed more processing than wild-type DNA-PKcs, but still much less than was seen in the presence of ATP with any of the DNA-PKcs proteins (open symbols in Fig. 4). Overall, it appears that phosphorylation of other sites must be at least as important as phosphorylation of the 2609–2647 cluster, which apparently promotes processing by TDP1 but is in itself neither necessary nor sufficient.
Figure 4.
Effect of mutations in the 2609–2647 serine/threonine cluster of DNA-PKcs on PG processing. A. The substrate shown was incubated in the absence of ATP for 10 min with 50 nM Ku and 20 nM either wild-type DNA-PKcs or DNA-PKcs having six S/T→A or six S/T→D mutations in the 2609–2647 cluster, then subjected to DNase footprinting. B. The same substrate was incubated with 50 nM Ku and 8 nM TDP1, in the presence (closed symbols) or absence (open symbols) of ATP, with either no DNA-PKcs (△), or 20 nM DNA-PKcs, either wild-type (●,○), or having the same six S/T→A (◆,◊) or S/T→D (▼,▿) mutations. PG processing was analyzed as in Fig. 3. B. Compilation of data from three replicate experiments. Symbols are the same as in (B.). The extent of processing under each condition has been normalized to that seen with wild-type DNA-PKcs in the presence of ATP. The mutated residues in DNA-PKcs are T2609, S2612, T2620, S2624, T2638 and T2647.
3.4 Inhibition of DNA-PK blocks most but not all TDP1-mediated end processing in cell extracts
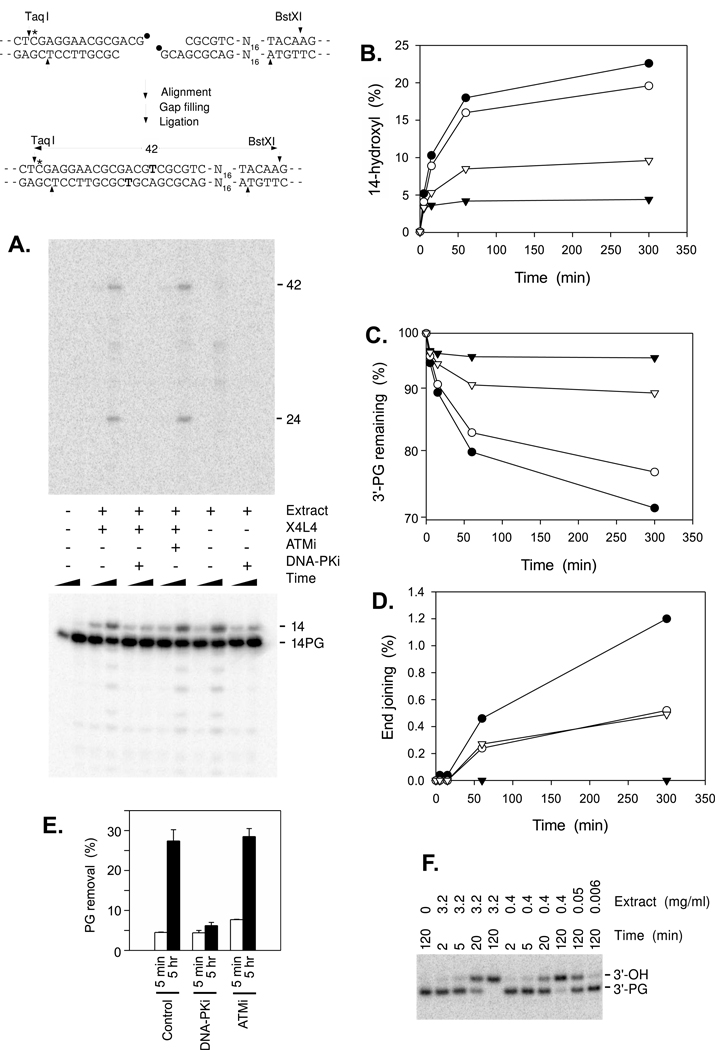
When model DSB substrates bearing protruding 3′-PG termini are incubated in human whole-cell extracts, a substantial fraction of the termini are converted to 3′-hydroxyls [4]. The absence of such conversion in SCAN1 extracts [11] and in Tdp1-/- mouse fibroblast extracts [32] strongly suggests that this reaction is catalyzed by TDP1 and PNKP. When the same substrate was incubated in HeLa nuclear extracts, a small fraction (~5%) of PG termini were removed within 5 min (Fig. 5A–C). PG removal continued for several hours, but at a steadily decreasing rate, so that even after 5 hr, ~70% of PG ends remained intact (closed circles in Fig. 5C). This decrease in the rate of processing did not appear to be due to loss of TDP1 activity over time, as a substrate added to extract that had been preincubated for 2 hr was likewise subject to an initial burst of rapid processing (open inverted triangles in Fig. 5C). Moreover, when a single-stranded 3′-PG substrate was incubated in the same diluted extracts, most of the glycolate was removed within 20 min and the reaction proceeded to completion by 2 hr even at an eightfold lower concentration (0.4 mg/ml) of extract (Fig. 5F). These results suggest that TDP1 in HeLa extracts is sufficiently abundant and stable to effect complete processing of exposed 3′-PG termini, and that the failure to completely process 3′ overhangs is due to protection of these ends by other cellular components.
Figure 5.
Effect of kinase inhibitors on processing of 3′-PG ends in HeLa extracts. A. The substrate shown, bearing PG termini on 3-base 3′ overhangs, was incubated in HeLa nuclear extracts (3.2 mg/ml) for 5 min or 5 hr, with or without DNA-PKi, ATMi and exogenous recombinant X4L4 as indicated. The 14-base fragment represents the free 3′-hydroxyl derived from removal of 3′-PG by TDP1 and PNKP. The 24-base fragment represents intermolecular head-to-head joining, by the same mechanism shown for head-to-tail joining. The top portion of the gel is shown at tenfold higher contrast to visualize end-joining bands. B.-D. Quantitation of data from an experiment similar to that shown in A., with incubations in the presence of DNA-PKi (▼), its solvent (DMSO) alone (○) or no addition (●). In a fourth sample (▿), the substrate was added after pre-incubation of the extract for 2 hr. The amount of each species is expressed as a fraction of the total radioactivity in the lane, excluding a small fraction that remained in the well. End joining (D.) is the sum of the 42- and 24-base products. E. Compilation of PG removal data from three such experiments, showing mean + SEM. F. Processing of 3′-PG termini on a single-strand 14-mer by various dilutions of HeLa extract, in the presence of 5 mM EDTA; samples were then denatured and treated with PNKP to remove 3′-phosphates.
To determine whether the levels of Ku and DNA-PKcs in HeLa nuclear extracts would be sufficient to block TDP1-mediated processing, Western blots were performed with various quantities of nuclear extract and of purified recombinant Ku and DNA-PKcs (Supplemental Fig. 3). These titrations indicated that HeLa extracts contained 560 ± 90 nM DNA-PKcs and 4.1 ± 1.4 µM Ku, which would result in concentrations of approximately 110 nM DNA-PKcs and 820 nM Ku in end joining reactions. Judging from the in vitro studies (Fig. 3), in the absence of DNA-PKcs phosphorylation, these concentrations would be more than adequate to block all processing. To test this prediction, reactions were performed in extracts containing the DNA-PK inhibitor KU-57788 (DNA-PKi).
When added 10 min before the substrate, DNA-PKi had little if any effect on PG removal in the first 5 min, but processing at later times was blocked completely (Fig. 5B, 5C and 5E). In the absence of inhibitor, there was also a detectable level of end joining, as indicated by the formation of diagnostic 42- and 24-base fragments (Fig. 5A), and this joining was also completely blocked by DNA-PKi, as expected (Fig. 5A, 5D and 5E). It has been reported that in intact cells, most if not all DNA-PKcs phosphorylation in the 2609–2647 cluster is catalyzed by ATM rather than DNA-PK itself [33]. However, an ATM inhibitor (ATMi) did not prevent end joining (Fig. 5A). Moreover, it did not inhibit processing of PG ends; if anything, ATMi appeared to slightly stimulate PG removal during the first 5 min (Fig. 5E). Overall, the results suggest that, when its catalytic activity is blocked, DNA-PK forms a complex on DNA ends that completely protects the ends from processing by TDP1. Thus, the rapid, DNA-PKi-resistant processing in the first 5 min probably occurred before the repair complex had fully formed on the DNA end.
3.5 TDP1-mutant SCAN1 cells show chromosomal hypersensitivity to calicheamicin
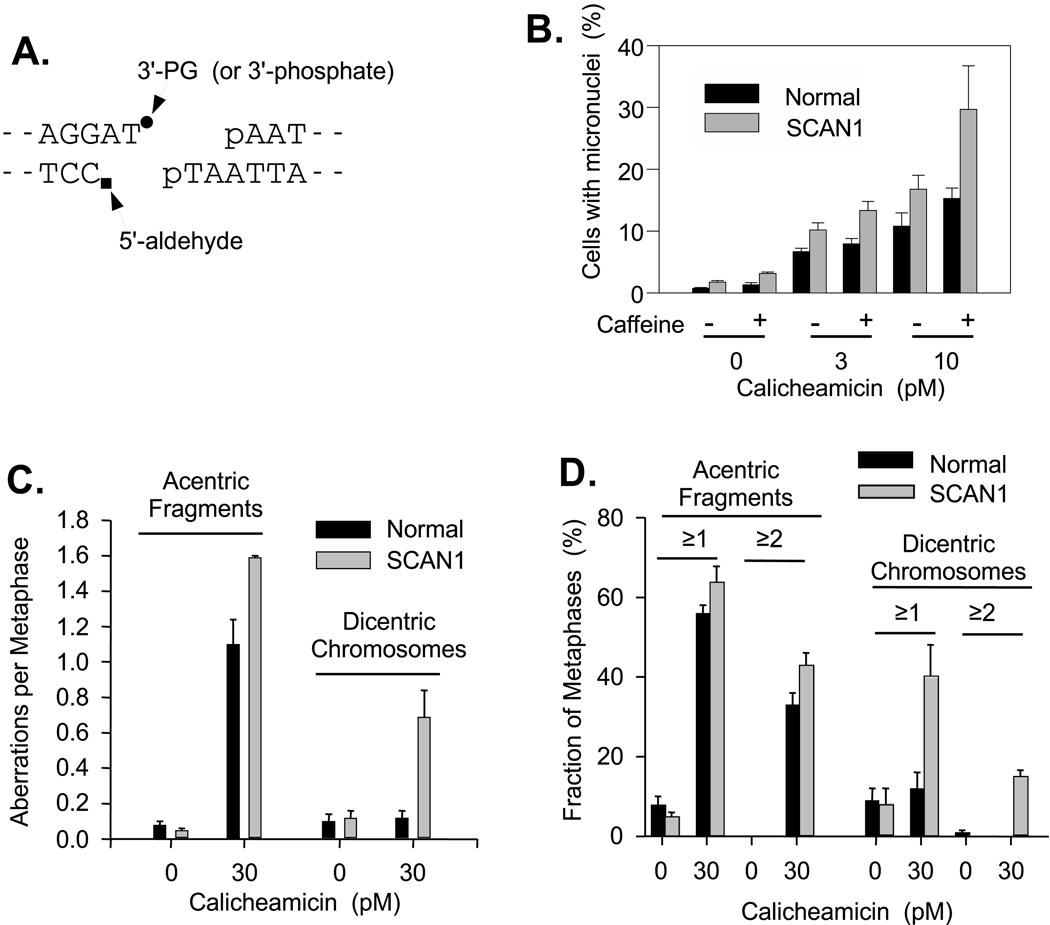
Calicheamicin is a natural product which upon activation forms a diradical species that simultaneously attacks both strands of DNA on a 3-base 3′ stagger [34]. One end of each calicheamicin-induced DSB has a 5′-phosphate, and 3-base 3′ overhang with a 3′-phosphate. The opposite DSB end has a 5′ aldehyde and a 2-base 3′ overhang with either a 3′-phosphate or a 3′-PG (Fig. 6A). Thus, the calicheamicin-induced DSBs that bear 3′-PG termini are similar to the synthetic substrates that require TDP1 for processing in cell extracts.
Figure 6.
Chromosomal sensitivity of SCAN1 cells to calicheamicin. A. Structure of calicheamicin-induced DSBs. B. SCAN1 (#1664) or normal (#1668) cells were treated with the indicated concentrations of calicheamicin and then stained with DAPI, and the fraction of cells with micronuclei was determined. Approximately 100 cells were scored in each of three independent experiments (mean ± SEM). C. Following calicheamicin treatment, metaphase cells were collected for 1 hr with colchicine, and then metaphase spreads were prepared and hybridized with a fluorescent (Cy3) centromeric probe (typical metaphase shown in Supplemental Fig. 4). Approximately 50 metaphases were photographed and scored for acentric fragments and dicentric chromosomes, and the number of each type of aberration per cell was calculated. D. Same data as (C.), but expressed as the fraction of cells having at least one, or more than one aberration of each type. The mitotic index was between 2% and 4% for all samples.
To determine the possible role of TDP1 in repair of these DSBs in intact cells, TDP1-mutant SCAN1 lymphoblastoid cells were treated with calicheamicin, and then assayed for formation of micronuclei (Fig. 6B). Consistently, either with or without 1 mM caffeine (to minimize G2 arrest), the fraction of cells containing micronuclei was about 1.5-fold higher for SCAN1 cells than for identically treated normal cells derived from unaffected relatives. Similarly, levels of acentric chromosome fragments as measured on metaphase spreads with fluorescent-stained centromeres, were ~1.5-fold higher for SCAN1 cells than for normal cells (Fig. 6C). 4). Furthermore, numbers of dicentric chromosomes were much greater in SCAN1 than in normal cells, and there was a particularly dramatic increase in the number of cells having more than one dicentric (Fig. 6D). Elevated levels of calicheamicin-induced dicentric chromosomes were detected consistently in two other SCAN1 lines as well (Supplemental Fig. 5). These results suggest that calicheamicin-induced DSBs were repaired less accurately and perhaps less efficiently or more slowly in SCAN1 than in normal cells. However, there was little if any chromosomal hypersensitivity to ionizing radiation (Supplemental Fig. 6).
3.6 Knockdown of TDP1 by siRNA in HeLa cells confers calicheamicin sensitivity
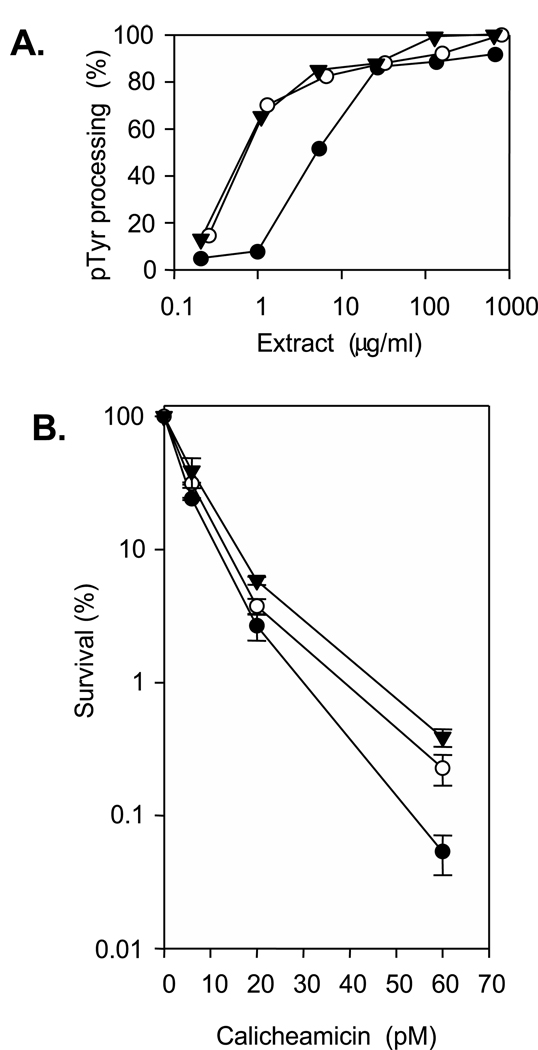
Attempts to assess calicheamicin cytotoxicity toward SCAN1 cells by a cytometry-based growth assay [35] yielded inconclusive results; although calicheamicin hypersensitivity was seen in some experiments, there was variability among individual cell lines that was comparable to differences between normal and SCAN1 lines (data not shown). Therefore, clonogenic survival assays were performed on HeLa cells following knockdown of TDP1 with a pool of four siRNAs (Fig. 7). Assays based on removal of tyrosine from a 3′-pTyr 18-mer indicated an 8-fold reduction in TDP1 activity in extracts of the siRNA-treated cells, compared to an unrelated control siRNA (Fig. 7A). TDP1 knockdown was associated with a small but reproducible increase in cytotoxicity, with a 1.25-fold increase in log cell kill at 60 pM calicheamicin (Fig. 7B).
Figure 7.
Effect of TDP1 knockdown on calicheamicin cytotoxicity toward HeLa cells. Cells were transfected with TDP1 siRNA (●), control siRNA (○) or transection reagent alone (▼) for 24 hr, incubated for 48 hr, treated for 4 hr with calicheamicin and then either lysed for determination of TDP1 activity (A), or plated for determination of clonogenic survival (B). Error bars represent the SEM for three survival determinations from two independent experiments. TDP1 activity titrations indicated a 7.9-fold decrease following TDP1 knockdown; a second independent experiment indicated an 8.0-fold decrease (not shown).
4. DISCUSSION
Previous work showed that TDP1-mutant SCAN1 cells, as well as extracts from these cells, are deficient in the rejoining of SSBs induced by topoI inhibitors [14,36], clearly implicating TDP1 in repair of topoI-mediated breaks. Moreover, TDP1 binds directly to DNA ligase III, the primary ligase in the SSB repair pathway [36]. In vitro, TDP1 can also resolve 3′-PG termini, which are typical of free radical-mediated DNA breaks, and extracts of SCAN1 cells as well as Tdp1-/- murine fibroblasts are completely deficient in processing of protruding 3′-PG termini at DNA DSB ends [11,32]. While these results suggest a possible role for TDP1 in resolving 3′ blocks at DSB termini, direct assays for repair of radiation-induced DSBs in intact cells did not reveal any apparent DSB repair defect in either SCAN1 cells or Tdp1-/- mouse fibroblasts [13,14]. However, interpretation of these data is complicated by the fact that the frequency of 3′-PG termini at these DSB ends is uncertain. Although early studies suggested that about half of radiation-induced DNA breaks had 3′-PG termini [12], a more recent estimate by mass spectrometry yielded a much lower estimate of ~10% [37]. If this latter value is correct, then a 3′-PG processing defect might not confer either significant radiosensitivity or a noticeable defect in DSB rejoining.
To further assess the possible role of TDP1 in repair of DSBs with blocked 3′ termini, we examined the interplay between TDP1 and the core NHEJ component DNA-PK, at DNA ends. Given the high levels of Ku and DNA-PKcs in human cells [25], it is highly likely that free DNA ends, regardless of their source, would rapidly recruit both Ku and DNA-PKcs. Thus, any putative end processing by TDP1 would take place in the context of ends sequestered by DNA-PK.
Experiments with purified proteins suggest that when Ku and unphosphorylated DNA-PKcs are bound to a DNA end bearing a PG on a short 3′ overhang, the PG is completely protected from TDP1-mediated glycolate removal (Fig. 3). Moreover, whereas phosphorylation of the 2609–2647 serine/threonine cluster in DNA-PKcs has been proposed to induce a conformational change that promotes end accessibility [31], S/T→D mutations that mimic such phosphorylation increased 3′-PG processing in the absence of ATP only very slightly. This result is somewhat at odds with a previous finding that the same S/T→D DNA-PKcs mutant allowed digestion of DNA ends by exonuclease V in the absence of ATP, under conditions where the wild-type enzyme protected the ends almost completely when phosphorylation was blocked by wortmannin [31]. This disparity may reflect differences in the details of binding of the individual enzymes to DNA, for example the proposed requirement that DNA become at least transiently single-stranded in order to properly bind to TDP1 [28]. Nevertheless, addition of ATP strongly promoted 3′-PG processing, suggesting that DNA-PK-mediated phosphorylation, of either itself or other proteins, does make ends more accessible. In whole-cell extracts, the S/T→A mutant form of DNA-PK still supports some end processing and gap filling, while both these processes are blocked completely by DNA-PKi, even in the presence of the S/T→D mutant. These results, like those reported above, suggest that there are other DNA-PK-catalyzed phosphorylations that are at least as important as the 2609–2647 cluster in promoting end accessibility and processing. However, none of these data distinguish whether this accessibility reflects dissociation of DNA-PKcs from some ends, or only a conformational change in DNA-PKcs that still remains bound.
In an attempt to distinguish between these alternatives, DNase footprinting was performed. If DNA-PKcs underwent a major conformational change without dissociation, a qualitatively different footprint might result. Instead, the DNase cleavage pattern seen after incubation in the presence of ATP was weaker but otherwise similar to that seen in the absence of DNA-PK (Fig. 3F), consistent with the view that a fraction of the DNA ends was free of any DNA-PK while the rest remained fully sequestered. There are at least 12 potential sites of autophosphorylation on DNA-PKcs, and extensive autophosphorylation of DNA end-bound DNA-PKcs can result in complete dissociation from the DNA [38]. Since in our experiments DNA-PKcs is present in at least a 10-fold excess with respect to DNA ends, it is expected that any such dissociated, phosphorylated DNA-PKcs would be replaced by a fresh unphosphorylated molecule. If reloading of DNA-PKcs onto the DNA end were sufficiently rapid, then there would be little or no processing until the entire pool of unphosphorylated DNA-PKcs was exhausted, at which point the rate of PG processing would abruptly increase. On the contrary, however, PG processing in the presence of DNA-PK and ATP appeared to proceed linearly from time zero (Fig. 3B and 3E), consistent with a model [39] in which each DNA-PKcs dissociation is followed by an interval when the DNA end is relatively accessible to TDP1, before the end is again sequestered by a new molecule of DNA-PKcs.
A similar situation appears to prevail in nuclear extracts (Fig. 5), where PG removal likewise proceeds continuously from time zero despite high concentrations of DNA-PKcs (~0.1 µM) and Ku (~0.8 µM). The fact that there is significant PG removal in the first 5 min even when DNA-PK kinase activity is blocked, also suggests that end sequestration by DNA-PKcs is not immediate and that there is opportunity for end processing before the end-bound repair complex is fully formed. Moreover, the experiments with cell extracts indicate that such processing can occur in the presence of not only DNA-PK, but of all proteins required for efficient NHEJ.
In intact cells, the chromosomal sensitivity of SCAN1 cells to calicheamicin provides additional evidence of a role for TDP1 in DSB repair. The calicheamicin-induced DSB is unique in that one end of the break often bears a 3′-PG and a 5′-aldehyde [34,40]; thus, at least one of these blocking groups must be removed in order for end joining to proceed. Most free radical-mediated DSBs resulting from deoxyribose oxidation at positions other than C-4′ will bear 5′-phosphate and 3′-phosphate termini [41,42], and the latter can be readily converted to 3′-hydroxyls by PNKP, which binds to the core NHEJ protein XRCC4. Thus, the vast majority of DSBs induced by diffusible free radicals could presumably be rejoined in at least one strand without any additional enzymes for removal of modified ends. The presence of abnormal termini in both strands at one end of the calicheamicin-induced DSB may explain, in part, the extraordinary cytotoxicity of this compound, as well as the apparent requirement for TDP1 for optimal repair. These doubly blocked ends are likely to be resistant to many of the 3′→5′ and 5′→3′ exonucleases that otherwise might initiate resection-based end joining [43]. Their resolution may instead require either a specific end-processing enzyme such as TDP1, or an endonucleolytic cleavage near the terminus, perhaps preceded by limited helicase-mediated unwinding of the terminal base pairs.
Thus, taken together, the results suggest that despite lack of demonstrable specific interactions with core NHEJ proteins, TDP1 can participate in resolution of blocked 3′ DSB termini in the context of NHEJ. Although Artemis nuclease plus DNA-PK can also process blocked termini on 3′ overhangs, this processing is slow and inefficient for very short overhangs [35]. Therefore, in SCAN1 cells, 3′-PG-terminated DSBs may tend to persist longer than in normal cells, potentially increasing the probability that they will be incorrectly joined. The very small enhancement of calicheamicin cytotoxicity seen in TDP1-knockdown cells, as well as the finding that in SCAN1 cells calicheamicin-induced dicentrics are enhanced more than chromosome breaks, suggest that TDP1 deficiency confers a marked increase in misrepair but perhaps only a slight defect in the extent of overall rejoining.
Supplementary Material
ACKNOWLEDGMENTS
We thank Graeme Smith and KUDOS Pharmaceuticals for providing KU-57788 and KU-55933, Wyeth Pharmaceuticals for supplying calicheamicin, Michael Weinfeld for supplying PNKP and Sarah Elsea (Dept. of Human Genetics) for advice on cytogenetics. This work was supported by Grants AG023783 and CA40615 from the National Institutes of Health, USDHHS. Confocal microscopy was supported in part by Grant P30 CA16059 to the Massey Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 2.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia. Cell. 2003;112:7–10. doi: 10.1016/s0092-8674(02)01247-3. [DOI] [PubMed] [Google Scholar]
- 4.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double-strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002;276:24323–24330. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 5.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DM. Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J. Mol. Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- 7.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 8.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 9.Raymond AC, Staker BL, Burgin AB., Jr Substrate specificity of tyrosyl-DNA phosphodiesterase I (Tdp1) J. Biol. Chem. 2005;280:22029–22035. doi: 10.1074/jbc.M502148200. [DOI] [PubMed] [Google Scholar]
- 10.Suh D, Wilson DM, III, Povirk LF. 3′-Phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma-Ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J. Biol. Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 13.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair. 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Katyal S, El-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, Northrop JL, Salih MA, Inoue K, Armstrong DL, Champoux JJ, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodarzi AA, Lees-Miller SP. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair. 2004;3:753–767. doi: 10.1016/j.dnarep.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 17.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes M, Rosenberg E, Valerie K. Adenovirus expressing p53. Methods Mol. Biol. 2003;234:1–16. doi: 10.1385/1-59259-408-5:1. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Hannis JC, Flora JW, Muddiman DC, Charles K, Yu Y, Povirk LF. Homogeneous preparations of 3′-phosphoglycolate-terminated oligodeoxynucleotides from bleomycin-treated DNA as verified by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Biochem. 2001;289:274–280. doi: 10.1006/abio.2000.4936. [DOI] [PubMed] [Google Scholar]
- 20.Bennett RAO, Gu X-Y, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Intl. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: Effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 22.Povirk LF, Zhou RZ, Ramsden DA, Lees-Miller SP, Valerie K. Phosphorylation in the serine/threonine 2609–2647 cluster promotes but is not essential for DNA-dependent protein kinase-mediated nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2007;5:3869–3878. doi: 10.1093/nar/gkm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollick JJ, Rigoreau LJ, Cano-Soumillac C, Cockcroft X, Curtin NJ, Frigerio M, Golding BT, Guiard S, Hardcastle IR, Hickson I, Hummersone MG, Menear KA, Martin NM, Matthews I, Newell DR, Ord R, Richardson CJ, Smith GC, Griffin RJ. Pyranone, thiopyranone, and pyridone inhibitors of phosphatidylinositol 3-kinase related kinases. Structure-activity relationships for DNA-dependent protein kinase and inhibition, and identification of the first potent and selective inhibitor of the ataxia telangiectasia mutated kinase. J. Med. Chem. 2007;50:1958–1972. doi: 10.1021/jm061121y. [DOI] [PubMed] [Google Scholar]
- 24.Hardcastle IR, Cockcroft X, Curtin NJ, El-Murr MD, Leahy JJ, Stockley M, Golding BT, Rigoreau L, Richardson C, Smith GC, Griffin RJ. Discovery of potent chromen-4-one inhibitors of the DNA-dependent protein kinase (DNA-PK) using a small-molecule library approach. J. Med. Chem. 2005;48:7829–7846. doi: 10.1021/jm050444b. [DOI] [PubMed] [Google Scholar]
- 25.Anderson CW, Lees-Miller SP. The nuclear serine/threonine protein kinase DNA-PK. Crit. Rev. Eukaryotic Gene Expression. 1992;2:283–314. [PubMed] [Google Scholar]
- 26.Chan DW, Mody CH, Ting NS, Lees-Miller SP. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem. Cell Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- 27.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 28.Davies DR, Interthal H, Champoux JJ, Hol WG. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure (Camb) 2002;10:237–248. doi: 10.1016/s0969-2126(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 29.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J. Biol. Chem. 2004;279:39408–39413. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- 32.Hawkins AJ, Subler MA, Akopiants K, Wiley JL, Taylor SM, Rice AC, Windle JJ, Valerie K, Povirk LF. In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair. 2009 doi: 10.1016/j.dnarep.2008.12.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Yajima H, Löbrich M, Chen DJ. ATM is essential for DNA-PKcs phosphorylations at T2609 cluster upon DNA double-strand break. J. Biol. Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 34.Dedon PC, Goldberg IH. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem. Res. Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 35.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 36.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 37.Chen B, Zhou X, Taghizadeh K, Chen J, Stubbe J, Dedon PC. GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3′-phosphoglycolate pathways of 4′ oxidation of 2-deoxyribose in DNA: application to DNA damage produced by gamma radiation and bleomycin. Chem. Res. Toxicol. 2007;20:1701–1708. doi: 10.1021/tx700164y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 39.Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- 40.Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin neocarzinostatin and other enediynes. Mutat. Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- 41.Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog. Nucleic Acids Res. Mol. Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 42.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid. Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 43.Harrigan JA, Fan J, Momand J, Perrino FW, Bohr VA, Wilson DM. WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech. Ageing Dev. 2007;128:259–266. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.