Abstract
The closely related microRNA (miRNA) and RNAi pathways have emerged as important regulators of virus–host cell interactions. Although both pathways are relatively well conserved all the way from plants to invertebrates to mammals, there are important differences between these systems. A more complete understanding of these differences will be required to fully appreciate the relationship between these diverse host organisms and the various viruses that infect them. Insights derived from this research will facilitate a better understanding of viral pathogenesis and the host innate immune response to viral infection.
Keywords: RNAi, innate immunity, microRNAs, viruses
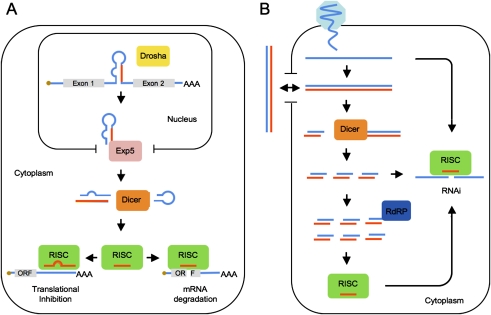
MicroRNAs (miRNAs) are small regulatory RNAs ∼22 nucleotides (nt) in length that are typically derived from a single arm of imperfect, ∼80-nt long RNA hairpins located within polymerase II (pol II)-derived transcripts referred to as primary miRNAs (pri-miRNAs) (Fig. 1A; for review, see Bartel 2004; Cullen 2004). Pri-miRNAs are capped and polyadenylated and may be almost any size, ranging from hundreds to thousands of nucleotides, and may encode a single miRNA or a cluster of several miRNAs (Lagos-Quintana et al. 2001; Lim et al. 2003). Pri-miRNA stem–loops, which consist of an ∼32-base-pair (bp) imperfect stem and a ≥10-nt terminal loop, are cleaved by the RNase III enzyme Drosha, acting together with its cofactor DGCR8, ∼22 bp from the stem/loop junction (Han et al. 2004; Zeng et al. 2005), thereby excising the ∼60-nt precursor miRNA (pre-miRNA) hairpin. Since the flanking 5′ and 3′ arms of the pri-miRNA are degraded following Drosha cleavage, pre-miRNAs are typically found within the exonic regions of noncoding RNAs or in the introns of protein-coding or noncoding transcripts (Fig. 1A).
Figure 1.
Schematic outline of miRNA and siRNA biogenesis. (A) miRNAs are transcribed as long pri-mRNAs and may be located in either noncoding RNAs or, as shown here, in an intron. After excision by Drosha, the pre-miRNA hairpin is exported to the cytoplasm for further processing by Dicer. One strand of the resulting miRNA duplex intermediate is incorporated into RISC, where it acts as a guide RNA to target RISC to either partially or fully complementary mRNAs. This primarily results in translational inhibition or mRNA degradation, respectively. (B) The antiviral RNAi pathway in plants and invertebrates is triggered by long, perfect dsRNAs that are generated by viral replication. These dsRNAs are cleaved by Dicer to generate siRNA duplexes, one strand of which is incorporated into RISC either directly or after being used as a primer by cellular RdRP to generate additional dsRNA substrates. When targeted to a viral mRNA, these viral siRNAs induce transcript cleavage and degradation. Viral dsRNAs may also be actively or passively released by virus-infected cells to induce a systemic antiviral state after capture by other, noninfected cells.
Cleavage by Drosha leaves the pre-miRNA hairpin with a 2-nt 3′ overhang. This is recognized by Exportin 5, which transports the pre-miRNA to the cytoplasm (Yi et al. 2003; Lund et al. 2004). There, the same 2-nt 3′ overhang is recognized by Dicer, another RNase III enzyme, and its cofactor TRBP. Binding of Dicer/TRBP to the base of the pre-miRNA is followed by cleavage to release the terminal loop, yielding an RNA duplex of ∼20 bp flanked by 2-nt 3′ overhangs (Fig. 1A; Chendrimada et al. 2005). The RNA strand that is less tightly base-paired at the 5′ end is loaded into the RNA-induced silencing complex (RISC) and forms the mature miRNA (Khvorova et al. 2003; Schwarz et al. 2003). The miRNA then guides RISC to mRNAs bearing complementary target sites (Hammond et al. 2000).
Although the vast majority of miRNAs are generated as described above, some exceptions exist. For example, some miRNAs are derived from short, excised introns, called mirtrons, which resemble pre-miRNA hairpins, bypassing the need for Drosha, and only require Dicer for maturation (Berezikov et al. 2007; Ruby et al. 2007). A small number of miRNAs are transcribed as shRNAs with a 5′ single-stranded tail (Babiarz et al. 2008). Similar to mirtrons, these “tailed” pre-miRNAs are Drosha-independent, but still require Dicer for final processing.
RISCs, which are minimally composed of a mature miRNA and an Argonaute protein, usually, but not invariably, bind to the 3′ untranslated region (UTR) of targeted transcripts. Functional targets are generally fully complementary to nucleotides 2–7, preferably 2–8, at the 5′ end of the miRNA, referred to as the miRNA seed (Bartel 2009). There are a few examples known, however, where base-pairing between the target transcript and the 3′ half of the miRNA can compensate for mismatches in the seed. Perhaps the best example of this phenomenon is provided by the miRNA lin-4 and a well-characterized target mRNA encoding lin-14, where robust inhibition of lin-14 expression is conferred by target sites lacking full seed homology (Ha et al. 1996).
RISCs bound to partially complementary mRNA targets induce the translational repression of that mRNA by a mechanism that remains to be fully defined (Fig. 1A; for review, see Filipowicz et al. 2008). Translational repression in turn often induces a modest destabilization of the target mRNA (Bagga et al. 2005). RISCs tend to function in a cooperative manner; the greater the number of RISCs bound to a target transcript, the greater the inhibitory effect (Doench et al. 2003; Doench and Sharp 2004).
While RISCs bound to partially complementary mRNA targets primarily induce translational inhibition, mRNAs bearing perfectly complementary miRNA targets are cleaved by RISC and degraded in a process referred to as RNAi (Fig. 1A; Hutvagner and Zamore 2002; Zeng et al. 2003). Unlike translational inhibition, which requires stable binding of RISC to its target mRNA, RNAi permits a single RISC to act enzymatically to irreversibly inhibit the expression of multiple mRNA molecules.
miRNAs are expressed by plants and metazoans (Bartel 2004), where they have been found to play a role in a wide variety of cellular processes including cellular proliferation, regulation of development, apoptosis, homeostasis, and tumor formation (Ambros 2004; He and Hannon 2004; Lai 2005). Humans encode >500 different miRNAs (Griffiths-Jones et al. 2008), which tend to be expressed in a developmental or tissue-specific manner (Landgraf et al. 2007). It has been demonstrated that individual miRNAs are capable of directly down-regulating the expression of hundreds of different mRNAs, and >30% of all mammalian mRNAs are thought to be regulated by miRNAs (Lewis et al. 2005).
RNAi as an innate antiviral mechanism
Infection by all RNA viruses, except retroviruses, leads to the generation of long dsRNAs during the virus life cycle. DNA viruses also produce dsRNAs by convergent transcription of their compact viral genomes. Unlike the short, imperfectly base-paired RNA stem–loop structures typically found in cellular transcripts, viral dsRNAs are generally both long and perfectly complementary. In plants and invertebrates, Dicer cleavage of such dsRNAs generates a pool of ∼22 bp dsRNAs, called siRNA duplexes, that are structurally similar to the miRNA duplex intermediate described above (Fig. 1A,B). As with miRNAs, one strand of the siRNA duplex is loaded into RISC, where it is used to guide RISC to complementary mRNAs. In this context, the relevant targets are viral mRNAs and genomic RNAs. As these are fully complementary to any viral siRNAs, RISC binding results in their cleavage and degradation (Fig. 1B), thus potently inhibiting virus replication. In plants and nematodes, this antiviral response is further amplified through a secondary wave of siRNAs generated by RNA-dependent RNA polymerases (RdRPs), which greatly increases the pool of siRNAs available to RISC (Aoki et al. 2007; Diaz-Pendon et al. 2007). Although insects do not share the ability of plants and nematodes to amplify the antiviral siRNA response using RdRPs, all of these organisms do mount systemic antiviral RNAi responses that are critical for effective antiviral immunity through uptake of viral dsRNAs either secreted or released by virus-infected cells (Feinberg and Hunter 2003; Saleh et al. 2009).
Plant and invertebrate viruses can block antiviral RNAi
The generation of antiviral siRNAs has been observed during viral infection in several plant and invertebrate systems (Hamilton and Baulcombe 1999; Zamore et al. 2000). To counter antiviral RNAi responses, many plant and invertebrate viruses have evolved proteins that act as suppressors of RNA silencing (SRS) (Voinnet et al. 1999; Li and Ding 2005). For example, the tomato bushy stunt virus p19 protein inhibits RNAi through direct binding of ∼21 bp dsRNAs with 2-nt 3′ overhangs, thus preventing the siRNAs from being loaded into RISC (Scholthof 2006). Another plant virus SRS, the cucumber mosaic cucumovirus 2b protein, is able to inhibit RNAi by three distinct mechanisms (Brigneti et al. 1998; Diaz-Pendon et al. 2007; Goto et al. 2007). This protein not only interferes with RISC to inhibit cleavage activity, but also impedes delivery of the systemic siRNA signal and inhibits the production of RdRP-derived viral siRNAs, thereby limiting the amplification and spread of the RNAi response.
Some insect viruses also encode SRS proteins. For example, the flock house virus (FHV) B2 protein inhibits RNAi through direct binding of dsRNAs, regardless of length, thus preventing Dicer processing (Li et al. 2002). B2 also inhibits viral siRNA production through a direct interaction with the viral RdRP during viral RNA synthesis (Aliyari et al. 2008). FHV is a member of the alphanodavirus subfamily of positive-strand RNA insect viruses that spread directly from insect to insect. In addition to FHV B2, SRS proteins have been identified in multiple members of the nodavirus superfamily (Li et al. 2002; Sullivan and Ganem 2005; Fenner et al. 2006), suggesting that inhibition of the antiviral RNAi response is an important requirement for effective propagation of these viruses. This is perhaps due to the fact that high virus titers must be produced within infected insect hosts in order to ensure that sufficient amounts are shed to continue the transmission cycle.
Arboviruses and antiviral RNAi
Although arboviruses also infect insects, there are as yet no reports of any arbovirus-encoded SRS proteins. Arboviruses, or arthropod-borne viruses, use vertebrate reservoirs as an essential component of their life cycle and are transmitted through mosquito, fly, or tick bites (Weaver and Barrett 2004). While arboviruses can be highly pathogenic in their vertebrate hosts, these viruses normally establish a persistent, nonpathogenic infection in their insect vectors. Successful virus propagation requires an adequate pool of infected insects to ensure continued transmission to subsequent vertebrate hosts (Black and Moore 2005). From an evolutionary standpoint, maintaining an infected, but healthy insect vector pool is of paramount importance to these viruses. In particular, increased virus replication in the insect vector is not a desired attribute if such an increase results in vector mortality or even significant pathogenicity. Therefore, it is possible that arboviruses have evolved to lack SRS proteins in order to ensure that their pathogenic potential within their insect vectors is limited. In contrast, and as discussed in more detail below, there is currently no evidence supporting the hypothesis that arboviruses, such as yellow fever virus (YFV), induce a protective RNAi response in mammalian cells (Pfeffer et al. 2005), and the lack of an SRS would therefore not prevent high-level replication and pathogenicity in these hosts.
Recent research on Sindbis virus (SINV) supports this hypothesis. SINV is a member of the alphavirus family of plus-strand RNA viruses infecting both mosquitoes and birds. Infection of mosquitoes with a recombinant SINV carrying a heterologous SRS protein produced higher viral titers and reduced levels of antiviral siRNAs when compared with wild-type virus (Myles et al. 2008). However, this SINV variant also induced a dramatic increase in mosquito mortality. Similar results were also obtained in parallel experiments using O'nyong-nyong virus, a mosquito-borne arbovirus that uses primates as a reservoir (Myles et al. 2008).
Mammalian innate antiviral defenses
In mammalian cells, viral infection is a potent trigger of the interferon (IFN) response (for review, see Sen 2001; Katze et al. 2002; Sadler and Williams 2008). Triggering of the IFN system is initiated by pattern-recognition receptors (PRRs) that recognize the presence of viral molecules. Some PRRs, including MDA5, RIG-I, and TLR3, recognize dsRNAs such as those generated during viral infection. MDA5 and RIG-I are cytoplasmic PRRs that, upon activation, initiate a signal cascade that results in the expression and secretion of IFN. TLR3, which is primarily located in endosomes, can trigger a similar signaling cascade after dsRNA binding, also leading to IFN production. The IFN response is then further amplified and spread to surrounding cells by a positive feedback loop involving the JAK/STAT pathway, thus generating a systemic antiviral state. Binding of IFNs to their cell surface receptors results in the production of numerous IFN-stimulated genes, including OAS/RNaseL and PKR. OAS proteins recognize viral dsRNAs and tag them with 2′,5′-adenosine oligomers. These oligomers then activate RNaseL, which degrades the modified RNAs. The resulting degradation products can also serve as triggers for MDA5 and RIG-I, inducing further expression of IFN in a positive feedback loop.
Upon recognition of viral dsRNA, PKR phosphorylates the translation factor eIF2α, thereby causing a global translational arrest and potentially leading to apoptosis of the infected cell. In this way, mammalian cells use multiple innate defense mechanisms, activated by dsRNAs, to inhibit virus replication in both a cell-autonomous and systemic fashion. Therefore, while both mammalian and invertebrate cells are primed to mount antiviral responses upon encountering long dsRNAs, the systemic components of this response appear to differ in being nucleic acid-based in invertebrates and protein-based in mammals.
As one would predict, mammalian viruses have evolved a diverse array of countermeasures to block the protein-based innate antiviral mechanisms delineated above. For example, the adenovirus E1A protein interferes with the JAK/STAT signaling pathway (Bhattacharya et al. 1996). Other viral proteins directly block PKR activation; e.g., hepatitis C virus (HCV) NS5A and herpes simplex virus type 1 (HSV-1) ICP34.5 (Gale et al. 1997; Leib et al. 2000). PKR can also be inhibited by high concentrations of viral RNA “decoys,” such as the adenoviral VAI and the Epstein-Barr virus (EBV) EBER RNAs (Goodbourn et al. 2000). Moreover, simian virus 40 (SV40) and encephalomyocarditis virus block the OAS/RNaseL pathway (Vilcek and Sen 1996). In this way, IFN response inhibitors evolved by mammalian viruses mirror the diverse SRS proteins encoded by plant and invertebrate viruses.
Do mammalian cells generate an antiviral RNAi response?
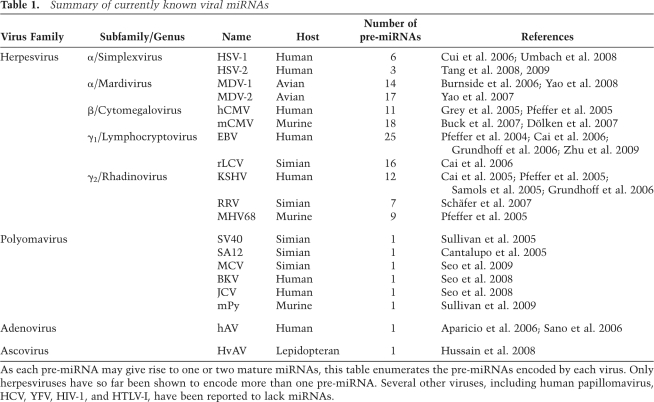
Whether or not viral infection of mammalian cells ever induces an effective antiviral RNAi response remains to be fully determined. In the most comprehensive examination of this issue performed to date, Pfeffer et al. (2005) cloned small RNAs from cell lines infected with a range of viruses including Kaposi's sarcoma-associated herpesvirus (KSHV), mouse γ-herpesvirus 68 (MHV68), human cytomegalovirus (hCMV), human immunodeficiency virus (HIV-1), YFV, and HCV. Although hundreds of virally derived short RNAs were recovered from the three herpesvirus samples, which were then all determined to be novel viral miRNAs (Table 1), no siRNAs of viral origin were recovered from any of the infected cells analyzed. Moreover, HIV-1, YFV, and HCV were found to produce neither viral miRNAs nor siRNAs in infected cells. Other groups, using small RNA cloning as well as deep sequencing, have observed a similar absence of viral siRNAs in cells infected by HIV-1, human T-cell leukemia virus type I (HTLV-I), HCV, hCMV, murine cytomegalovirus (mCMV), EBV, and influenza virus (Cai et al. 2006; Buck et al. 2007; Dölken et al. 2007; Lin and Cullen 2007; Randall et al. 2007; JL Umbach and BR Cullen, unpubl.). While one group has reported the existence of a single HIV-1 siRNA (Bennasser et al. 2005), this small RNA, if genuine, appears to be expressed at very low levels.
Table 1.
Summary of currently known viral miRNAs
As each pre-miRNA may give rise to one or two mature miRNAs, this table enumerates the pre-miRNAs encoded by each virus. Only herpesviruses have so far been shown to encode more than one pre-miRNA. Several other viruses, including human papillomavirus, HCV, YFV, HIV-1, and HTLV-I, have been reported to lack miRNAs.
In addition to the inability to detect viral siRNAs in virally infected mammalian cells reported by several groups, analysis of small RNAs expressed in mammalian somatic cells using deep sequencing has also failed to detect siRNAs of any kind, including the repeat-associated siRNAs (rasiRNAs) commonly seen in plants and invertebrates (Tam et al. 2008). However, endogenous siRNAs have been cloned from mouse oocytes and embryonic stem (ES) cells, demonstrating that mammalian Dicer is capable of generating siRNAs through the progressive cleavage of long dsRNAs (Babiarz et al. 2008; Tam et al. 2008). These dsRNAs were derived from several sources, including bidirectional transcription of a single locus, endogenous hairpin RNAs, and pairing of near-complementary pseudogene RNAs, substrates similar to those used to generate antiviral siRNAs in plants and invertebrates. It should be noted, however, that oocytes and ES cells are unusual in that the IFN pathway is not functional in these cells (Wianny and Zernicka-Goetz 2000; Yang et al. 2001).
Do mammalian viruses encode SRS proteins?
There have also been reports arguing that SRS proteins are encoded by some mammalian viruses, including influenza NS1 (Li et al. 2004), vaccinia virus E3L (Li et al. 2004), Ebola virus VP35 (Haasnoot et al. 2007), HIV-1 Tat (Bennasser et al. 2005), and primate foamy virus (PFV) Tas (Lecellier et al. 2005). It is difficult to determine the precise role that these putative SRS activities play in the viral life cycle, as all of these proteins also have other well-defined and essential functions. Additionally, there is the question of specificity, since it has been demonstrated that prokaryotic proteins containing a dsRNA-binding domain can also exhibit an SRS phenotype when overexpressed (Lichner et al. 2003). The question of specificity and physiological relevance is especially pertinent since the SRS activities of NS1, E3L, and VP35 have all been mapped to their dsRNA-binding domains (Li et al. 2004; Haasnoot et al. 2007). Currently, there are no studies demonstrating that any of these putative mammalian virus SRS proteins affect viral siRNA production, for the simple reason that no viral siRNAs have yet been identified in infected mammalian cells.
Although not a protein, the adenoviral VA1 RNA has been reported to have SRS properties in addition to its well-established role in PKR inhibition (Lu and Cullen 2004; Andersson et al. 2005). VA1 is a small, highly structured RNA with a short 3′ overhang that is recognized and bound by Exportin 5. Since VA1 is produced at extremely high levels during adenoviral infection (up to 108 copies per cell), VA1 is able to saturate the Exportin 5 pathway, thereby inhibiting the nuclear export of pre-miRNAs. Additionally, due to the fact that its secondary structure resembles that of pre-miRNAs, VA1 is also able to bind Dicer. Yet, VA1 is a poor substrate for Dicer cleavage and, in fact, functions as a competitive inhibitor of Dicer function (Andersson et al. 2005). Nevertheless, a small fraction (∼1%) of VA1 is cleaved by Dicer to give rise to a reported adenovirus miRNA (Table 1; Aparicio et al. 2006; Sano et al. 2006). It is currently unclear whether inhibition of the miRNA/siRNA pathway by VA1 is an important function of this molecule, or if it is simply an incidental consequence of its secondary structure and the extraordinary abundance of the molecule in adenovirus-infected cells.
In conclusion, although it appears that the cellular machinery required to generate an antiviral siRNA response in plants and invertebrates is largely conserved in mammals, it does not seem that this mechanism is used, at least in somatic cells. Additionally, mammalian cells lack the RdRPs used by plants and nematodes to amplify the siRNA population and have not been shown to be able to mount a systemic antiviral RNAi response. Together with the lack of convincing evidence for a mammalian virus SRS, these results collectively suggest it is unlikely that mammals use RNAi as an antiviral innate immune response. Rather, it appears that the RNAi response has been replaced by the perhaps more versatile IFN system. In this way, mammalian cells also avoid any potential nonspecific, off-target effects pools of viral siRNAs might exert on the expression of partially complementary cellular mRNAs.
Viral miRNAs
miRNAs have several features that may make them particularly useful to viruses. First, and most obviously, they can be used to target specific genes for down-regulation in order to establish a favorable environment for virus replication. Second, the evolution of a miRNA complementary to a new target gene can presumably be accomplished much more easily than the evolution of a novel regulatory protein. Third, miRNAs are small; the minimal size requirement for a functional pri-miRNA is <200 nt, ideal for the tight space constraints characteristic of viral genomes. Finally, unlike proteins, miRNAs are not antigenic.
Just as viruses have co-opted many other cellular functions for their own benefit, a growing body of research has demonstrated that many viruses encode miRNAs. All herpesviruses examined to date have been found to encode miRNAs, as have human adenovirus, Heliothis virescens ascovirus (HvAc), and several members of the polyomavirus family, (Table 1; Cantalupo et al. 2005; Sullivan et al. 2005, 2009; Aparicio et al. 2006; Hussain et al. 2008; Seo et al. 2008, 2009). That these are all nuclear DNA viruses likely reflects the fact that Drosha and DGCR8, required for the initial pre-miRNA excision event, localize to the nucleus—viruses that replicate exclusively in the cytoplasm would not have ready access to this machinery. In addition, miRNA processing can result in the destruction of the entire pri-miRNA transcript, except for the mature miRNA itself, so it is unlikely that viruses with RNA genomes would encode a miRNA, unless it conferred a very significant survival advantage. Nevertheless, some groups have proposed that HIV-1 encodes a viral miRNA (Omoto et al. 2004; Klase et al. 2007; Ouellet et al. 2008), although this has been disputed by others (Pfeffer et al. 2005; Lin and Cullen 2007).
Another consideration is the fact that miRNAs act on RNA transcripts. Even with complete translational inhibition, the existing pool of proteins would likely have to turn over before a phenotype could be observed. Depending on the stability of the protein in question, this delay may limit the potential benefits of miRNAs for viruses with short life cycles. (While it is possible that a viral miRNA could target a novel cellular transcript induced by infection, such a scenario has yet to be reported.) Although exceptions may exist, these considerations suggest that the viruses most likely to encode miRNAs would be nuclear DNA viruses capable of persistent or latent infection, as has indeed been observed.
Viral mRNA targets of viral miRNAs
Thus far it has been easier to identify viral targets of viral miRNAs rather than cellular targets. This is partly due to the fact that a viral mRNA transcribed antisense to a viral miRNA makes an obvious target. In addition, viral genomes are significantly smaller than host genomes, so candidate mRNA targets are fewer and easier to test. Interestingly, most of the viral mRNA targets of viral miRNAs identified thus far are either targets of the host immune response and/or viral regulatory proteins. For example, SV40 miR-S1 down-modulates production of the viral T antigen (TAg) (Sullivan et al. 2005). During infection, SV40 TAg is expressed as an early protein, where it regulates viral transcription and DNA replication (Sullivan and Pipas 2002). During late viral replication, TAg is no longer required, but its continued presence can elicit a cytotoxic T-cell (CTL) response against SV40-infected cells. miR-S1, which is located antisense to the TAg mRNA and is expressed as a viral late gene product, cleaves and degrades TAg mRNA. Although miR-S1 does not affect virus replication per se, down-regulation of TAg expression late in the viral life cycle was proposed to reduce the susceptibility of SV40-infected cells to CTLs.
The genomic location of SV40 miR-S1 is conserved in other members of the polyomavirus family, including the human viruses JCV and BKV, mouse polyomavirus (mPy), and the primate SA12 and Merkel cell virus (MCV). The function of these miRNAs also appears to be conserved as several also down-regulate TAg expression in their cognate viruses (Cantalupo et al. 2005; Seo et al. 2008, 2009; Sullivan et al. 2009). Experimental infection of mice with a mutant form of mouse polyomavirus that does not express this miRNA did not, however, result in an appreciable decrease in replication when compared with wild-type virus; infection was established with equal efficiency by both viruses, and both were cleared from the host with approximately the same kinetics (Sullivan et al. 2009). Indeed, no difference in T-cell-mediated immune responses could be detected. Therefore, it was hypothesized that this miRNA may function in a way that is not fully recapitulated by this experimental system; e.g., during virus transmission.
The first viral target of a viral miRNA to be described was the mRNA encoding the EBV DNA polymerase, BALF5, which is down-regulated by EBV miR-BART2 (Barth et al. 2008). miR-BART2 is located antisense to BALF5, and thus acts as an siRNA cleaving the target transcript in the center of its perfect binding site. (Interestingly, the BALF5 cleavage product was first mapped in 1993, well before the discovery of the EBV miRNAs [Furnari et al. 1993].) In a possible case of convergent evolution, it was reported recently that the insect virus HvAc also encodes a miRNA that down-regulates the viral DNA polymerase (Hussain et al. 2008). Unlike EBV miR-BART2, this HvAc miRNA is not encoded antisense to the polymerase transcript and binds the target imperfectly, although it does appear to destabilize the transcript. How down-regulation of the viral DNA polymerase gene facilitates viral replication is currently unclear, although in the case of EBV it could potentially stabilize viral latency.
Two reports have demonstrated that the hCMV miRNA miR-UL112-1 is able to target the viral gene IE1 (Grey et al. 2005; Murphy et al. 2008). IE1, also known as IE72, is an immediate-early gene product required for activating transcription of hCMV early genes. Although IE1 is not essential, lack of this protein is detrimental at low multiplicities of infection. During hCMV infection, IE1 is initially expressed at high levels. As replication progresses, IE1 levels decline as levels of miR-UL112-1, which is expressed late in infection, increase. When miR-UL112-1 was prematurely expressed early in hCMV infection, a decrease in viral replication was observed (Grey et al. 2007). Although it is currently unknown whether miR-UL112-1 is also expressed during hCMV latency, it has been suggested that miR-UL112-1 could play a role in either the establishment or maintenance of latency (Murphy et al. 2008). Conversely, miR-UL112-1 may play a role similar to that proposed for SV40 miR-S1 (Sullivan et al. 2005) by removing a viral early transcription factor late in the replication cycle to prevent cytotoxicity or CTL killing of infected cells.
Herpes Simplex Virus Type 1 (HSV-1) establishes long-term latent infections in neurons of the trigeminal ganglia in vivo, from where it occasionally reactivates to cause a localized productive infection. HSV-1 was shown recently to express two miRNAs in latently infected neurons that are capable of down-regulating ICP0 and ICP4, immediate early proteins important for initiating productive replication (Umbach et al. 2008). One of these miRNAs, miR-H2, is transcribed antisense to ICP0, while a second HSV-1 miRNA, miR-H6, targets ICP4 mRNA through binding to an extended miRNA seed region. Two other HSV-1 miRNAs, miR-H3 and miR-H4, are both transcribed antisense to a viral mRNA encoding ICP34.5, a neurovirulence factor. Although down-regulation of ICP34.5 by these HSV-1 miRNAs has yet to be directly demonstrated, previous mutational studies strongly indicate that this is the case.
These results are congruent with recent reports demonstrating that HSV-2, a close relative of HSV-1, expresses several functionally homologous miRNAs in latently infected neurons (Tang et al. 2008, 2009). HSV-2 miR-H3 and miR-H4 are similar to HSV-1 miR-H3 and miR-H4 in their genomic location and sequence (Fig. 2B) and have been shown to inhibit ICP34.5 expression. The conserved down-regulation of ICP34.5 by miRNAs encoded by both HSV-1 and HSV-2 may protect neurons from the known neurovirulent effects of ICP34.5 during virus reactivation (Bolovan et al. 1994). Moreover, HSV-2 miR-H2 is also homologous to HSV-1 miR-H2 in both its genomic location and its ability to down-regulate ICP0, which may aid in the establishment and/or maintenance of the latent state. This would be similar to the proposed role for miR-UL112-1 in hCMV latency noted above; ICP0 is similar to IE1 in that both are immediate–early transcription factors that induce viral early gene expression. Additionally, the fact that HSV-1 miR-H6, which is again primarily expressed during latency, down-regulates ICP4, a second immediate–early protein required for productive replication, suggests that these viral miRNAs indeed act to stabilize HSV-1 and HSV-2 latency in vivo (Umbach et al. 2008; Tang et al. 2009).
Figure 2.
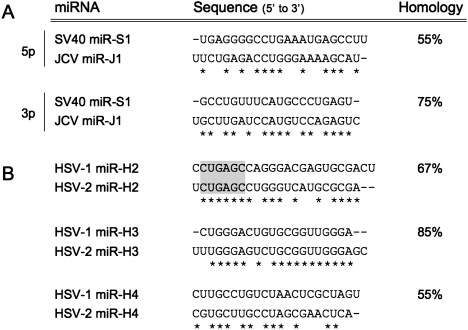
Several viral miRNAs show sequence similarity but lack a conserved miRNA seed region. Alignment of miRNAs encoded by the related polyomaviruses SV40 and JCV (A) or by the related herpesviruses HSV-1 and HSV-2 (B). Only in the case of HSV-1 miR-H2 and HSV-2 miR-H2 is conservation of the miRNA seed region observed (in gray), although there is considerable conservation outside of the seed in the other miRNAs listed. See the text for detailed discussion.
Overall, these reports demonstrate that viral miRNA regulation of viral gene expression is a common strategy used by several DNA viruses. Using viral miRNAs to regulate the expression of viral proteins may facilitate viral replication by inhibiting the ability of the host antiviral immune response to identify and eliminate infected cells and may also play a key role in the temporal regulation of the virus life cycle.
Conservation of viral miRNAs and their target sequences
While we have so far exclusively described viral mRNA targets of viral mRNAs, it is obviously possible for a single viral miRNA to inhibit both viral and cellular mRNAs. Indeed, at least one example of such a “dual function” miRNA has been reported; i.e., hCMV miR-UL112-1, which inhibits both viral IE1 expression and cellular MICB expression (see below for further discussion). As cellular mRNA targets, unlike viral mRNA targets, are not likely to coevolve with a viral miRNA, targeting a cellular mRNA should lead to miRNA sequence conservation, at least in the critical seed region. Conversely, viral miRNAs that target viral mRNAs might not be evolutionarily conserved, especially if the viral mRNA target lies directly antisense to the miRNA. It is therefore interesting that, although the genomic location and viral mRNA target of polyomavirus miRNAs are conserved among members of this family, the sequences of the miRNA seed regions are not (Fig. 2A). This suggests that the sole target of these miRNAs is the antisense TAg mRNA, as they seem unlikely to target the same cellular transcripts (Seo et al. 2008). This also may be true for HSV-1 and HSV-2 miR-H3 and miR-H4 (Fig. 2B). Although both the genomic locations and down-regulation of the antisense ICP34.5 mRNA are conserved, there is only partial sequence identity between the miRNA seed regions, although there is significant sequence similarity overall (Fig. 2B). Again, it seems unlikely that these viral miRNAs target the same cellular transcripts. Therefore, it is possible that down-regulation of a single viral gene product—i.e., ICP34.5—may be the sole physiological function of these two viral miRNAs. On the other hand, the seed regions of HSV-1 and HSV-2 miR-H2 do seem to be conserved (Fig. 2B). Therefore, it is possible that these two viral miRNAs down-regulate not only the antisense viral ICP0 mRNA, but also a similar set of cellular transcripts.
While the polyomavirus and herpes virus miRNAs described above share similar or identical genomic locations and function and at least partial sequence similarity, this is not invariably the case in other viruses. For example, comparison of the related herpesviruses MDV-1 and MDV-2, hCMV and mCMV, or KSHV and RRV reveals conservation of miRNA location in some instances, but no evidence for conservation of miRNA sequence. This may imply that these viruses have evolved to target distinct cellular mRNA targets and/or that these viral miRNAs are rapidly coevolving with a presumably viral mRNA target. The single current exception to this lack of miRNA sequence conservation is provided by EBV and its rhesus equivalent, rhesus lymphocryptovirus (rLCV), where eight of the viral miRNAs encoded by rLCV are highly similar in sequence to EBV miRNAs encoded at a similar genomic location (Cai et al. 2006). This implies the existence of shared cellular mRNA targets that are down-regulated by miRNAs encoded by both these primate γ-herpesviruses.
Although a lack of viral miRNA sequence conservation between related viruses would seem to imply the evolution of distinct functions, it is important to note that this is not necessarily the case; i.e., apparently “nonconserved” viral miRNAs could target different regions of the same target mRNA or even two different gene products in the same cellular pathway. This is certainly a possibility given the apparent evolutionary flexibility of viral miRNAs. However, direct evidence for this scenario has not been reported thus far, although this may reflect the scarcity of known viral miRNA targets.
Cellular targets of viral miRNAs
In addition to “autoregulation” of viral targets, viral miRNAs targeting cellular mRNAs have also been described. Most cellular mRNA targets identified thus far play a role in either regulation of apoptosis or modulation of the host antiviral immune response.
Examples of apoptosis regulators include PUMA, a proapoptotic factor, which was identified recently as a target of EBV miR-BART5 (Choy et al. 2008). PUMA plays a role in apoptosis as a direct downstream target of p53 in addition to being able to initiate apoptosis via a p53-independent mechanism (Han et al. 2001). Down-regulation of PUMA by miR-BART5 may protect EBV-infected cells from virus-induced apoptosis.
BCLAF1, another apoptotic factor, was identified recently as a cellular target of the KSHV miRNAs miR-K5, miR-K9, and miR-K10 (Ziegelbauer et al. 2009). BCLAF1 can apparently function as a pro- or anti-apoptotic factor, depending on its cellular context. Regardless, inhibition of miR-K5, miR-K9, or miR-K10 function in latently KSHV-infected cells resulted in elevated levels of BCLAF1 and decreased virus production after induction of productive viral replication, suggesting that BCLAF1 stabilizes KSHV latency.
Another study reported that THBS1 is down-regulated by multiple KSHV miRNAs, including miR-K1, miR-K3-3p, miR-K6-3p, and miR-K11 (Samols et al. 2007). THBS1 is a multifunctional protein that regulates a variety of processes including cell adhesion, migration, and angiogenesis (Lawler 2002). It has also been reported to function as a chemoattractant, involved in the recruitment of monocytes and T cells to sites of infection (Narizhneva et al. 2005). Down-regulation of THBS1 by KSHV miRNAs may aid KSHV-infected cells in avoiding detection by the host immune system.
Another example of a virally regulated immunomodulator is provided by the chemokine CXCL-11, which was identified as a target of EBV miR-BHRF1-3 (Xia et al. 2008). As CXCL-11 is a potent T-cell chemoattractant (Cole et al. 1998), down-regulation might allow infected cells to avoid T-cell detection and killing.
The hCMV miRNA miR-UL112-1 was reported to inhibit expression of MICB through binding sites within the 3′UTR of this mRNA (Stern-Ginossar et al. 2007). MICB is a stress-induced cell surface ligand recognized by the NKG2D receptor found primarily on natural killer (NK) cells. Exposure of cells to a stressor such as virus infection induces expression of MICB, which is followed by binding to the NKG2D receptor, resulting in NK cell killing of the affected cell. Thus, down-regulation of MICB by miR-UL112-1 may allow hCMV to escape NK cell immune surveillance.
In conclusion, most cellular targets of viral miRNAs identified so far fall into two related categories; i.e., apoptosis regulators and immunomodulators. By interfering with apoptosis or evading host immune responses, viruses are able to prolong the life of the infected cell and maximize their own replication potential.
Antiviral cellular miRNAs?
It has been suggested that, in lieu of an antiviral RNAi response, mammalian cells might inhibit virus infection by targeting viral transcripts with cellular miRNAs (Lecellier et al. 2005; Otsuka et al. 2007; Pedersen et al. 2007). However, several considerations suggest that it is unlikely that cellular miRNAs have evolved specifically as an antiviral defense mechanism. First, many miRNAs are conserved across a wide variety of vertebrate species, all the way from birds to humans. It is therefore unlikely that a given miRNA could have specifically evolved to counter a particular virus, since viral host ranges are typically much more limited. Second, viruses have much shorter life cycles than their hosts, and their polymerases often have a high mutation rate. As a consequence, viruses tend to evolve much more quickly than their host counterparts. This would make it very difficult for a cellular miRNA to inhibit a virus effectively, given that a single nucleotide mutation in the seed region of the viral mRNA target has the potential to block down-regulation. This is particularly true for RNA viruses such as HIV-1 and HCV, which exist as highly diverse “pseudo-species” rather than as homogeneous populations. Third, many viruses appear to have evolved relatively recently, long after their current host species. Therefore, cellular miRNAs encoded by that host could not have evolved to specifically inhibit these viruses.
Nevertheless, it has been reported that mice exhibiting a reduced level of Dicer activity become hypersusceptible to infection by vesicular stomatitis virus (VSV) due to reduced levels of cellular miR-24 and miR-93, which down-regulate VSV protein expression (Otsuka et al. 2007). This suggests that cellular miRNAs can indeed have an antiviral effect. It should be noted however, that these experiments were done with a laboratory strain of VSV, and that strains more representative of VSV field isolates do not share the reported binding sites for miR-24 or miR-93 (Muller and Imler 2007). In addition, VSV is an arbovirus that is normally transmitted from black flies to livestock (Mead et al. 2000). Although mice can be experimentally infected with VSV, they represent nonphysiological, dead-end hosts, and are not capable of transmitting VSV to uninfected flies. Therefore, these observations may be accurate for the system used, but the importance of the conclusions drawn must be tempered with knowledge that this experimental system may not be that physiologically relevant (Mahajan et al. 2009).
Another report demonstrated that several cellular miRNAs, including miR-28, miR-125b, miR-150, miR-223, and miR-382, are capable of inhibiting HIV-1 replication via binding sites located within the viral genome (Huang et al. 2007). As these miRNAs are primarily expressed in resting CD4+ T cells, as opposed to the activated CD4+ T cells that support HIV-1 replication in vivo, it was proposed that HIV-1 has evolved to utilize these cellular miRNAs to facilitate entry into viral latency. Latently HIV-1-infected T cells are clinically important, as they provide the only long-lived viral reservoir in individuals undergoing treatment with highly active anti-retroviral therapy (HAART). However, current evidence indicates that latent HIV-1 is transcriptionally inert, so there would not actually be any viral mRNAs available for these miRNAs to target (Han et al. 2007). Moreover, the pool of latently infected T cells comprises <0.1% of all infected cells within the host; the primary targets for infection are activated T cells and macrophages, and these infections persist at a high level for the life of the host through the continuous infection of naive cells. Therefore, latently infected T cells do not make a significant contribution to overall virus replication kinetics within the host in the absence of HAART; i.e., they confer no advantage to the virus under normal physiological conditions. Instead, these latently infected T cells likely arise due to the propensity of a small percentage of HIV-1-infected, activated T cells to cycle into a resting memory state; latent infection could result if HIV-1 infection occurs immediately prior to this step (Han et al. 2007).
It was also reported that cellular miR-32 serves as an antiviral defense against PFV, another retrovirus, in infected Hela cells (Lecellier et al. 2005). It seems unlikely that this miRNA has specifically evolved to counter PFV infection, given that this miRNA is conserved from chickens to human, while PFV has a narrow host range limited to chimpanzees (Meiering and Linial 2001). Additionally, the miR-32 target site is not well conserved among the primate foamy virus species, and it remains to be determined whether miR-32 is even expressed in the salivary glands, where PFV normally establishes infection.
Although convincing evidence demonstrating that individual cellular miRNAs are capable of acting as an antiviral defense mechanism in a physiologically relevant setting has yet to be presented, these reports do collectively indicate that cellular miRNAs can inhibit viral replication when target sites are presented by the infecting virus. In fact, in vitro studies have demonstrated that RNAi can, at least temporarily, suppress members of virtually every class of virus after the introduction of exogenous siRNAs, or shRNA expression vectors, that target viral transcripts (Dykxhoorn and Lieberman 2006; Leonard and Schaffer 2006). However, when these siRNAs were introduced individually, viruses were able to rapidly evolve escape mutants that avoided further inhibition (Boden et al. 2003; Das et al. 2004; Gitlin et al. 2005; Wilson and Richardson 2005). The simultaneous inclusion of multiple, distinct siRNAs, however, made viral escape more difficult to achieve. Although the ability of viruses to escape from inhibition by RNAi is clearly a serious problem, antiviral siRNAs are nevertheless under development or in clinical trials for several important human pathogens, including HCV, HIV-1, and Respiratory Syncytial Virus (Castanotto and Rossi 2009).
Influence of cellular miRNAs on viral tropism
Viral tropism is a complex issue influenced by many factors. At the molecular level such factors include, but are not limited to the availability of cell surface receptors, the presence or absence of required cellular cofactors, or innate antiviral resistance factors, and the state of the infected cell; e.g., whether the cell is quiescent or actively replicating. In addition to these considerations, cellular miRNA profiles are likely to play a role in determining the tissue tropism of a given virus; i.e., the expression of certain miRNAs may limit viral replication in certain cell types through targeting and inhibition of viral transcripts. Conversely, the selective expression of miRNAs that have the potential to boost viral replication would tend to favor virus replication in that cell type.
Compelling evidence demonstrating that cellular miRNAs can influence viral tropism is provided by HCV, which contains two adjacent binding sites for cellular miR-122 within its 5′UTR (Jopling 2008). Unlike canonical 3′UTR miRNA-binding sites, which inhibit translation of target transcripts, binding of miR-122 to the viral 5′UTR facilitates HCV replication (Jopling et al. 2005). The exact mechanism underlying this phenomenon has yet to be fully elucidated, as conflicting reports have suggested that miR-122 may act by increasing the level of HCV RNA replication in infected cells, and/or by increasing the translation of the HCV polyprotein (Henke et al. 2008; Jopling 2008). Regardless, the fact that miR-122 is exclusively expressed in the liver (Jopling et al. 2005), the primary replication site of HCV, suggests that this cellular miRNA plays a key role in determining the tropism of HCV for this tissue.
It has been proposed that cellular mRNAs show “selective avoidance” of specific cellular miRNAs; i.e., transcripts present in a particular cell type have evolved to lack binding sites for miRNAs expressed in the same cell (Farh et al. 2005). For example, as cells differentiate and the expression of specific miRNAs is induced, concurrently induced mRNAs tend to lack binding sites for these miRNAs, presumably to avoid inappropriate down-regulation. In addition, it has been found that metazoan housekeeping genes typically have relatively short 3′UTRs compared with orthologous genes in plants, where 3′UTR targeting by miRNAs is less commonly observed (Stark et al. 2005). These short metazoan 3′UTRs presumably limit the number of potential miRNA-binding sites.
It is likely that viruses have also evolved to “selectively avoid” binding sites for inhibitory miRNAs that are expressed in their normal target tissue (e.g., activated CD4+ T cells for HIV-1) but may still retain target sites for inhibitory miRNAs present in cells that they do not infect under physiological conditions (e.g., HeLa cells for PFV). Experiments that force a virus to infect a nonphysiological host or target cell may therefore uncover inhibition of viral replication by specific cellular miRNAs, but these are, in essence, artifacts. Interestingly, it has been demonstrated that viruses can be engineered to be incapable of replicating in a given tissue by insertion into their genome of artificial target sites for cellular miRNAs selectively expressed in that tissue (Barnes et al. 2008; Kelly et al. 2008; Ylösmäki et al. 2008). This observation has clear clinical potential, as it not only provides a novel way of attenuating viruses for the purpose of developing novel vaccines (Barnes et al. 2008), but also provides a way of tailoring the tissue tropism of viral vectors so as to avoid infection of inappropriate tissues during gene therapy (Kelly et al. 2008; Ylösmäki et al. 2008). However, the observation of viral revertants that had either partially or completely lost the inserted miRNA target sites in two of these studies (Barnes et al. 2008; Kelly et al. 2008) again demonstrates that it is difficult to use miRNAs or siRNAs to stably restrict the replication of these genetically plastic pathogens in an otherwise permissive cellular environment. Conversely, positive effects of a miRNA on virus replication, as seen with miR-122 and HCV, are likely subject to positive selection and hence may become fixed in a given viral population.
Viral induction of cellular miRNAs
Several publications have reported changes in the cellular miRNA expression profile following viral infection (Triboulet et al. 2007; Cameron et al. 2008; Wang et al. 2008). On the one hand, these changes could represent one aspect of the host cell innate immune response triggered by viral infection and might therefore act to inhibit virus replication (Pedersen et al. 2007). On the other hand, changes in cellular miRNA expression may be specifically induced by a given virus in order to create a more favorable intracellular environment for viral replication. Viral infection of a cell is a major disturbance, with almost all cellular processes subverted at some level and/or hijacked to produce foreign molecules that may trigger systemic warnings and responses. It is not surprising, therefore, that the expression of many cellular gene products, including miRNAs, is affected. Whether or not any of these changes are promoted by the virus to enhance virus replication, or instead act to inhibit virus replication, is currently unclear.
At present, the most compelling evidence in favor of functionally relevant changes in cellular miRNA expression after virus infection involves the EBV-induced up-regulation of cellular miR-155 (Yin et al. 2008). MiR-155 is classified as an oncomir, or oncogenic miRNA, since increased expression of miR-155 is linked to the development of B-cell lymphomas and miR-155 is elevated in several types of cancer (Kluiver et al. 2005). Intriguingly, KSHV, a related oncogenic γ-herpesvirus, encodes a viral miRNA, miR-K11, that not only has the same seed as miR-155, but also targets many of the same cellular mRNAs for down-regulation (Gottwein et al. 2007; Skalsky et al. 2007). Moreover, the oncogenic chicken α-herpesvirus virus Marek's disease virus type 1 (MDV-1) also encodes a miR-155 ortholog, while MDV-2, a related nononcogenic virus, does not (Zhao et al. 2009). Together, these data suggest that miR-155 and its viral homologs play a significant role in some aspect of the life cycle of these diverse herpesviruses, presumably by down-regulating cellular mRNAs that remain to be identified. An unintentional consequence of the virus-induced expression of miR-155, or its viral orthologs, is that EBV-, KSHV-, or MDV-1-infected cells may be more likely to undergo oncogenic transformation.
Concluding remarks
The discovery of the miRNA and siRNA pathways has begun to revolutionize our understanding of virus–host cell interactions. Although our knowledge of the role of miRNAs and siRNAs in virally infected cells is still far from complete, it is already evident from the degree that both viruses and host cells utilize these systems that they are likely to play an important role in both host innate antiviral defense and virus replication. Continued study of these interactions in plants, invertebrates, and mammals, and careful comparisons of how these systems are both alike and different will continue to further our understanding of the interplay of viruses with the host antiviral innate immune response.
Acknowledgments
Our research was supported by the NIH (R01-GM071408, R01-AI067968, and T32-CA009111).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1793309.
References
- Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston DM. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- Black WC, Moore CG. Population biology as a tool to study vector-borne diseases. In: Marquardt WC, editor. Biology of disease vectors. Elsevier Acadmeic; Burlington, MA: 2005. pp. 187–206. [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolovan CA, Sawtell NM, Thompson RL. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J Virol. 1994;68:48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–266. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP, Lin W, Boyd JG, Moser B, Wood DE, et al. Interferon-inducible T cell α chemoattractant (I-TAC): A novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Lieberman J. Silencing viral infection. PLoS Med. 2006;3:e242. doi: 10.1371/journal.pmed.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- Fenner BJ, Thiagarajan R, Chua HK, Kwang J. Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J Virol. 2006;80:85–94. doi: 10.1128/JVI.80.1.85-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Adams MD, Pagano JS. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MJ, Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Stone JK, Andino R. Poliovirus escape from RNA interference: Short interfering RNA-target recognition and implications for therapeutic approaches. J Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S, Didcock L, Randall RE. Interferons: Cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81:2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48:1050–1060. doi: 10.1093/pcp/pcm074. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ, Zhu L, Chittenden T. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82:9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jopling CL. Regulation of hepatitis C virus by microRNA-122. Biochem Soc Trans. 2008;36:1220–1223. doi: 10.1042/BST0361220. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Katze MG, He Y, Gale M., Jr Viruses and interferon: A fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. miRNAs: Whys and wherefores of miRNA-mediated regulation. Curr Biol. 2005;15:R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J. Thrombospondin-1 as an endogenous inhibitor of angiogenesis and tumor growth. J Cell Mol Med. 2002;6:1–12. doi: 10.1111/j.1582-4934.2002.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JN, Schaffer DV. Antiviral RNAi therapy: Emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li HW, Ding SW. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichner Z, Silhavy D, Burgyan J. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J Gen Virol. 2003;84:975–980. doi: 10.1099/vir.0.18987-0. [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Mahajan VS, Drake A, Chen J. Virus-specific host miRNAs: Antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DG, Ramberg FB, Besselsen DG, Mare CJ. Transmission of vesicular stomatitis virus from infected to noninfected black flies co-feeding on nonviremic deer mice. Science. 2000;287:485–487. doi: 10.1126/science.287.5452.485. [DOI] [PubMed] [Google Scholar]
- Meiering CD, Linial ML. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol Rev. 2001;14:165–176. doi: 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Imler JL. Dicing with viruses: microRNAs as antiviral factors. Immunity. 2007;27:1–3. doi: 10.1016/j.immuni.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc Natl Acad Sci. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narizhneva NV, Razorenova OV, Podrez EA, Chen J, Chandrasekharan UM, DiCorleto PE, Plow EF, Topol EJ, Byzova TV. Thrombospondin-1 up-regulates expression of cell adhesion molecules and promotes monocyte binding to endothelium. FASEB J. 2005;19:1158–1160. doi: 10.1096/fj.04-3310fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–350. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553–1564. doi: 10.1016/j.febslet.2006.01.085. [DOI] [PubMed] [Google Scholar]
- Schäfer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNAs encoded by the primate γ-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364:21–27. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof HB. The Tombusvirus-encoded P19: From irrelevance to elegance. Nat Rev Microbiol. 2006;4:405–411. doi: 10.1038/nrmicro1395. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Pipas JM. T antigens of simian virus 40: Molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev. 2002;66:179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J Virol. 2005;79:7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009 doi: 10.1016/j.virol.2009.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek J, Sen GC. Interferons and other cytokines. In: Knipe D, et al., editors. Fields virology. Lippincott-Raven; Philadelphia, PA: 1996. pp. 375–399. [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J Virol. 2007;81:7164–7170. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: Predominance of virus-encoded microRNAs. J Virol. 2008;82:4007–4015. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylösmäki E, Hakkarainen T, Hemminki A, Visakorpi T, Andino R, Saksela K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, Kgosana L, Wang X, Nair V. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]