Abstract
The chitinase (EC 3.2.1.14) of the human malaria parasite Plasmodium falciparum, PfCHT1, has been validated as a malaria transmission-blocking vaccine (TBV). The present study aimed to delineate functional characteristics of the P. vivax chitinase PvCHT1, whose primary structure differs from that of PfCHT1 by having proenzyme and chitin-binding domains. The recombinant protein rPvCHT1 expressed with a wheat germ cell-free system hydrolyzed 4-methylumbelliferone (4MU) derivatives of chitin oligosaccharides (β-1,4-poly-N-acetyl glucosamine (GlcNAc)). An anti-rPvCHT1 polyclonal antiserum reacted with in vitro-obtained P. vivax ookinetes in anterior cytoplasm, showing uneven patchy distribution. Enzymatic activity of rPvCHT1 shared the exclusive endochitinase property with parallelly expressed rPfCHT1 as demonstrated by a marked substrate preference for 4MU-GlcNAc3 compared to shorter GlcNAc substrates. While rPvCHT1 was found to be sensitive to the general family-18 chitinase inhibitor, allosamidin, its pH (maximal in neutral environment) and temperature (max. at ~25 °C) activity profiles and sensitivity to allosamidin (IC50=6 μM) were different from rPfCHT1. The results in this first report of functional rPvCHT1 synthesis indicate that the P. vivax chitinase is enzymatically close to long form Plasmodium chitinases represented by P. gallinaceum PgCHT1.
Keywords: Chitinase, Transmission-blocking, Plasmodium vivax, Allosamidin
1. Introduction
To complete transmission between a vertebrate host and an anopheline mosquito, Plasmodium male and female gametes merge in the mosquito midgut to form zygotes that elongate into the invasive motile form, the ookinete. The ookinete must traverse the chitin-containing peritrophic matrix (PM) en route to invading the midgut epithelium to become a sporozoite-forming oocyst. The ookinete secretes a chitinase [1] that facilitates this process, as has been shown in gene knockout studies [2,3], membrane feeding assays with a chitinase inhibitor allosamidin [4], and with chitinase-specific antibodies [5,6]. Therefore, the chitinase is a potential target for transmission blocking of malaria with chemical or immunological strategies.
Genes encoding chitinases have been identified from several Plasmodium species, but functional analysis including studies on enzymatic activity has only been done with the chitinases PfCHT1 of Plasmodium falciparum [7] and PgCHT1 of P. gallinaceum [8]. P. gallinaceum is the only malaria parasite species in which more than one chitinase genes have been identified. Although both PgCHT1 [4,8] and PgCHT2 [5,8-10] are members of the family 18 chitinases, these enzymes differ significantly in their enzymatic properties including pH optima and quantitative sensitivity to allosamidin [8]. The short form PgCHT2 lacks two structure characteristics present in the long form PgCHT1; 1) a “repeat/insert” region to form a proenzyme domain, between N-terminal signal peptide and a catalytic domain, and 2) a putative chitin-binding domain at the C-terminus. The P. falciparum PfCHT1 and the chitinase of the chimpanzee malaria parasite P. reichenowi PrCHT1 are short forms; the enzymatic characteristics of PfCHT1 has been experimentally demonstrated to be more similar to the orthologous PgCHT2 than to PgCHT1 [7].
Detailed functional analysis including enzymatic characterization of PvCHT1, the chitinase of the other major human malaria parasite P. vivax is fundamental to assessing the potential of Plasmodium chitinase as a candidate transmission-blocking target. The deduced amino acid sequence from cloned pvcht1 [11] indicates that this enzyme is orthologous to the long form of Plasmodium chitinase represented by PgCHT1. An unrooted phylogenetic tree of the conserved catalytic domain supports the clustering of PvCHT1 and PgCHT1 (TBLASTN amino acid identity values=81%/positives=92%) distant from another clustering of PgCHT2 (I38%/P61%), PfCHT1 (I37%/P62%) and PrCHT1 (I37%/P61%) [10]. No other chitinase appears to be present in the P. vivax genome [12]. Indirect immunofluorescence assay (IFA) using antiserum raised against the synthetic peptide corresponding to the catalytic active site of PgCHT1 detected the protein in P. vivax ookinetes [11]. Since it is difficult to obtain sufficient native PvCHT1 protein for functional analysis, analysis of recombinant PvCHT1 (rPvCHT1) in vitro is necessary. Given the difficulties of expressing rPvCHT1 in Escherichia coli (J. Vinetz, unpublished data), we have utilized a cell-free expression system using wheat germ extracts [13,14] which has been demonstrated to be useful for making recombinant Plasmodium proteins [15,16] for obtaining functional rPvCHT1. Use of this system included production not only of the common catalytic domain but also of the chitin-binding domain specific to PgCHT1 and PvCHT1 (long form). The system was used as well for the P. falciparum counterpart PfCHT1 (short form having a catalytic domain only) to allow for direct enzymatic comparison [7].
2. Materials and methods
2.1. Plasmid constructs
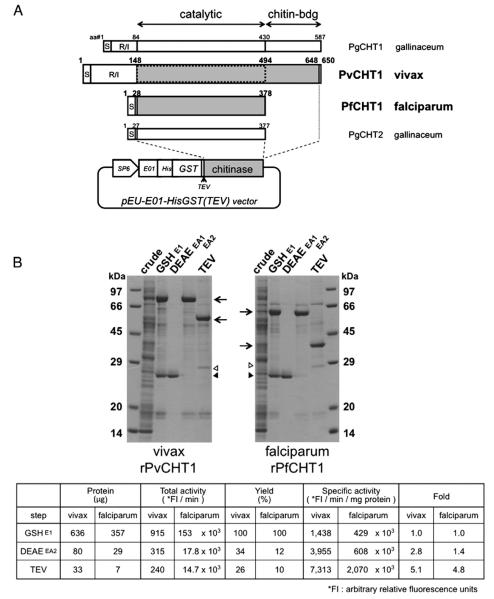
The P. vivax PvCHT1 expression construct was prepared by PCR-amplifying the native coding sequence from P. vivax SalI genomic DNA excluding N-terminal signal peptide and repeat/insert region of pvcht1 (GenBank accession no. AB106896) (Fig. 1A). The XhoI-containing 5′ primer was GAG ACT CGA GAT GGG AGG TAG CCA TGA TAG AAA ACC TCC (the restriction site underlined, and a start methionine codon typed in italic). The BamHI-containing 3′ primer was GAG AGG ATC CTC ACG CGT CTT CCT CTT GCT CAT AGG G. The deduced PvCHT1 protein begins at Gly-148 immediately downstream from the repeat/insert region, and terminated at Ala-648, the third residue upstream from the C terminus (501 residues). PCR products were restriction-digested, ligated in-frame into the XhoI and BamHI sites of the pEU-E01-HisGST(TEV) expression vector for wheat germ cell-free translation system, a slight modification from original pEU-E01-MCS (CellFree Sciences, Japan. http://www.cfsciences.com/eg/). This vector gives an N-terminal hexa-histidine and glutathione S-transferase (GST) tag followed by a tobacco etch virus (TEV) protease cleavage site upstream of cloned pvcht1 (Fig. 1A). The construct was transfected into JM109 E. coli cells and the correct clone was verified by automated sequencing.
Fig. 1.
Synthesis and purification of recombinant P. vivax chitinase (PvCHT1) and P. falciparum PfCHT1 proteins. A. Schematic representation and translated sequence of PvCHT1 and PfCHT1 constructs in a cell-free expression vector pEU-E01-HisGST(TEV). The diagram of two P. gallinaceum chitinases PgCHT1 and PgCHT2 as well as the positions of signal peptide (S), repeat/insert region (R/I), and catalytic or chitin-binding domains are placed for comparison. The shaded regions are cloned to obtain recombinant (r) proteins. The second rPvCHT1 mimicking such a short form species as PfCHT1 is bordered by dotted line (see Section 2.1). The locations of SP6 promoter (SP6), translational enhancer (E01), hexahistidine (His), glutathione S-transferase (GST), and rPvCHT1 or rPfCHT1 (chitinase) on the vector are shown. B. A representative result after sequential purification steps (see Sections 2.3 and 2.4). 2.4 μl of total cell-free synthesis products (crude) or 10 μl of the samples at various purification steps were analyzed by SDS-PAGE under reducing condition. The protein products at each step are shown by arrows. 1) A contaminant glutathione (GSH)-binding protein in the eluates E1 after GSH/GST affinity chromatography, or in EA1 after DEAE anion exchange chromatography, and 2) the remnants of the N-terminal His-GST portion and possibly AcTEV protease itself (28–29 kDa) after AcTEV treatment and dialysis, are respectively shown by closed and open triangles. The chitinase specific activities of rPvCHT1 and rPfCHT1 samples are shown in the table format below; in the eluates E1, EA2, and in the final products after AcTEV treatment and dialysis.
The second PvCHT1 expression construct was established by changing 3′ primer to GAG AGG ATC CTC ACC ATA CAT TCG TAA GGA ACG CAT CTA TAT, which gives shorter PvCHT1 terminating at Trp-494 (347 residues). This shorter construct mimicking such a short form chitinase as PfCHT1 covers only the common domain excluding putative chitin-binding domain (dotted on PvCHT1 in Fig. 1A).
The single copy gene pfcht1 (accession no. AF172445) of P. falciparum chitinase (PfCHT1) was cloned into the same pEU-E01-HisGST(TEV) vector. This clone confers the identical PfCHT1 amino acid sequence, Gly-28 through His-378 (351 residues) to that used for previous study with conventional E. coli system [7] (Fig. 1A).
2.2. Recombinant protein synthesis by wheat germ cell-free system
We employed the wheat germ cell-free protein expression system for protein production using the bilayer translation reaction method described previously [13,17]. Briefly, 1.2 ml of transcription mixture containing 120 μg of the pEU vector construct, 80 mM HEPES-KOH, pH 7.8, 16 mM magnesium acetate, 2 mM spermidine, 10 mM dithiothreitol, 2.5 mM each of nucleotide triphosphates, 1 U/μl of SP6 RNA polymerase (Promega), and 1 U/μl of RNasin (Promega) was incubated 6 h at 37 °C. After the incubation, the solution with mRNA was centrifuged at 11,000 ×g for 5 min at 4 °C and supernatant was recovered and mixed with an equal volume (1.2 ml) of wheat germ extract (240 A260 units) (CellFree Sciences) supplemented with 2.4 μl of 20 mg/ml creatine kinase to prepare translation mixture. On top of 0.4 ml-each mixture aliquot in six-well plate, the 4.4 ml substrate mix containing 30 mM HEPES-KOH, pH 7.8, 100 mM potassium acetate, 2.7 mM magnesium acetate, 0.4 mM spermidine, 2.5 mM dithiothreitol, 0.3 mM each of 20 amino acids,1.2 mM rATP, 0.25 mM rGTP, and 16 mM creatine phosphate were added and then incubated at 17 °C for 16 h.
2.3. Purification by affinity and anion-exchange chromatography
A 300 μl-bed volume of glutathione-coupled gel (Glutathione Sepharose 4B, GE Healthcare) was pre-equilibrated with 20 mM Tris–HCl (pH 8.0). The whole 28.8 ml of cell-free protein synthesis products was mixed with the gel and incubated for 16 h at 4 °C with gentle agitation. The suspension was then transferred into an empty column (Bio-Spin #732-6008, Bio-Rad Laboratories) and centrifuged at 500 ×g for 5 min. After washing the matrix pellet with 20 bed volumes (6 ml) of 20 mM Tris–HCl (pH 8.0), 3 and then 1 bed volume of elution buffer (10 mM reduced glutathione in the Tris-buffer above) were added to collect corresponding eluates E1 and E2.
Anion-exchange column chromatography was conducted on a 150 μl-bed volume of TOYOPEARL DEAE-650-M gel (TOSOH) in Bio-Spin column, pre-equilibrated with 20 mM Tris–HCl (pH 8.0). The E1 eluate above was added to the column and the gel matrix was washed primarily with 1.5 ml of 20 mM Tris–HCl (pH 8.0). The gel was next washed by 0.1 M and then 0.18 M sodium chloride in the same buffer, 0.3 ml each, into the combined eluates EA1. Sodium chloride was concentrated further to 0.5 M (0.75 ml) to collect EA2 eluate.
2.4. AcTEV protease treatment to remove His-GST portion
The 2.5 times EA2 concentrate (0.3 ml) by ultrafiltration (Ultra-free™ #UFV5BGC, 10 kDa molecular mass cutoff, Millipore) was mixed with 10 mU/μl (final) of AcTEV protease (Invitrogen) and incubated for 3 h at 30 °C. 0.2 ml of the product was further mixed with two different matrices, 9 μl of Glutathione Sepharose 4B and 3 μl of nickel-charged resin (Ni-NTA Superflow, Qiagen), pre-equilibrated with 20 mM Tris–HCl (pH 8.0). The mixture was incubated overnight at 4 °C and the supernatant including cleaved PvCHT1 molecules was separated by 0.22 μm microfiltration unit (Ultrafree™ #UFC30GV, Millipore) from His-GST portion on matrices. The solution was dialyzed overnight at 4 °C in the buffer containing 40 mM Tris–HCl, pH 7.5, 400 mM sodium chloride, 2 mM EDTA, and 0.2% TritonX-100. These recombinant PvCHT1 (rPvCHT1) or rPfCHT1 protein samples were finally mixed with an equal volume of 100% glycerol, either for immediate use or for storage at −25 °C until further use.
2.5. Immunization of mice and rabbit with rPvCHT1 and Pvs25
The purified rPvCHT1 proteins were suspended in 1× phosphate buffered saline (PBS). Groups of female BALB/c mice were subcutaneously immunized three times on days 0, 21, and 42 with 10 μg of the rPvCHT1 emulsified in Freund's adjuvant (complete for priming, incomplete Freund's adjuvant for subsequent immunizations). Antiserum was prepared as described elsewhere [18]. For obtaining polyclonal serum for counterstaining, the recombinant ookinete surface protein Pvs25 was administered subcutaneously (250 μg, 3 times) into a Japanese White rabbit following the same protocol [19].
2.6. Indirect immunofluorescence assay (IFA)
Blood specimens from gametocytemic vivax malaria patients were used as a source of zygotes/ookinetes produced in vitro [20]. The use of all human materials in this study was reviewed and approved by institutional Ethics committee of the Thai Ministry of Public Health and the Human Subjects Research Review Board of the United States Army. Peripheral blood was collected with heparinized syringes with written informed consent from patients who came to the malaria clinics in Mae Sod, Thailand. Infection with P. vivax was confirmed by microscopic observation of Giemsa-stained thick and thin blood smears.
Indirect immunofluorescence assays (IFAs) were performed with P. vivax zygotes/ookinetes acetone-fixed onto glass slides. After blocking in phosphate-buffered saline (PBS) containing 5% nonfat skim milk, the specimens were incubated 1 h at 37 °C with two primary antibodies in the same buffer, mouse anti-PvCHT1 antiserum (1:50 dilution) and rabbit anti-Pvs25 (1:200). After washing with 1× PBS, slides were then incubated 30 min at 37 °C with Alexa 488-labeled goat anti mouse IgG (1:500 dilution; Invitrogen), Alexa 546-labeled goat anti rabbit IgG (1:500; Invitrogen), and 4,6-diamidino-2-phenyl-indole HCl (DAPI, 1 μg/ml; Wako Pure Chemical). The slides were mounted with Prolong Gold® (Invitrogen) and examined with LSM5 Pascal confocal laser microscope system (Carl Zeiss MicroImaging).
2.7. Assessment of chitinase activity
Chitinase activity of purified rPvCHT1 or rPfCHT1 aliquots was assayed by microfluorimetry using 4-methylumbelliferone (4MU) derivatives of chitin oligosaccharides (β-1,4-poly-N-acetyl glucosamine (GlcNAc)). The reaction volume of 200 μl in 96-well black round bottom plates (Corning) includes 0.1 mM Tris–HCl (pH 8.0), 1 mM sodium chloride, 5 μM EDTA, 0.005% (w/v) Triton X-100, and 19 μM of 4-methylumbelliferyl-N,N′,N″-β-d-triacetyl chitotrioside (4MU-GlcNAc3) (Sigma-Aldrich). The production of 4MU during continuous incubation at 26 °C was monitored (Wallac ARVO™ MX 1420 multilabel counter (Perkin Elmer); excitation at 355 nm; emission at 450 nm). The resultant enzymatic activity was reported as initial rates of substrate hydrolysis per minute using a standard curve to transform relative fluorescent units. Quantification of proteins was done using Advanced Protein Assay™ reagent #ADV101 (Cytoskeleton).
2.8. Determination of substrate specificity, pH profile, temperature profile, and allosamidin inhibition curves
To assess substrate specificity of the recombinant Plasmodium chitinases, mono-, di-, and tri-GlcNAc derivatives of 4-methylumbelliferone (4MU-GlcNAc1–3) (Sigma-Aldrich) were used in enzyme assays under the same conditions as described in Section 2.7. Only 4MU-GlcNAc3 was used for the other three sets of test below. The pH activity profile was monitored at 26 °C using a potassium phosphate buffer (40 mM final with respect to phosphate) system, instead of Tris–HCl, with pH at 4.3 and ranging from 4.5 to 8.0 in 0.5 pH unit increments. The temperature activity profile was tested from 10 to 50 °C in 5 °C increments using standard Tris–HCl (pH 8.0) buffer. An inhibitor against family-18 chitinases, allosamidin (courtesy of Dr. Shohei Sakuda, University of Tokyo, Japan), was prepared as a 4 mM stock in 0.1 M acetate that was diluted just before assay into the standard reaction mixture in the previous section to estimate IC50 at fixed 19 μM 4MU-GlcNAc3. The stock allosamidin solution was concentrated so that the solvent acetate did not cause pH fluctuation among working solutions at various allosamidin concentrations (data not shown). These experiments were repeated three times each.
3. Results and discussion
Recombinant protein of a chitinase from P. vivax (rPvCHT1) was successfully produced using the wheat germ cell-free translation system (Fig. 1A). rPvCHT1 shared general characteristics with the other Plasmodium family-18 chitinases, yet enzymatic characters varied in some details compared to P. falciparum recombinant PfCHT1 (rPfCHT1), as described below.
3.1. Enzymatically active PvCHT1 recombinant (r) protein, covering both catalytic and chitin-binding domains, was obtained using the wheat germ cell-free system (Fig. 1B)
A series of rPvCHT1 purification steps produced a prominent protein band with apparent mass of ~80 kDa in the eluate E1 after glutathione (GSH)/glutathione S-transferase (GST) affinity purification, which remained ~80 kDa after anion-exchange chromatography in the eluate EA2, and finally changed to ~60 kDa after AcTEV protease treatment (arrows in Fig. 1B). Another major band with less intensity was at 26–27 kDa in the E1 and separated into EA1 eluate (closed triangle) from the dominant band shown above. This profile is also the case with rPfCHT1; the dominant at 60–65 kDa in the E1 going down to ~40 kDa after AcTEV treatment, while the other band into EA1 at an identical 26–27 kDa indicating a contaminant GSH-binding protein from the wheat germ extract. The profile fits predicted molecular masses: full-length products of 85 kDa (rPvCHT1) or 57 kDa (rPfCHT1) from the constructs in the pEU-E01-HisGST(TEV) should be AcTEV-digested into common 28 kDa N-terminal His-GST portion (open triangle in Fig. 1B) and chitinase part with different sizes (arrows). Using the full long form of rPvCHT1, the chitinase-specific activities digesting 4-methylumbelliferyl-N,N′,N″-β-d-triacetyl chitotrioside (4MU-GlcNAc3) were detected compared to mock reactions using protein synthesis products without template mRNA. The rPfCHT1 hydrolyzed 4MU-GlcNAc3 as well, whereas the second (shortened) PvCHT1 construct lacking the putative chitin-binding domain did not show any hydrolyzing activity (data not shown). These results demonstrate for the first time that P. vivax pvcht1 chitinase can be expressed successfully as an active recombinant protein, and that a putative chitin-binding domain which the P. falciparum paralog PfCHT1 lacks at its C-terminus is important for rPvCHT1 to exhibit its enzymatic activity.
Further confirmation of the activity of rPvCHT1 was demonstrated by assays during sequential purification steps. rPvCHT1 chitinase-specific activity increased with purity, but that of rPfCHT1 protein prepared in parallel was <100 times higher than rPvCHT1 at corresponding purification steps. This comparatively weak activity by rPvCHT1 on an artificial substrate 4MU-GlcNAc3 led us to the other test for substrate specificity described in Section 3.3.
3.2. PvCHT1 was localized in cytoplasmic organelle of zygote/ookinete (Fig. 2)
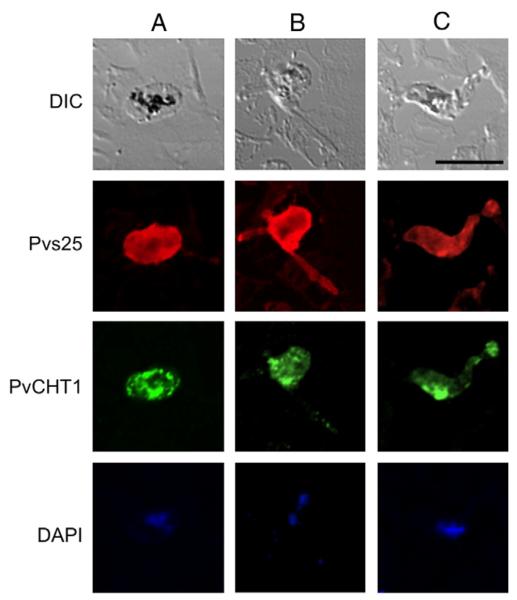
Fig. 2.
Immunofluorescence localization of P. vivax chitinase PvCHT1 in in vitro-developed mosquito midgut stage parasites. Column A (left), zygote; B, immature ookinete; C, mature ookinete. Each of 3 columns consists of 4 panels showing from top to bottom; differential interference contrast (DIC) image, staining with rabbit anti-Pvs25 antiserum (red), with mouse anti-PvCHT1 (green), and DAPI-stained nuclei (blue). Bar = 10 μm. See Section 2.6. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To demonstrate the presence of PvCHT1 in P. vivax mosquito midgut stage parasites, immuno-localization assay was carried out. Confocal immunofluorescence microscopy with anti-rPvCHT1 on P. vivax zygote/ookinetes demonstrated a nondiffuse, lumpy-granular, and uneven staining (green) throughout the different developmental stages. The PvCHT1 distributed in zygote cytoplasm as granular pattern (Fig. 2, column A), while in immature ookinetes, it was concentrated into zygote remnant space out of which the ookinete is emerging (B). In mature ookinetes there was the patchy appearance of the immunofluorescent signals of PvCHT1 mainly in anterior cytoplasm (C), suggesting the micronemal localization of PvCHT1. This localization was distinct from parasite surface membranes, as delineated by red staining of zygote/ookinete surface antigen Pvs25. The localization pattern of PvCHT1 is consistent with previous IFA studies on P. gallinaceum and P. falciparum [21] as well as that on P. vivax ookinete using anti-P. gallinaceum CHT1 active-site antiserum [11], indicating that recombinant PvCHT1 protein with the same immunological properties was successfully prepared.
3.3. The enzymatic characteristics of rPvCHT1, unlike rPfCHT1, were close to plasmodial long form chitinase PgCHT1 (Fig. 3)
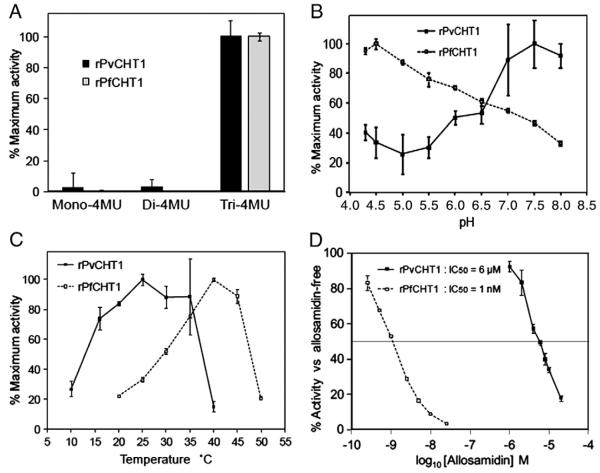
Fig. 3.
Detailed analyses of chitinase activities by rPvCHT1 and rPfCHT1. Data with standard errors are displayed as the mean of three separate experiments. See Section 2.8. A. The substrate specificity. Quantification of fluorescence at 26 °C/pH 8.0 on 3 substrates with different number of N-acetyl-d-glucosamine (GlcNAc), 4MU-GlcNAc1–3. Results are shown as relative rates of activities. B and C. pH or temperature activity profiles. Relative rates of activities at different (B) pH (at a constant temperature of 26 °C) or (C) temperature (at pH 8.0) levels. D. Allosamidin sensitivity on 4MU-GlcNAc3 at 26 °C/pH 8.0. Chitinase activity is expressed as percent activity of enzyme in the absence of allosamidin.
We found that rPvCHT1 hydrolyses only 4MU-GlcNAc3 but not 4MU-GlcNAc1–2 (Fig. 3A). This substrate preference is shared not only with the rPfCHT1 prepared here with the wheat-germ cell-free system, but is also consistent with previous reports using E. coli-produced recombinant PfCHT1 [7] and recombinant PgCHT1 [8]; the same was observed with P. gallinaceum native chitinase activity by ookinete crude extracts [8]. These data indicate that PvCHT1 maintains the specificity unique to Plasmodium chitinases; an endochitinase with a preference for longer chitin oligosaccharides substrates.
We examined the rPvCHT1 pH activity profile, which demonstrated that optimal activity occurs under neutral condition (pH at 7.0–8.0 in Fig. 3B). It is unique among plasmodial chitinases examined so far, and is consistent with the pH 8.0–9.5 in the adult Anopheles mosquito midgut [22] and further with the pH 7.5–7.6 after bloodmeal ingestion [23]. As a control, rPfCHT1 produced in the wheat germ system was most active at pH 4.5, consistent with previous reports (optimal pH at 4.0–5.0) using native or recombinant proteins PfCHT1, PgCHT1 and PgCHT2 [7,8]. Although the physiological implications in this preference for lower pH by control rPfCHT1 is not instantly clear, it should be noted that the chitinase activity of rPfCHT1 at pH 8.0 still maintains <30% of its maximum at pH 4.5, suggesting that these Pf or Pg chitinases do as well work in the adult mosquitoes midgut. Furthermore, studies on chitinases from microbial sources including bacteria and fungi (Table 1 in review article [24] and references therein) suggest that the difference in optimum pH to this extent is not quite exceptional. The temperature activity profile examined here for the first time even for rPfCHT1 as well as rPvCHT1 (Fig. 3C) showed a preference of rPvCHT1 for relatively lower temperature (maximum at 25 °C and <80% max. at 20–35 °C) than rPfCHT1 (max. at 40 °C). The result is consistent with geographical prevalence of falciparum/vivax malaria; the former is in tropical region and the latter prefers more temperate environment. And it should be noted again that the studies on chitinases from microbial sources (Table 1 in [24]; the optimum Tm ranging from 30 to 60 °C) suggest that the results here by rPvCHT1 and rPfCHT1 are not quite exceptional.
rPvCHT1 was <1,000-fold less sensitive (IC50 = 6 μM) to the chitinase inhibitor allosamidin than rPfCHT1 (1 nM), when measured at a fixed concentration of substrate 4MU-GlcNAc3 (19 μM) (Fig. 3D). If Plasmodium chitinases are grouped following the P. gallinaceum nomenclature into a long form (PgCHT1) and a short form (PgCHT2 lacking a putative chitin-binding domain), this difference was observed previously; the IC50 values were 12 μM and 7 μM respectively for recombinant and native PgCHT1 preparation, while as low as 0.3 μM for native PgCHT2 [8]. P. falciparum recombinant PfCHT1 produced in E. coli showed IC50 values around 0.1 μM (40–150 nM) under different pH conditions of 5.0, 6.0 and 7.0 [7], a sensitivity much closer to its apparent ortholog PgCHT2 than to its paralog PgCHT1. The sensitivity of rPfCHT1 here produced by the wheat germ cell-free system (IC50 = 1 nM) is even more remarkable, whereas the IC50 (6 μM) of rPvCHT1 here is closer to its ortholog PgCHT1. However, it should also be noted that the 1 mM and 0.1 mM concentrations of allosamidin used in previous study to block oocyst development [4] far exceed this value, and they would completely inhibit P. vivax chitinase activity in vivo.
Collectively, these data in this first report of the functional property of the P. vivax chitinase, PvCHT1, indicate that it has most characteristics of Plasmodium chitinases. However more in enzymatic detail it is close to long forms represented by P. gallinaceum PgCHT1. Also taken into account the difference in the catalytic domain sequence between PvCHT1 and PfCHT1 (Introduction and Fig. 1A), it has yet to be determined whether a single strategy targeting both chitinases is feasible for human malaria transmission-blocking.
Acknowledgements
The authors are grateful for vivax malaria patients at Mae Sod district, Tak Province, Thailand for providing blood specimens, as well as the staff of the Vector Borne Disease Training Center, Pra Budhabat, Saraburi, Thailand, for assistance in setting up the field sites and the staff of the Department of Entomology, AFRIMS, Bangkok, Thailand. A chitinase specific inhibitor allosamidin is a kind gift from Dr. Shohei Sakuda, University of Tokyo, Japan. This work was supported in part by Grants-in-Aid for Scientific Research (17790277, 18390129 and 19406009) and Scientific Research on Priority Areas (19041053) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and grants from the U.S. National Institutes of Health (JMV).
References
- [1].Huber M, Cabib E, Miller LH. Malaria parasite chitinase and penetration of the mosquito peritrophic membrane. Proc Natl Acad Sci U S A. 1991;88:2807–10. doi: 10.1073/pnas.88.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai YL, Hayward RE, Langer RC, Fidock DA, Vinetz JM. Disruption of Plasmodium falciparum chitinase markedly impairs parasite invasion of mosquito midgut. Infect Immun. 2001;69:4048–54. doi: 10.1128/IAI.69.6.4048-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dessens JT, Mendoza J, Claudianos C, Vinetz JM, Khater E, Hassard S, et al. Knockout of the rodent malaria parasite chitinase pbCHT1 reduces infectivity to mosquitoes. Infect Immun. 2001;69:4041–7. doi: 10.1128/IAI.69.6.4041-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahabuddin M, Toyoshima T, Aikawa M, Kaslow DC. Transmission-blocking activity of a chitinase inhibitor and activation of malarial parasite chitinase by mosquito protease. Proc Natl Acad Sci U S A. 1993;90:4266–70. doi: 10.1073/pnas.90.9.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer RC, Li F, Popov V, Kurosky A, Vinetz JM. Monoclonal antibody against the Plasmodium falciparum chitinase, PfCHT1, recognizes a malaria transmission-blocking epitope in Plasmodium gallinaceum ookinetes unrelated to the chitinase PgCHT1. Infect Immun. 2002;70:1581–90. doi: 10.1128/IAI.70.3.1581-1590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Templeton TJ, Popov V, Comer JE, Tsuboi T, Torii M, et al. Plasmodium ookinete-secreted proteins secreted through a common micronemal pathway are targets of blocking malaria transmission. J Biol Chem. 2004;279:26635–44. doi: 10.1074/jbc.M401385200. [DOI] [PubMed] [Google Scholar]
- 7.Vinetz JM, Dave SK, Specht CA, Brameld KA, Xu B, Hayward R, et al. The chitinase PfCHT1 from the human malaria parasite Plasmodium falciparum lacks proenzyme and chitin-binding domains and displays unique substrate preferences. Proc Natl Acad Sci U S A. 1999;96:14061–6. doi: 10.1073/pnas.96.24.14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinetz JM, Valenzuela JG, Specht CA, Aravind L, Langer RC, Ribeiro JM, et al. Chitinases of the avian malaria parasite Plasmodium gallinaceum, a class of enzymes necessary for parasite invasion of the mosquito midgut. J Biol Chem. 2000;275:10331–41. doi: 10.1074/jbc.275.14.10331. [DOI] [PubMed] [Google Scholar]
- 9.Langer RC, Li F, Vinetz JM. Identification of novel Plasmodium gallinaceum zygoteand ookinete-expressed proteins as targets for blocking malaria transmission. Infect Immun. 2002;70:102–6. doi: 10.1128/IAI.70.1.102-106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Patra KP, Vinetz JM. An anti-Chitinase malaria transmission-blocking single-chain antibody as an effector molecule for creating a Plasmodium falciparum-refractory mosquito. J Infect Dis. 2005;192:878–87. doi: 10.1086/432552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuboi T, Kaneko O, Eitoku C, Suwanabun N, Sattabongkot J, Vinetz JM, et al. Gene structure and ookinete expression of the chitinase genes of Plasmodium vivax and Plasmodium yoelii. Mol Biochem Parasitol. 2003;130:51–4. doi: 10.1016/s0166-6851(03)00140-3. [DOI] [PubMed] [Google Scholar]
- 12.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–63. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawasaki T, Ogasawara T, Morishita R, Endo Y. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci U S A. 2002;99:14652–7. doi: 10.1073/pnas.232580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc Natl Acad Sci U S A. 2000;97:559–64. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuboi T, Takeo S, Iriko H, Jin L, Tsuchimochi M, Matsuda S, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect Immun. 2008;76:1702–8. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mudeppa DG, Pang CK, Tsuboi T, Endo Y, Buckner FS, Varani G, et al. Cell-free production of functional Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Mol Biochem Parasitol. 2007;151:216–9. doi: 10.1016/j.molbiopara.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Sawasaki T, Hasegawa Y, Tsuchimochi M, Kamura N, Ogasawara T, Kuroita T, et al. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 2002;514:102–5. doi: 10.1016/s0014-5793(02)02329-3. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa T, Komesu A, Otsuki H, Sattabongkot J, Udomsangpetch R, Matsumoto Y, et al. Nasal immunization with a malaria transmission-blocking vaccine candidate, Pfs25, induces complete protective immunity in mice against field isolates of Plasmodium falciparum. Infect Immun. 2005;73:7375–80. doi: 10.1128/IAI.73.11.7375-7380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sattabongkot J, Tsuboi T, Hisaeda H, Tachibana M, Suwanabun N, Rungruang T, et al. Blocking of transmission to mosquitoes by antibody to Plasmodium vivax malaria vaccine candidates Pvs25 and Pvs28 despite antigenic polymorphism in field isolates. Am J Trop Med Hyg. 2003;69:536–41. [PubMed] [Google Scholar]
- 20.Suwanabun N, Sattabongkot J, Tsuboi T, Torii M, Maneechai N, Rachapaew N, et al. Development of a method for the in vitro production of Plasmodium vivax ookinetes. J Parasitol. 2001;87:928–30. doi: 10.1645/0022-3395(2001)087[0928:DOAMFT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 21.Langer RC, Hayward RE, Tsuboi T, Tachibana M, Torii M, Vinetz JM. Micronemal transport of Plasmodium ookinete chitinases to the electron-dense area of the apical complex for extracellular secretion. Infect Immun. 2000;68:6461–5. doi: 10.1128/iai.68.11.6461-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Pilar Corena M, VanEkeris L, Salazar MI, Bowers D, Fiedler MM, Silverman D, et al. Carbonic anhydrase in the adult mosquito midgut. J Exp Biol. 2005;208:3263–73. doi: 10.1242/jeb.01739. [DOI] [PubMed] [Google Scholar]
- 23.Billker O, Miller AJ, Sinden RE. Determination of mosquito bloodmeal pH in situ by ion-selective microelectrode measurement: implications for the regulation of malarial gametogenesis. Parasitology. 2000;120(Pt 6):547–51. doi: 10.1017/s0031182099005946. [DOI] [PubMed] [Google Scholar]
- 24.Dahiya N, Tewari R, Hoondal GS. Biotechnological aspects of chitinolytic enzymes: a review. Appl Microbiol Biotechnol. 2006;71:773–82. doi: 10.1007/s00253-005-0183-7. [DOI] [PubMed] [Google Scholar]