Abstract
Inhibitors of heat-induced heat shock protein 70 (HSP70)a expression have the potential to enhance the therapeutic effectiveness of heat induced radiosensitization of tumors. Among known small molecule inhibitors, quercetin has the advantage of being easily modified for structure-activity studies. Herein, we report the ability of five mono-methyl and five carbomethoxymethyl derivatives of quercetin to inhibit heat-induced HSP70 expression and enhance HSP27 phosphorylation in human cells. While quercetin and several derivatives inhibit HSP70 induction and enhance HSP27 phosphorylation at Ser78, other analogs selectively inhibit HSP70 induction without enhancing HSP27 phosphorylation that would otherwise aid in cell survival. We also show that good inhibitors of HSP70 induction are also good inhibitors of both CK2 and CamKII, kinases that are known to activate HSP70 expression by phosphorylation of heat shock transcription factor 1. Derivatives that show poor inhibition of either or both kinases are not good inhibitors of HSP70 induction, suggesting that quercetin’s effectiveness is due to its ability to inhibit both kinases.
Keywords: heat shock response, heat shock proteins, heat shock element, quercetin, HSP70, HSP27, inhibitors, kinases, heat induced radiosensitizers
Introduction
Heat shock proteins (HSPs) are stress proteins that are found in nearly all living organisms and are highly induced by various environmental and pathophysiological stimuli.1-5 The heat shock protein family consists of several different molecular weight classes, of which HSP60s, HSP70s and HSP90s function as highly conserved molecular chaperones6 that assist nascent polypeptide folding, assembly of protein complexes and protein transport through cellular membranes.7 While HSP90 is known to stabilize multiple signaling proteins,8 HSP70s are reported to promote cell survival by inhibiting lysosomal membrane permeabilization9 and interfering with the cell apoptotic cascade.10 Heat shock proteins are also found to be overexpressed in cancer cells and participate in oncogenesis and resistance to chemotherapeutic agents.11-14 Over-expression of HSP90s and HSP70s in many tumors15 correlates with the proliferation and survival of oral,16 gastrointestinal,17 colorectal,18 histiocytic lymphoma,19 and breast cancers.20, 21 Thus heat shock proteins have recently become of interest as targets for anticancer therapy.22 Small molecular inhibitors of HSP90 like 17-allylamino-17-demethoxy-geldanamycin (17AAG)23, 24 and 17-(dimethylaminoethylamino)-17-demethoxygeldanamycin (17DMAG)25 have been found to enhance tumor killing by ionizing radiation. Recently it has been shown that inhibition of HSP70 in tumor cells by either a gene knockout26 or by antisense agents27 enhances heat-induced radiosensitization (HIR). It would therefore be highly desirable to have a small molecular inhibitor of heat induced HSP70 that could be administered prior to HIR to further enhance killing.
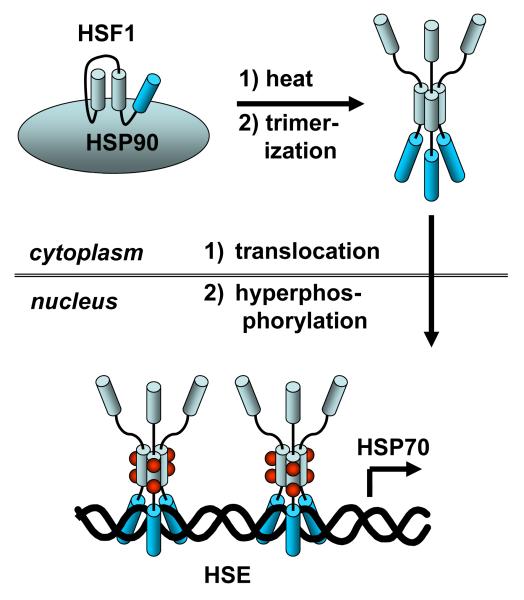
Heat induction and regulation of HSP70 is a rather complicated process, involving initial release of the heat shock transcription factor 1 (HSF1) from HSP90, followed by trimerization, translocation into the nucleus, and then binding to the heat shock element (HSE) (Figure 1).28-30 For transcription to occur the trimer must first be activated by phosphorylation at serine 230 with calcium/calmodulin kinase II (CamKII)31 and/or threonine 142 by casein kinase 2 (CK2).32 In contrast, phosphorylation at other sites down regulates the transcriptional activity of HSF1. For example, phosphorylation at serine 121 by MAPK-activated kinase 2 (MK2) causes unfolding of HSF1 and rebinding to HSP90.33 Likewise, phosphorylation of serine 307 by ERK or serine 363 by JNK inactivates HSF1.34
Figure 1.
Schematic of the pathway for heat shock induction of HSP70 expression. See text for a discussion.
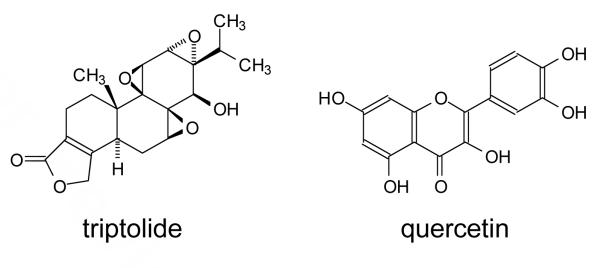
A number of small molecule inhibitors of heat-induced HSP70 expression have been reported, the most effective of which are triptolide and quercetin (Figure 2).35 Triptolide is a highly functionalized molecule that has been found to prevent heat shock HSF1 from activating transcription in the steps following trimerization, phosphorylation and DNA binding.36 Quercetin, on the other hand, is a structurally much simpler natural product of the flavone family, that has been reported to have a wide variety of biological activities,37, 38 one of which is inhibition of heat-induced HSP70 expression by as yet unknown mechanisms.39, 40 Quercetin inhibits a number of kinases,41 including CK2 which has been shown to enhance HSP70 induction through phosphorylation of HSF1 threonine 142.32 It is therefore possible that quercetin inhibits HSP70 induction by blocking phosphorylation of HSF1 by CK2 and/or CamKII.31 Quercetin is also metabolized in vivo and it is possible that some of the metabolites are the active pharmaceutical agents.42
Figure 2.
Two known inhibitors of heat induced HSP70 expression.
To guide the design and synthesis of biotinylated probes for identifying the target of quercetin inhibition of heat-induced HSP70 expression, and to enhance its specificity, we synthesized all of the mono-methyl and selected carbomethoxymethyl derivatives of quercetin. Systematic methylation (“methyl scanning”) has been previously proposed and validated as a general method for mapping the interactions of biologically active molecules with receptors and for designing affinity probes.43, 44 The quercetin derivatives were tested for their ability to inhibit heat-induced HSP70 expression and modulate HSP27 phosphorylation in human Jurkat cells and human HeLa cells, respectively, by Western blot analysis. Two classes of active agents were discovered, one that both inhibits HSP70 induction and enhances HSP27 phosphorylation, and another that inhibits HSP70 induction without enhancing HSP27 phosphorylation. Quercetin and derivatives that inhibited HSP70 induction were also found to inhibit both CK2 and CamKII kinases. On the other hand, derivatives that showed poor inhibition of either or both kinases did not inhibit HSP70 induction, indicating that quercetin’s effectiveness is due to its ability to inhibit more than one enzyme.
Chemistry
Synthesis of O-alkylated quercetin derivatives
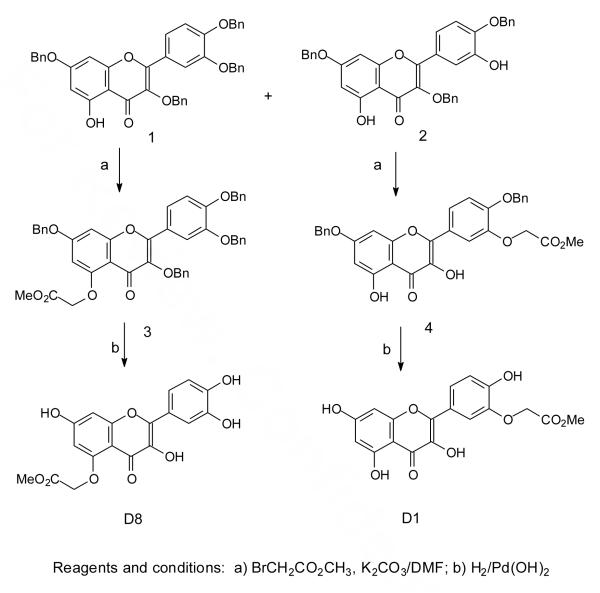
To determine the importance of individual hydroxyl groups on the inhibition of heat-induced HSP70 expression and other biological activities, we synthesized the mono-methylated quercetin derivatives D2, D3, D4, D6, and D7 according to previously reported procedures.45 Because we ultimately intend to attach affinity tags such as biotin to quercetin to help in identifying the protein target(s) of quercetin activity, we also synthesized a number of carboxymethylated derivatives by the same synthetic route (Scheme 1). Thus, quercetin was benzylated to give 20% of the tribenzyl derivative 1 and 60% of the tetrabenzyl derivative 2. The tribenzyl derivative was selectively carbomethoxymethylated at the 3′-OH to give 4, whereas the tetrabenzyl derivative was carbomethoxymethylated at the 5-OH to give 3. The products were debenzylated with palladium hydroxide and hydrogen to afford the methyl esters D8 and D1 respectively.
Scheme 1.
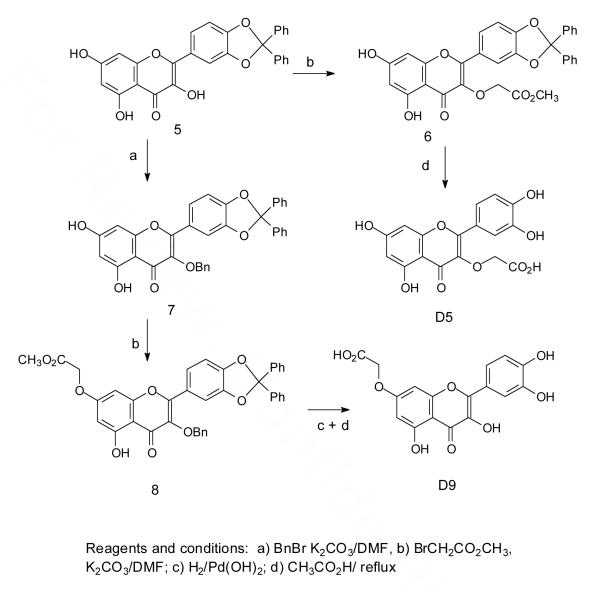
To carbomethoxymethylate positions 3 and 7, we utilized the 3′,4′-diphenylmethylene derivative of quercetin, compound 5 (Scheme 2). This derivative reacted with methylbromoacetate under basic conditions at the 3-OH to give 6, which then afforded the carboxymethyl derivative D5 following removal of the diphenylmethylene group by refluxing with acetic acid, which also hydrolyzed the ester. To obtain the O7 derivative, the 3-OH of compound 5 was first benzylated to give 7 which was then carbomethoxymethylated at the 7 position. Removal of the benzyl and diphenylmethylene groups was carried out in two steps by hydrogenolysis with palladium hydroxide followed by hydrolysis with acetic acid and water which also unexpectedly hydrolyzed the ester to the carboxymethyl product D9.
Scheme 2.
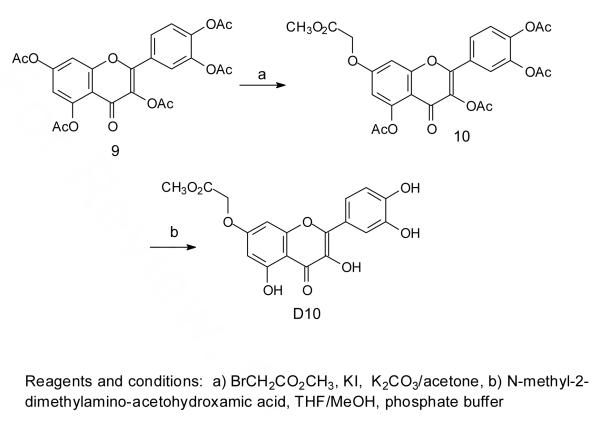
Many attempts to methylate D9 to produce D10 were unsuccessful and so other synthetic routes were investigated. The best of these involved a previously reported method for selectively methylating or benzylating the 7-OH of quercetin by treatment of quercetin pentaacetate with the alkylating agent in the presence of potassium carbonate and potassium iodide in acetone.46 Thus treatment of quercetin pentaacetate47 under these conditions with methyl bromoacetate yielded compound 10 which could be deacetylated in high yield under neutral conditions with N-methyl-2-dimethylaminoacetohydroxamic acid48 to give D10 (Scheme 3). This deacetylation method was chosen as more basic conditions, such as sodium methoxide and methanol at 0 °C, tend to result in air oxidation of the product,49 and more acidic conditions lead to hydrolysis of the methyl ester.
Scheme 3.
Structural characterization
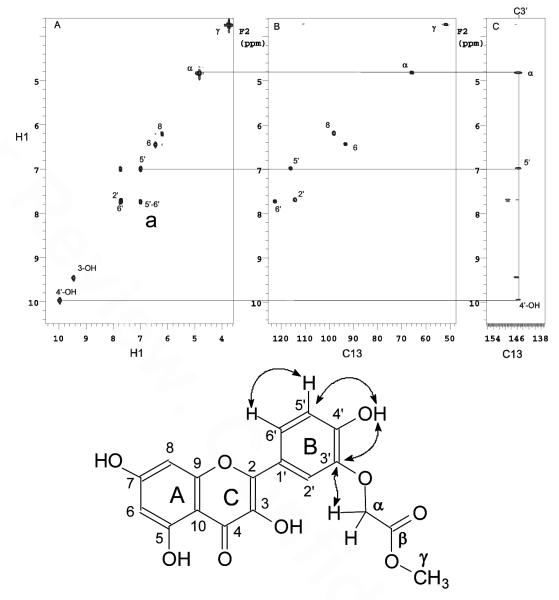
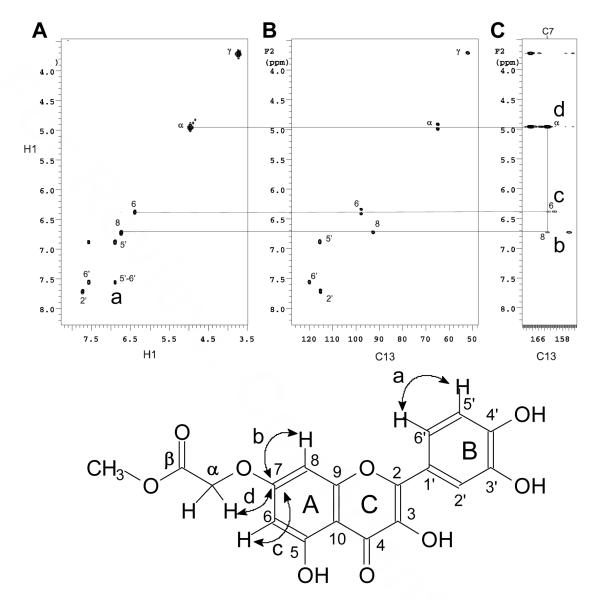
The sites of alkylation in all the compounds were confirmed by analysis of the coupling patterns in the 1D 1H NMR spectra together with correlations in the COSY, HMQC and HMBC spectra. As an illustration, the B ring hydrogens H5′ and H6′ of D1 (Figure 3) and D10 (Figure 4) were assigned by the large ortho coupling, and the strong crosspeak in the COSY spectrum (crosspeak a, panel A). These assignments led to the assignment of H2′ through meta coupling with H6′ that could be detected in the 1D 1H NMR spectrum. The carbons directly attached to these protons could then be assigned through the HMQC spectra (Panel B). The hydrogen signal at 10 ppm in D1 could then be assigned to the 4′-OH by the presence of a long range correlation to C5′ at 116 ppm (Figure 3, crosspeak e) and to a quaternary carbon signal at 146 ppm (crosspeak b) together with the absence of a correlation to C2′ in the HMBC spectrum (panel C). The signal at 146 ppm could therefore be assigned to C3′ which in turn showed a correlation to the α hydrogen of the carbomethoxymethylene group (crosspeak d, panel C). The position of the carbomethoxymethylene group in D10 could be assigned to O7 via a correlation between the α-methylene protons and a quaternary carbon signal at 162 ppm (Figure 4, crosspeak d in panel C). The signal at 162 ppm could be assigned to C7 via multiple bond correlations to the two meta coupled protons in the A ring in the HMBC spectrum (crosspeaks b & c in panel C).
Figure 3.
Structure assignment of D1. (A) COSY, (B) HMQC and (C) HMBC of D1 in DMSO-d6
Figure 4.
Structure assignment of D10. (A) COSY, (B) HMQC and (C) HMBC of D10 in DMSO-d6.
Biological Studies
Inhibition of heat induced HSP70 expression by quercetin derivatives
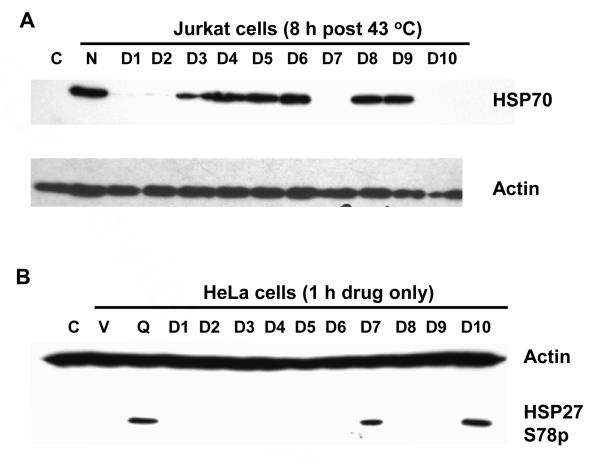
To examine the ability of the quercetin and its derivatives to inhibit heat induced HSP70 expression, we chose human Jurkat cells due to their low basal level of HSP70 expression and the large induction upon heat shock. Western blot analysis of the cellular proteins in Jurkat cells following heat shock revealed that pre-heat treatment with the quercetin derivatives D1, D2, D7 and D10 all had a similar inhibitory effect on HSP70 induction as did quercetin, indicating that the 3′ and 7 hydroxyls are important for activity (Figures 5A, Table 1). Compound D1 which bears a carbomethoxymethyl group at O3′ is not as good an inhibitor as D2, which bears a methyl group, indicating that the 3′ hydroxyl group occupies a more sterically constrained site in the target protein than does the 7 hydroxyl group. Compound D9, which bears a carboxylic acid group at O7, does not inhibit HSP70 like D10 which bears an ester group, presumably because it forms a salt at neutral pH that greatly reduces its membrane permeability.
Figure 5.
Effect of quercetin and its derivatives on heat induced HSP70 expression and phosphorylation of HSP27. Panel A. Inhibition of heat induced HSP70 expression. Jurkat cells pre-treated 1 h with 50 μg/ml (approx. 150 μM) of the quercetin derivatives D1-D10 were heat shocked as indicated then allowed to recover at 37 °C for 8 h before protein isolation and analysis. HSP70 and actin protein levels were determined by Western blotting: Lane C: non-heated cells; N: heated no drug; D1-D10, quercetin derivatives. Panel B. Drug induced phosphorylation of human HSP27 Ser78. HeLa cells were treated for 1 h with drug (50 μg/ml, approx. 150 μM) before cellular proteins were isolated for Western blot analysis with actin and HSP27 Ser78P antibodies: Lane C: no drug; V: DMSO vehicle only; Q: quercetin; D1-D10: quercetin derivatives.
Table 1.
Quercetin derivatives. The substituent is an H unless otherwise noted.
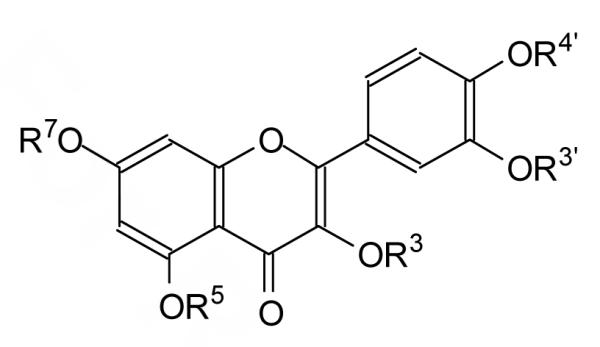 | |||||||
|---|---|---|---|---|---|---|---|
| Cmpd | R3 | R5 | R7 | R3′ | R4′ | HSP70 inhibition |
HSP27 phosphorylation |
| Quercetin | +++ | +++ | |||||
| D1 | CH2CO2CH3 | ++ | - | ||||
| D2 | CH3 | +++ | - | ||||
| D3 | CH3 | + | - | ||||
| D4 | CH3 | - | - | ||||
| D5 | CH2CO2H | - | - | ||||
| D6 | CH3 | - | - | ||||
| D7 | CH3 | +++ | +++ | ||||
| D8 | CH2CO2CH3 | - | - | ||||
| D9 | CH2CO2H | - | - | ||||
| D10 | CH2CO2CH3 | +++ | +++ | ||||
Enhancement of HSP27 phosphorylation
In examining the effects of quercetin treatment on other heat shock proteins, we found that quercetin enhances HSP27 phosphorylation at Ser78 in the absence of heat shock. Phosphorylated HSP27 has anti-apoptotic activity2 which would tend to counteract any lethal effects achieved from inhibiting HSP70 induction. Thus, it would be highly desirable to inhibit HSP70 without enhancing phosphorylation of HSP27. The HSP27 studies were carried out in HeLa cells, which unlike Jurkat cells, have a high basal level of HSP27, making it easier to detect drug-induced phosphorylation of HSP27 by Western analysis. HeLa cells, however, are not as suitable for concurrent inhibition studies of HSP70 induction since they have high basal level of HSP70. Only D7 and D10 showed enhancement of HSP27 phosphorylation in HeLa cells, and both of these compounds also showed good to strong inhibition of HSP70 induction in Jurkat cells (Figure 5B). On the other hand, D1 and D2 which were also good inhibitors of HSP70 induction, did not activate HSP27 phosphorylation.
Effect of quercetin on HSF1 binding to the HSE
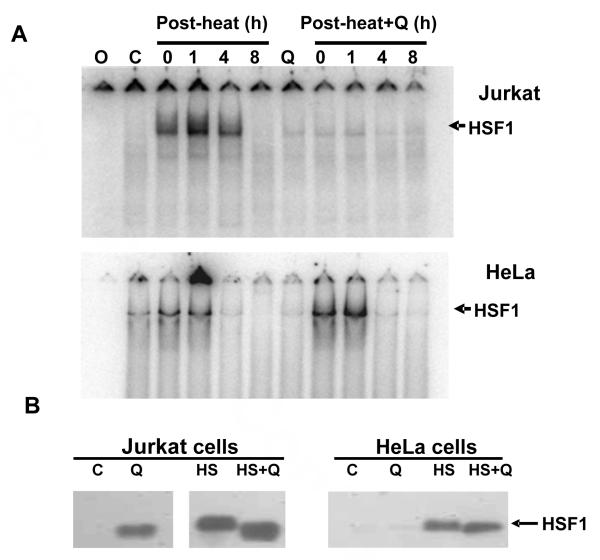
In order to gain insight into the mechanism by which quercetin inhibits HSP70 induction, gel mobility shift analysis was carried out to examine binding of heat shock element (HSE) DNA by heat shock transcription factor 1 (HSF1), the transcription factor responsible for heat induced HSP70 gene transcription (Figure 6A upper panel). No HSF1/HSE complex was detected with nuclear extracts prepared from control, non-heated Jurkat cells but the complex was readily detected with extracts prepared immediately following a 43 °C 30 min heat shock. The amount of complex increased by 1 h post-heat then declined by 4 h post-heat and returned to control levels by 8 h post-heat. In Jurkat cells treated 1 h prior to heat shock with quercetin, however, only a very low level of complex formed in extracts from heated cells. Since an identical low level of complex was also detected in extracts from quercetin-treated but non-heated cells, this response appears to be due to drug induced cell stress not heat. In contrast to Jurkat cells, HeLa cells displayed an accelerated lost of HFS1/HSE complex during recovery from heat shock with nearly undetectable levels by 4 h post-heat (Figure 6A, lower panel). Moreover, quercetin treatment did not inhibit HSF1/HSE complex formation in heated HeLa cells as it did in Jurkat cells suggesting a complex interaction between the drug and possibly multiple cell specific factors.
Figure 6.
Effect of quercetin on HSF1 binding to HSE DNA. Jurkat (Panel A-upper) or HeLa cells (Panel A-lower) were heated or pre-treated with quercetin (50 μg/ml, approx. 150 μM) 1 h prior to heating and allowed to recover at 37 °C for 0-8 h at 37 °C. Cells were isolated and nuclear extracts prepared for EMSA analysis with a 5′-radiolabeled HSE oligodeoxynucleotide. Position of the HSF1-HSE complex is indicated by the arrows. Lane O: HSE oligonucleotide without extract; C: unheated extract; Q: quercetin treated, no heat. HSF1 content of EMSA nuclear extracts (Panel B). Nuclear extracts utilized in Panel A were subjected to Western blot analysis with anti-HSF1 antibody to determine the HSF1 levels and relative size. Lane C: nuclear extract from unheated cells; Q: quercetin treated for 2.5 h; HS: 1 h after a 43 °C/30 min heat shock; HS+Q: 1 h quercetin treatment, 43 °C/30 min heat shock and 1 h recovery.
Further information about the mechanism by which quercetin inhibits heat-induced HSP70 expression was obtained by Western blot analysis of the nuclear extracts for the presence and size of the HSF1 protein (Fig. 6B). In non-heated Jurkat control cells, HSF1 is not detected in nuclear extracts while only a very low level can be detected in HeLa cells. Surprisingly, nuclear extracts from quercetin treated Jurkat cells contained significant levels of HSF1 while similar HeLa cell extract contained the very low levels detected in untreated extracts. In both Jurkat and Hela cells, heating increased HSF1 nuclear extract levels and induced a corresponding decrease in gel mobility that reflects the increased level of heat-induced phosphorylation required for transcriptional activity.31, Soncin, 2003 #6321 While quercetin treatment did not block the heat-induced increase in nuclear HSF1 levels in either Jurkat and HeLa cells, the drug did inhibit the decrease in HSF1 mobility, presumably by inhibiting the critical phosphorylation modifications required for transcription activation. Thus in Jurkat cells, quercetin treatment appears to affect at least two steps in heat induced HSP70 expression, HSF1 post translational modification and DNA binding activity. Quercetin treatment of HeLa cells does not result in loss of HSF1 DNA binding activity but does appear to inhibit heat induced phosphorylation.
Kinase inhibition assays
We then determined whether or not the activity of the quercetin analogs correlated with their ability to inhibit either or both CKII and CamKII kinases that have previously been shown to phosphorylate HSF1. We used an enzyme-linked inhibition assay that utilizes luciferase to quantify the amount of ATP remaining in a kinase reaction.50 The IC50 values for quercetin and D1-D10 are listed in Table 2 in comparison to their relative ability to inhibit HSP70 induction. Good inhibition of HSP70 induction appears to correlate with good inhibition of both CK2 and CamKII. Quercetin derivatives that showed poor inhibition (IC50 > 100 μM) of either or both kinases showed little to no ability to inhibit heat induction of HSP70. The only exception was D9 which was a very good inhibitor of both kinases, but showed no ability to inhibit HSP70 induction, which we attribute to the presence of a carboxylate which reduces membrane permeability.
Table 2.
IC50 values (μM) for inhibition of CK2 and CamKII by quercetin and derivatives D1-D10.
| Q | D1 | D2 | D3 | D4 | D5 | |
|---|---|---|---|---|---|---|
| CK2 | 5.6 ± 1.2 | 53 ± 3.7 | 0.50 ± 0.01 | >100 | 0.83 ± 0.10 | 2.45 ± 0.25 |
| CamKII | 3.3 ± 0.6 | 6.3 ± 1.3 | 0.91 ± 0.25 | 16.4 ± 1.9 | >100 | >100 |
| HSP70 | +++ | ++ | +++ | + | - | - |
| D6 | D7 | D8 | D9 | D10 | ||
|---|---|---|---|---|---|---|
| CK2 | >100 | 9.7 ± 1.4 | >100 | 9.7 ± 3.6 | 0.85 ± 0.30 | |
| CamKII | 4.0 ± 1.1 | 8.9 ± 0.3 | 69 ± 6.0 | 9.3 ± 1.8 | 9.1 ± 0.1 | |
| HSP70 | - | +++ | - | - | +++ |
Discussion
Inhibition of heat-induced HSP70 expression has been shown to enhance radiosensitization by heat26, 27 and thus small molecule inhibitors of HSP70 induction have potential therapeutic value. Quercetin has long been known to inhibit heat-induced HSP70 expression, though the exact mechanism is unclear. The results herein suggest that quercetin treatment blocks heat-induced phosphorylation of the HSF1 transcription factor that is responsible for inducing HSP70 expression. In Jurkat, but not HeLa cells an additional level of inhibition appears to be loss of HSF1 HSE element binding capacity, which has also been reported in quercetin/heated human COLO 320DM cells.51 Unfortunately, quercetin is also well known to affect many additional biological processes, probably as a result of its ability to inhibit a number of different kinases,41 which diminishes its potential as a therapeutic. On the other hand it seems reasonable that the specificity for inhibiting HSP70 induction could be increased by selective modification of quercetin. Also identifying the target of quercetin responsible for inhibiting HSP70 induction could aid in the development of more selective inhibitors. To these ends we have been able to show that the C7 and C3′ hydroxyls on quercetin can be derivatized with a bulky carbomethoxymethyl group without affecting its ability to inhibit heat induced HSP70, and could therefore be used to attach a biotin affinity probe.
The finding that the 3′ and 7 hydroxyls of quercetin can be modified without affecting inhibition of heat-induced HSP70 expression suggested modification of these hydroxyls might also increase the selectivity by interfering with binding to other protein targets. Quercetin has been crystallized with a number of kinases, and inspection of the crystal structures reveals that the 7-OH of quercetin is buried in the Hck kinase complex52 as well as in the F1-ATPase53 and therefore O7 derivatives would not be expected to inhibit these enzymes. The O7 position of quercetin is not buried, however, in the phosphoinositide 3-kinase complex,54 but the O3′-position is, suggesting that quercetin derivatized at both the O7 and O3′ positions would also not inhibit any of these enzymes, but would still inhibit HSP70 induction. This class of doubly derivatized quercetin remains to be synthesized and examined for its ability to inhibit HSP70 induction.
Given that quercetin is a well known kinase inhibitor, and that the pathway for heat induction of HSP70 expression involves phosphorylation of the heat shock factor 1 (HSF1) by CK2 and CamKII kinases, we investigated whether the two might be correlated. It has previously been established that quercetin is an inhibitor or CK2 kinase, but its ability to inhibit CamKII kinase has not been reported. We now show that quercetin is also a good inhibitor of CamKII and that quercetin derivatives D1, D2, D7 and D10, that are good inhibitors of HSP70 induction, are also good inhibitors of both CK2 and CamKII kinases. Quercetin derivatives that were good inhibitors of only one or neither of the two kinases, however, were not good inhibitors of HSP70 induction. It would seem that full activation of HSP70 by HSF1 may only require phosphorylation by either CK2 or CamKII, both of which quercetin and derivatives D1, D2, D7 and D10 are able to inhibit due to their broader specificity.
The ability of quercetin to enhance phosphorylation of HSP27 has not been previously recognized. Phosphorylation of serine 78 has been shown to be carried out by the MK2 kinase,55 which in turn is activated by MAP kinase (MAPK). It is not clear how quercetin activates phosphorylation of HSP27, as inhibiting any of the kinases in the pathway should abolish activation. Activation of HSP27 has been associated with poor prognosis and resistance to chemotherapy or radiation56 and so the quercetin analogs D1 and D2 which retain the ability to inhibit HSP70 induction without inducing HSP27 phosphorylation may have superior therapeutic properties to D7 and D10.
Conclusion
The original goal of this study was to find derivatives of quercetin that could be elaborated into affinity probes to identify the targets responsible for inhibition of heat-induction of HSP70. We were able to identify two such derivatives, which are now in the process of being converted to biotinylated probes. In the course of evaluating these derivatives, we discovered that their ability to inhibit HSP70 induction correlated with their ability to inhibit both CK2 and CamKII, kinases that are known to activate HSP70 transcription by phosphorylating HSF1. Derivatives that show poor inhibition of either or both kinases are poor inhibitors of HSP70 induction, suggesting that HSP70 expression can be activated by either kinase. This would appear to be a case in which making the drug more specific reduces its effectiveness, suggesting that future screens for HSP70 inhibitors be carried out for activity against both kinases. We have also discovered that quercetin enhances the phosphorylation of HSP27 required for its anti-apoptotic action, which would serve to diminish the radiosensitizing effects of inhibiting HSP70 induction. Two derivatives of quercetin, D1 and D2, were found, however, that could inhibit HSP70 induction without enhancing HSP27 phosphorylation, but the degree to which they may be able to enhance heat-induced radiosensitization remains to be determined. In addition to probing the mechanism of inhibition of HSP70 induction by quercetin, this family of methylated quercetin derivatives might also be useful for mapping out interactions with its other targets.
Experimental Section
General
Reagents and solvents were obtained from either Sigma Aldrich or Alfa Aesar. Anhydrous solvents were distilled and then stored over activated 5 Å molecular sieves. All other commercial materials were used without further purification unless otherwise stated. Analytical thin-layer chromatography was performed on Aldrich Silica gel 60 F254 plates (0.25 mm) and compounds were visualized with UVG-54 mineral light UV lamp at 254 nm. Flash column chromatography was conducted using the indicated solvent on E. Merck silica gel 60 (40-63 μm).
NMR Spectroscopy
1H NMR and 13C NMR spectra were recorded with Mercury-300, Inova-500 and Inova-600 (Varian Assoc., Palo Alto, CA) spectrometers and the data were processed with VNMR software. Proton and carbon chemical shifts were measured in parts per million (ppm) downfield from an internal TMS standard. Proton spectra were obtained in DMSO-d6 with a 8200-Hz spectral width collected into 32K data points. Carbon spectra were obtained with a 34000-Hz spectral width collected into 64k data points. The gradient COSY experiments were collected with a spectral width of 8200 Hz and 2048 complex points in F2 dimension and 512 real points in F1 dimension. The proton-detected heteronuclear multiple quantum coherence (HMQC) spectra were recorded using a 0.3 s 1H-13C nulling period. The 90° 1H pulse width was 9.8 μs and the 90° 13C pulse width was 12.5 μs. Phase sensitive 2D spectra were obtained by employing the Hypercomplex method. A 2 × 256 × 2048 data matrix with 16 scans per t1 value was collected. Gaussian line broadening was used in weighting both the t2 and the t1 dimension. After two-dimensional Fourier transform, the spectra resulted in 512 × 2048 data points, which were phase and baseline, corrected in both dimensions.
Compound 4
A solution of 2 (0.60 g, 1.1 mmol) in DMF (20 ml) was mixed with potassium carbonate (0.19 g, 1.4 mmol) under nitrogen. Methyl bromoacetate (0.160 g, 1.05 mmol) was then slowly added. The resulting solution was stirred at room temperature for 12 h, diluted with 40 ml of water and extracted with 60 ml ethyl acetate. The organic phase was washed with water, dried over anhydrous sodium sulfate and concentrated under reduced pressure. The residue was purified by flash column chromatography using DCM/Hexane (85:15) to afford 4 in 90% yield (0.58 g). 1H NMR (CDCl3, 300 MHz) δ 12.74 (s, 1H, OH), 7.67 (d, J=2 Hz, 1H), 7.66 (dd, J=9, 2 Hz, 1H), 7.46-7.22 (m, 15H), 6.93 (d, J=9 Hz, 1H), 6.44 (d, J=2 Hz, 1H), 6.40 (d, J=2 Hz, 1H), 5.19 (s, 2H), 5.07 (s, 2H), 5.05 (s, 2H), 4.54 (s, 2H), 3.71 (s, 3H). 13C NMR(CDCl3, 300 MHz) δ 179.0 (C=O), 169.4 (COO), 164.8, 162.3, 156.9, 156.2, 151.3, 147.6, 137.8, 136.8, 136.6, 136.1, 129.0, 128.9, 128.6, 128.5, 128.4, 127.8, 127.6, 124.0, 123.6, 115.9, 114.0, 106.4, 98.9, 93.3, 74.7, 71.1, 70.7, 66.8 (OCH2), 52.4 (OCH3). MS (FAB) [M+Na+] m/z 667.2, HRMS (FAB) C39H32O9Na [M+Na+], calculated m/z 667.1944, found 667.1932.
Compound D1
A suspension of compound 4 (100 mg) was dissolved in a minimum amount of EtOH/THF (1:1) and treated with 10% palladium hydroxide (0.01 g). The suspension was stirred over night at room temperature under one atmosphere of hydrogen from a balloon. After filtration with Celite to remove the palladium, the filtrate was concentrated under reduce pressure, followed by a recrystallization with methanol and water to give D1 in 75% yield (43 mg). 1H NMR ([D6]DMSO, 300 MHz) δ 12.45 (s, 1H, OH), 10.77 (s, 1H, OH), 9.96 (s, 1H, OH), 9.45(s, 1H, OH), 7.72 (d, J=2 Hz, 1H), 7.68 (dd, J=8.5, 2 Hz, 1H), 6.96 (d, J=8.5 Hz, 1H), 6.42 (d, J=2 Hz, 1H), 6.17 (d, J=2 Hz, 1H), 4.81 (s, 2H), 3.72 (s, 3H). 13C NMR ([D6]DMSO, 500 MHz) δ 175.9 (C=O), 169.4 (COO), 163.9, 160.7, 156.1, 149.2, 146.2, 145.6, 135.9, 122.6, 121.9, 116.1, 114.2, 103.0, 98.2, 93.4, 65.9 (OCH2), 51.8 (OCH3). MS (ESI) [M+Na+] m/z 397.1, HRMS (ESI) C18H14O9Na [M+Na+], calculated m/z 397.0536, found 397.0543.
Compound 3
Two equivalents of methyl bromoacetate (0.46 g, 3.0 mmol) and anhydrous potassium carbonate (0.42 g, 3.0 mmol) were added to a solution of compound 1 (1.0 g, 1.5 mmol) in 35 ml DMF under nitrogen. After stirring for 24 h, the resulting mixture was worked up with 50 ml water and extracted with 100 ml EtOAc. The organic phase was washed with water, dried over anhydrous sodium sulfate and concentrated under reduced pressure. The red crude product was recrystallized with EtOAc to afford 3 in 92% yield (1.0 g). 1H NMR (CDCl3, 300 MHz) δ 7.75 (d, J=2 Hz, 1H), 7.57(dd, J=9, 2 Hz, 1H), 7.48-7.23 (m, 20H), 6.97 (d, J=9 Hz, 1H), 6.61 (d, J=2 Hz, 1H), 6.36 (d, J=2 Hz, 1H), 5.25 (s, 2H), 5.12 (s, 2H), 5.08 (s, 2H), 4.97 (s, 2H), 4.83 (s, 2H), 3.81 (s, 3H). 13C NMR (CDCl3, 300MHz) δ 174.0 (C=O), 169.0 (COO), 162.8, 159.2, 158.9, 153.5, 150.8, 148.4, 140.0, 137.3, 137.2, 137.0, 135.7, 129.2, 129.0, 128.8, 128.8, 128.7, 128.4, 128.3, 128.2, 128.1, 128.0, 127.6, 127.4, 124.0, 122.4, 115.4, 114.0, 110.3, 98.9, 95.1, 74.3, 71.3, 71.1, 70.9, 66.8 (OCH2), 52.7 (OCH3) MS (FAB) [M+Li]+ m/z 741.5. HRMS (FAB) C46H38O9Li [M+Li+], calculated m/z 741.2676, found 741.2677.
Compound D8
Compound 3 (255 mg, 0.35 mmol) was dissolved in a minimum amount of THF/EtOH (1:1). After addition of 10% Pd(OH)2 (38 mg), the suspension was stirred for 16 h under one atmosphere of hydrogen from a balloon. The suspension was filtered through Celite, diluted with EtOH and concentrated under vacuum. The residue was purified by recrystallization with MeOH/H2O to afford D8 as a green solid in 86% yield (111 mg). 1H NMR ([D6]DMSO, 300 MHz) δ 10.77 (s, 1H, OH), 9.50 (s, 1H, OH), 9.28 (s, 1H, OH), 8.76 (s, 1H, OH), 7.64 (d, J=2 Hz, 1H), 7.50 (dd, J=8.5, 2 Hz, 1H), 6.88 (d, J=8.5 Hz, 1H), 6.52 (d, J=2 Hz, 1H), 6.25 (d, J=2 Hz, 1H), 4.89 (s, 2H), 3.73 (s, 3H). 13C NMR ([D6]DMSO, 300MHz) δ 171.5 (C=O), 169.5 (COO), 162.7, 159.2, 158.5, 147.7, 145.7, 142.8, 137.8, 122.9, 119.9, 116.3, 115.2, 106.2, 98.4, 96.3, 66.4 (OCH2), 52.6 (OCH3) MS (FAB) [M+Na]+ m/z 397.1, HRMS (FAB) C18H14O9Na [M+Na+], calculated m/z 397.0536 found 397.0518.
Compound 6
A suspension of 5 (0.70 g, 1.50 mmol) was dissolved in 20 ml of DMF, followed by addition of anhydrous potassium carbonate (0.311 g, 2.25 mmol) and methyl bromoacetate (0.23 g, 1.5 mmol). The mixture was stirred overnight at room temperature, under nitrogen. The solution was diluted with 50 ml water, acidified with 1.0 N HCl and extracted with 100 ml ethyl acetate. The organic layer was washed with water, brine and dried over anhydrous sodium sulfate. The residue after concentration in vacuum was purified by flash column chromatography using 5% EtOAc in DCM, to afford 6 in 37% yield (0.30 g). 1H NMR (CDCl3, 300 MHz) δ7.78 (dd, J=8.5, 2 Hz, 1H), 7.71 (d, J=2 Hz, 1H), 7.64-7.61 (m, 4H), 7.45-7.41(m, 6H), 7.02 (d, J=8.5 Hz, 1H), 6.39 (d, J=2 Hz, 1H), 6.28 (d, J=2.0 Hz, 1H), 4.80 (s, 2H), 3.73 (s, 3H). 13C NMR (CDCl3, 600 MHz) δ 177.9 (C=O), 169.6 (COO), 162.4, 162.0, 156.6, 155.4, 149.5, 147.4, 139.7, 136.7, 129.3, 128.3, 126.2, 124.2, 123.8, 117.9, 108.9, 108.5, 105.6, 99.2, 93.9, 68.4 (OCH2), 52.0 (OCH3). MS (FAB) [M+Na+], m/z 561.1. HRMS(FAB) C31H22O9Na [M+Na+], calculated m/z 561.1162, found 561.1164.
Compound D5
Compound 6 (0.30 g, 0.56 mmol) was mixed with 50 ml CH3CO2H/H2O (4:1) and refluxed overnight. The solution is worked up by addition of 50 ml of water and then carefully neutralized by dropwise addition of saturated NaHCO3. After the extraction with 100 ml EtOAc, the organic layer was washed with brine, dried over anhydrous sodium sulfate and vacuum concentrated. The crude residue was recrystallized with methanol to afford 0.156 g of D5 as a red solid in 74% yield. 1H NMR ([D6]DMSO, 300 MHz) δ 12.61 (s, 1H, OH), 7.63 (d, J=2 Hz, 1H), 7.62 (dd, J=8, 2 Hz, 1H), 6.93 (d, J=8 Hz, 1H), 6.48 (d, J=2 Hz, 1H), 6.26 (d, J=2 Hz, 1H), 4.73 (s, 2H). 13C NMR ([D6]DMSO, 300 MHz) δ 177.4 (C=O), 171.5 (CO2H), 169.8, 164.1, 161.0, 156.1, 155.1, 148.6, 145.0, 135.7, 121.0, 120.6, 115.5, 103.9, 98.5, 93.4, 67.7 (OCH2-CO2H). MS (ESI) [M+H+], m/z 361.0. HRMS (ESI) C17H13O9 [M+H+], calculated 361.0560, found 361.0599.
Compound 8
Compound 7 (0.50 g, 0.90 mmol) in 30 ml DMF was mixed with methyl bromoacetate (0.138 g, 0.90 mmol), followed by addition of anhydrous potassium carbonate (0.152 g, 1.10 mmol). The mixture was stirred under nitrogen for 6 h at room temperature and then neutralized with 1.0 N HCl. The solution was extracted with 100 ml EtOAc, washed with water, and dried over anhydrous sodium sulfate. The solvent was removed under vacuum and the residue was flash chromatographed using EtOAc/petroleum ether (1:1) to afford 0.360 gm of the desired product 8 in 64% yield. 1H NMR (CDCl3, 300 MHz) δ 12.78 (s, 1H, OH), 7.62-7.15 (m, 17H), 6.92 (d, J=8.5 Hz, 1H), 6.40 (d, J=2 Hz, 1H), 6.36 (d, J=2 Hz, 1H), 5.04 (s, 2H), 4.70 (s, 2H), 3.83 (s, 3H). 13C NMR (CDCl3, 300MHz) δ 207.3, 179.0 (C=O), 168.7(COO), 163.6, 162.4, 157.1, 156.8, 149.6, 147.5, 140.0, 137.5, 136.4, 126.7, 129.2, 128.7, 128.5, 126.5, 124.4, 124.3, 118.1, 109.3, 108.6, 106.8, 98.5, 93.1, 74.7, 65.4 (OCH2), 52.8 (OCH3) MS (ESI) m/z 629.2 for [M+H+], m/z 651.2 for [M+Na+], m/z 667.1 for [M+K+], MS (FAB) [M+Na+], m/z 651.1. HRMS (ESI) C38H29O9[M+H+], calculated m/z 629.1830, found 629.1830.
Compound D9
Compound 8 (0.482 g) in 20 ml of EtOH/THF (1:2) was mixed with 10% Pd(OH)2 (10 mg) and stirred overnight under hydrogen from a balloon. The suspension was filtered through Celite and the clear green filtrate was diluted with 20 ml EtOH and concentrated under vacuum. The residue was refluxed with acetic acid as described in th preparation of D5 to give 0.224 g of D9 in 78% yield. 1H NMR ([D6]DMSO, 300 MHz) 12.46 (s, 1H, OH), 9.63 (s, 1H, OH), 9.50 (s, 1H, OH), 9.30 (s, 1H, OH), 7.69 (d, J=2 Hz, 1H), 7.55 (dd, J=8.5, 2 Hz, 1H), 6.86 (d, J=8.5 Hz, 1H), 6.66 (d, J=2 Hz, 1H), 6.32 (d, J= 2 Hz, 1H), 4.80 (s, 2H). 13C NMR ([D6]DMSO, 300 MHz) δ 175.8 (C=O), 169.5(CO2H), 163.2, 160.2, 155.7, 147.8, 147.2, 145.0, 136.0, 121.7, 120,0, 115.5, 115.1, 104.2, 97.7, 92.4, 64.8 (OCH2). MS (ESI) [M+H+], m/z 361.1. HRMS (ESI) C17H13O9 [M+H+], calculated m/z 361.0560, found m/z 361.0555.
Compound 10
Quercetin pentaacetate 9 (1.0 g, 1.95 mmol) in 50 ml dry acetone was mixed with methyl bromoacetate (1.0 g, 6.55 mmol), anhydrous potassium carbonate (2.0 g, 14.5 mmol) and 0.168 g potassium iodide. The suspension was refluxed until 9 was completely consumed as judged by TLC. The suspension was filtered and the filtrate was concentrated under vacuum, and flash chromatographed using EtOAc/Hexane (4:3). The product was recrystallized with acetone/hexane to afford 0.577 g of 10 as a white solid in 54% yield. 1H NMR (CDCl3, 300 MHz) δ 7.74 (dd, J=8.5, 2 Hz, 1H), 7.70 (d, J=2 Hz, 1H), 7.37 (d, J=2 Hz, 1H), 6.83 (d, J=2 Hz, 1H), 6.20 (d, J=2 Hz, 1H), 4.76 (s, 2H), 3.87 (s, 3H), 2.46 (s, 3H), 2.37 (s, 9H). 13C NMR (CDCl3, 300 MHz) δ 169.8 (C=O), 169.3 (COO), 168.0, 167.9, 167.8, 167.7 (4 CH3C=O), 161.8, 157.8, 153.3, 150.9, 144.2, 142.2, 133.9, 127.9, 126.4, 123.8, 123.7, 111.8, 109.0, 99.6, 65.4 (OCH2), 52.6 (OCH3), 21.0, 20.8, 20.6, 20.5 (4 CH3C=O) MS: ESI [M+Na+] 565.1, [M+H+] 543.0, [M-CH2CO+H+] 501.1, [M-2CH2CO+H+] 459.1, [M-3CH2CO+H+] 417.1 HRESI C26H22O13Na [M+Na+], calculated 565.0958, found 565.0948.
Compound D10
Twenty mg (0.036 mmol) of 10 was dissolved in 10 ml of THF/MeOH/pH 7 phosphate buffer (9:2:9), after which 5 equivalent of N-methyl-2-dimethylaminoacetohydroxamic acid (24 mg, 0.180 mmol) was added. The solution was stirred under nitrogen atmosphere at room temperature for 12 h after which the mixture was worked up with water, acidified to pH 6 by 1 M HCl and extracted with 50 ml of EtOAc. The organic phase was washed with water and dried over anhydrous sodium sulfate. The residue after concentration in vacuum was recrystallized from MeOH/H2O to afford 11 mg of D10 as a green solid in 83% yield. 1H NMR ([D6]DMSO, 300 MHz) δ 7.71 (d, J=2 Hz, 1H), 7.55 (dd, J=8.5, 2 Hz, 1H), 6.87 (d, J=8.5 Hz, 1H), 6.72 (d, J=2 Hz, 1H), 6.37 (d, J=2 Hz, 1H), 4.94 (s, 2H), 3.71 (s, 3H). 13C NMR ([D6]DMSO, 600MHz) δ 176.4 (C=O), 169.0 (COO), 163.5, 160.8, 156.2, 148.3, 147.9, 145.5, 136.5, 122.2, 120.4, 116.0, 115.7, 104.8, 98.2, 93.0, 65.3 (OCH2), 52.4 (OCH3). MS (ESI) [M+Na]+ 397.1, HRESI C18H14O9Na [M+Na]+, calculated 397.0536, found 397.0535.
HSP70 Inhibition Assays
Exponentially growing Jurkat cells, grown in RPMI media with 10% fetal bovine serum, were treated with 50 μg/ml of quercetin (165 μM) or its derivatives (methyl: 158 μM; carboxymethyl: 139 μM; carbomethoxymethyl: 134 μM) for 1 h prior to being heated at 43 °C for 30 min. The cells were allowed to recover at 37 °C for 8 h to allow for HSP70 expression and cells harvested for Western blot analysis with mouse monoclonal antibodies against inducible HSP70 (Assay Designs, SPA-810) and actin (ICN, clone C4). Secondary antibodies were goat anti-mouse conjugated horse radish peroxidase (Millipore) or alkaline phosphatase (Jackson ImmunoResearch). Protein bands were visualized by either chemiluminescence (SuperSignal West Femto Substrate, Thermo Scientific) or colorimetric (BCIP/NBT, Sigma) detection.
HSP27 Phosphorylation Assays
Phosphorylation of HSP27 was detected by Western blot analysis of total cell extracts prepared from exponentially growing HeLa cells. Cells were grown in DMEM media with 10% calf serum and treated for 1 hr with 50 μg/ml of quercetin or its derivatives before protein isolation. A rabbit antibody specific for human HSP27 phosphorylated on serine 78 (Assay Designs, SPA-523) coupled with goat anti-rabbit alkaline phosphatase secondary antibody (Jackson ImmunoResearch) was utilized for colorimetric detection.
HSF1 Electrophoretic Mobility Shift Assays (EMSA)
Analysis of HSF1 interactions with the HSE DNA promoter element was carried out by EMSA essentially as described previously (Hunt CR et al. 1999) except that nuclear extracts were utilized in place of whole cell extracts. The heat shock element probe (HSE) d(GCGAAACTGCTGGAAGATTCCT) was 5′-32P end labeled followed by annealing to the complementary oligonucleotide to produce the duplex DNA. Western blot analysis of the same nuclear extracts utilized for EMSA was carried out with rabbit anti-HSF1(Cell SignalingTechnology) followed by goat anti-rabbit HRP (Millipore) and chemiluminescent detection.
Kinase inhibition assays
Phosphate kinases CAMKII and CK2, substrates, and buffers were from New England BioLabs. Inhibition assays were carried out with the PKlight HTS Protein Kinase Assay kit (Lonza) according to the manufacturer’s procedure. Briefly, this assay involves determining in triplicate the extent of phosphorylation of a peptide substrate in the presence and absence of various concentrations of inhibitor by quantifying the amount of ATP consumed via a luciferase-based bioluminescence assay. A kinase concentration (EC50) is used that results in 50% consumption of ATP in the absence of inhibitor under the reaction time and conditions. The data was then fit via non-linear least squares fitting to a standard sigmoidal dose-response equation. CK2 inhibition assay: Casein kinase II (CK2) was incubated at 30 °C for 30 min with 300 μM substrate (RRRADDSDDDDD), 100 μM ATP, in a buffer containing 20 mM Tris-HCl, 50 mM KCl, 10 mM MgCl2, with or without the presence of inhibitor. CamKII inhibition assay: Ca2+/Calmodulin-dependent protein kinase II (CaMKII) supplemented with 200 μM ATP, 1.2 μM calmodulin and 2 mM CaCl2 was incubated for 10 min at 30 °C to preactivate the enzyme and then diluted to the required concentration. The activated CaMKII was then incubated at 30°C for 30 min with or without the inhibitor, along with 300 μM autocamtide-2 (KKALRRQETVDAL), 100 μM ATP, in a buffer containing 50 mM Tris-HCl, 10 mM MgCl2, 2 mM DTT, 0.1 mM Na2EDTA at pH 7.5.
Supplementary Material
Acknowledgment
This work is supported by NIH PO1 CA 104457 and by grants to the Washington University NIH Mass Spectrometry Resource (Grant No. P41 RR000954) and Washington University NMR facility (Grant No. RR1571501).
Footnotes
Abbreviations: CaMKII, Ca2+/calmodulin-dependent protein kinase II; CK2, casein kinase II; COSY, correlation spectroscopy; DCM, dichloromethane; DMEM, Dulbecco’s modified essential medium; DMF, dimethyl formamide; DMSO, dimethyl sulfoxide; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; ERK, extracellular signal-regulated protein kinase; ESI, electrospray ionization; EtOAc, ethyl acetate; EtOH, ethanol; FAB, fast-atom bombardment; HIR, heat-induced radiosensitization; HMBC, heteronuclear multiple bond coherence spectroscopy; HMQC, heteronuclear multiple quantum coherence spectroscopy; HPLC, high pressure liquid chromatography; HSE, heat shock element; HSF1, heat shock factor 1; HSP, heat shock protein; IC50, 50% inhibitory concentration; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MeOH, methanol; MK2 (MAPKAPK2), MAPK-activated protein kinase 2; THF, tetrahydrofuran; TLC, thin layer chromatography; Tris, tris(hydroxymethyl)aminomethane.
Supporting Information Available: HPLC purity analysis of compounds D1-D10. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 2.Arya R, Mallik M, Lakhotia SC. Heat shock genes - integrating cell survival and death. J. Biosci. 2007;32:595–610. doi: 10.1007/s12038-007-0059-3. [DOI] [PubMed] [Google Scholar]
- 3.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlesinger MJ. Heat shock proteins. J. Biol. Chem. 1990;265:12111–12114. [PubMed] [Google Scholar]
- 5.Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 6.Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 8.Yufu Y, Nishimura J, Nawata H. High constitutive expression of heat shock protein 90 alpha in human acute leukemia cells. Leuk. Res. 1992;16:597–605. doi: 10.1016/0145-2126(92)90008-u. [DOI] [PubMed] [Google Scholar]
- 9.Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Hoyer-Hansen M, Weber E, Multhoff G, Rohde M, Jaattela M. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J. Exp. Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aghdassi A, Phillips P, Dudeja V, Dhaulakhandi D, Sharif R, Dawra R, Lerch MM, Saluja A. Heat shock protein 70 increases tumorigenicity and inhibits apoptosis in pancreatic adenocarcinoma. Cancer Res. 2007;67:616–625. doi: 10.1158/0008-5472.CAN-06-1567. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood SK, Ciocca DR. Heat shock proteins: stress proteins with Janus-like properties in cancer. Int. J. Hyperthermia. 2008;24:31–39. doi: 10.1080/02656730701858305. [DOI] [PubMed] [Google Scholar]
- 12.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- 14.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanbu K, Konishi I, Komatsu T, Mandai M, Yamamoto S, Kuroda H, Koshiyama M, Mori T. Expression of heat shock proteins HSP70 and HSP90 in endometrial carcinomas. Correlation with clinicopathology, sex steroid receptor status, and p53 protein expression. Cancer. 1996;77:330–338. doi: 10.1002/(SICI)1097-0142(19960115)77:2<330::AID-CNCR16>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Kaur J, Ralhan R. Induction of apoptosis by abrogation of HSP70 expression in human oral cancer cells. Int. J. Cancer. 2000;85:1–5. doi: 10.1002/(sici)1097-0215(20000101)85:1<1::aid-ijc1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Maehara Y, Oki E, Abe T, Tokunaga E, Shibahara K, Kakeji Y, Sugimachi K. Overexpression of the heat shock protein HSP70 family and p53 protein and prognosis for patients with gastric cancer. Oncology. 2000;58:144–151. doi: 10.1159/000012091. [DOI] [PubMed] [Google Scholar]
- 18.Lazaris AC, Theodoropoulos GE, Davaris PS, Panoussopoulos D, Nakopoulou L, Kittas C, Golematis BC. Heat shock protein 70 and HLA-DR molecules tissue expression. Prognostic implications in colorectal cancer. Dis. Colon Rectum. 1995;38:739–745. doi: 10.1007/BF02048033. [DOI] [PubMed] [Google Scholar]
- 19.Marunouchi T, Hosoya H. Regulation of hsc70 expression in the human histiocytic lymphoma cell line, U937. Cell Struct. Funct. 1993;18:437–447. doi: 10.1247/csf.18.437. [DOI] [PubMed] [Google Scholar]
- 20.Ciocca DR, Clark GM, Tandon AK, Fuqua SA, Welch WJ, McGuire WL. Heat shock protein hsp70 in patients with axillary lymph node-negative breast cancer: prognostic implications. J Natl Cancer Inst. 1993;85:570–574. doi: 10.1093/jnci/85.7.570. [DOI] [PubMed] [Google Scholar]
- 21.Ciocca DR, Fuqua SA, Lock-Lim S, Toft DO, Welch WJ, McGuire WL. Response of human breast cancer cells to heat shock and chemotherapeutic drugs. Cancer Res. 1992;52:3648–3654. [PubMed] [Google Scholar]
- 22.Didelot C, Lanneau D, Brunet M, Joly AL, De Thonel A, Chiosis G, Garrido C. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr. Med. Chem. 2007;14:2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- 23.Neckers L, Neckers K. Heat-shock protein 90 inhibitors as novel cancer chemotherapeutic agents. Expert Opin. Emerg. Drugs. 2002;7:277–288. doi: 10.1517/14728214.7.2.277. [DOI] [PubMed] [Google Scholar]
- 24.Russell JS, Burgan W, Oswald KA, Camphausen K, Tofilon PJ. Enhanced cell killing induced by the combination of radiation and the heat shock protein 90 inhibitor 17-allylamino-17- demethoxygeldanamycin: a multitarget approach to radiosensitization. Clin. Cancer Res. 2003;9:3749–3755. [PubMed] [Google Scholar]
- 25.Dote H, Cerna D, Burgan WE, Camphausen K, Tofilon PJ. ErbB3 expression predicts tumor cell radiosensitization induced by Hsp90 inhibition. Cancer Res. 2005;65:6967–6975. doi: 10.1158/0008-5472.CAN-05-1304. [DOI] [PubMed] [Google Scholar]
- 26.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei YQ, Zhao X, Kariya Y, Teshigawara K, Uchida A. Inhibition of proliferation and induction of apoptosis by abrogation of heat-shock protein (HSP) 70 expression in tumor cells. Cancer Immunol. Immunother. 1995;40:73–78. doi: 10.1007/BF01520287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol. Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 31.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, Eriksson JE, Sistonen L. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soncin F, Zhang X, Chu B, Wang X, Asea A, Ann Stevenson M, Sacks DB, Calderwood SK. Transcriptional activity and DNA binding of heat shock factor-1 involve phosphorylation on threonine 142 by CK2. Biochem. Biophys. Res. Commun. 2003;303:700–706. doi: 10.1016/s0006-291x(03)00398-x. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Khaleque MA, Zhao MJ, Zhong R, Gaestel M, Calderwood SK. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. J. Biol. Chem. 2006;281:782–791. doi: 10.1074/jbc.M505822200. [DOI] [PubMed] [Google Scholar]
- 34.Dai R, Frejtag W, He B, Zhang Y, Mivechi NF. c-Jun NH2-terminal kinase targeting and phosphorylation of heat shock factor-1 suppress its transcriptional activity. J. Biol. Chem. 2000;275:18210–18218. doi: 10.1074/jbc.M000958200. [DOI] [PubMed] [Google Scholar]
- 35.Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581:3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 36.Westerheide SD, Kawahara TL, Orton K, Morimoto RI. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J. Biol. Chem. 2006;281:9616–9622. doi: 10.1074/jbc.M512044200. [DOI] [PubMed] [Google Scholar]
- 37.Pietta PG. Flavonoids as antioxidants. J. Nat. Prod. 2000;63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- 38.Choi JA, Kim JY, Lee JY, Kang CM, Kwon HJ, Yoo YD, Kim TW, Lee YS, Lee SJ. Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int. J. Oncol. 2001;19:837–844. doi: 10.3892/ijo.19.4.837. [DOI] [PubMed] [Google Scholar]
- 39.Zanini C, Giribaldi G, Mandili G, Carta F, Crescenzio N, Bisaro B, Doria A, Foglia L, di Montezemolo LC, Timeus F, Turrini F. Inhibition of heat shock proteins (HSP) expression by quercetin and differential doxorubicin sensitization in neuroblastoma and Ewing’s sarcoma cell lines. J. Neurochem. 2007;103:1344–1354. doi: 10.1111/j.1471-4159.2007.04835.x. [DOI] [PubMed] [Google Scholar]
- 40.Fujita M, Nagai M, Murata M, Kawakami K, Irino S, Takahara J. Synergistic cytotoxic effect of quercetin and heat treatment in a lymphoid cell line (OZ) with low HSP70 expression. Leuk. Res. 1997;21:139–145. doi: 10.1016/s0145-2126(96)00080-x. [DOI] [PubMed] [Google Scholar]
- 41.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DJ, Lamb JH, Verschoyle RD, Howells LM, Butterworth M, Lim CK, Ferry D, Farmer PB, Gescher AJ. Characterisation of metabolites of the putative cancer chemopreventive agent quercetin and their effect on cyclo-oxygenase activity. Br. J. Cancer. 2004;91:1213–1219. doi: 10.1038/sj.bjc.6602091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pirrung MC, Liu Y, Deng L, Halstead DK, Li Z, May JF, Wedel M, Austin DA, Webster NJ. Methyl scanning: total synthesis of demethylasterriquinone B1 and derivatives for identification of sites of interaction with and isolation of its receptor(s) J. Am. Chem. Soc. 2005;127:4609–4624. doi: 10.1021/ja044325h. [DOI] [PubMed] [Google Scholar]
- 44.Kim H, Deng L, Xiong X, Hunter WD, Long MC, Pirrung MC. Glyceraldehyde 3-phosphate dehydrogenase is a cellular target of the insulin mimic demethylasterriquinone B1. J. Med. Chem. 2007;50:3423–3426. doi: 10.1021/jm070437i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouktaib M, Lebrun S, Atmani A, Rolando C. Hemisynthesis of all the O-monomethylated analogues of quercetin including the major metabolites, through selective protection of phenolic functions. Tetrahedron. 2002;58:10001–10009. [Google Scholar]
- 46.Jurd L. Plant Polyphenols.V.Selective alkylation of the 7-hydroxyl group in polyhydroxyflavones. J.Am.Chem.Soc. 1958;80:6. [Google Scholar]
- 47.Picq M, Prigent AF, Nemoz G, Andre AC, Pacheco H. Pentasubstituted quercetin analogues as selective inhibitors of particulate 3′;5′-Cyclic-AMP phosphodiesteras from rat brain. J.Med.Chem. 1982;25:7. doi: 10.1021/jm00352a019. [DOI] [PubMed] [Google Scholar]
- 48.Ono M, Itoh I. N-Methyl-2-(dimethylamino)acetohydroxamic acid as a new reagent for the selective cleavage of active esters under neutral conditions. Tetrahedron Lett. 1989;30:207–210. [Google Scholar]
- 49.Li M, Han X, Yu B. Facile synthesis of flavonoid 7-O-glycosides. J. Org. Chem. 2003;68:6842–6845. doi: 10.1021/jo034553e. [DOI] [PubMed] [Google Scholar]
- 50.Worzella T, Gallagher A. Optimizing kinase assays for ultrahigh-throughput profiling using the Kinase-Glo Plus Assay. JALA. 2007;12:99–103. [Google Scholar]
- 51.Hosokawa N, Hirayoshi K, Kudo H, Takechi H, Aoike A, Kawai K, Nagata K. Inhibition of the activation of heat shock factor in vivo and in vitro by flavonoids. Mol. Cell Biol. 1992;12:3490–3498. doi: 10.1128/mcb.12.8.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 53.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 54.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 55.Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992;313:307–313. doi: 10.1016/0014-5793(92)81216-9. [DOI] [PubMed] [Google Scholar]
- 56.Garrido C, Ottavi P, Fromentin A, Hammann A, Arrigo AP, Chauffert B, Mehlen P. HSP27 as a mediator of confluence-dependent resistance to cell death induced by anticancer drugs. Cancer Res. 1997;57:2661–2667. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.