Abstract
Background
5’-Nucleotidases play a critical role in nucleotide pool balance and in the metabolism of nucleoside analogs such as gemcitabine and cytosine arabinoside (AraC). We previously performed an expression array association study with gemcitabine and AraC cytotoxicity using 197 human lymphoblastoid cell lines. One gene that was significantly associated with gemcitabine cytotoxicity was a nucleotidase family member, NT5C3. Very little is known with regard to the pharmacogenomics of this family of enzymes.
Methods
We set out to identify common genetic variation in NT5C3 by resequencing the gene and to determine the effect of that variation on NT5C3 protein function and potential effect on response to cytidine analogues. We identified 61 NT5C3 polymorphisms, 48 of which were novel, by resequencing 240 ethnically defined DNA samples. Functional studies were performed with one nonsynonymous (G847C, Asp283His) and 4 synonymous cSNPs (T9C, C276T, T306C, and G759A), as well as three combined variants (T276/His283, T276/C306, T276/C9).
Results
The His283 and T276/His283 constructs showed decreased levels of enzyme activity and protein. Substrate kinetic analysis showed no significant differences in Km values between WT and His283 when CMP, AraCMP and GemMP were used as substrates. An association study between SNPs and NT5C3 expression in the 240 cell lines from which DNA was extracted to resequence NT5C3 identified 4 SNPs that were significantly associated with NT5C3 expression. EMSAs showed that two of those SNPs, I4(-114) and I6(9), altered DNA-protein binding patterns. These findings suggest that genetic variation in NT5C3 might affect protein function and potentially influence drug response.
Keywords: Gemcitabine, cytosine arabinoside, cytosolic 5’-nucleotidase III, pharmacogenomics, single nucleotide polymorphisms (SNPs), functional genomics
Introduction
The 5’-nucleotidases are a family of enzymes that catalyze the dephosphorylation of various nucleoside 5’-monophosphates [1]. One family member, cytoplasmic 5-nucleotidase-III (NT5C3), mainly catalyzes the dephosphorylation of pyrimidine nucleoside monophosphates, including nucleoside analogs such as gemcitabine and AraC that are used to treat cancer. This reaction results in the deactivation of active phosphorylated metabolites [2, 3]. Large variations have been observed in response to these drugs, and one possible explanation is that genetic variation in genes involved in the activation and inactivation of these drugs might contribute to differences in drug response. We previously reported that NT5C3 gene expression was significantly associated with cytotoxicity for both gemcitabine and AraC in human lymphoblastoid cell lines [4]. We also showed that “knockdown” of NT5C3 altered cancer cell sensitivity to both of these drugs [4]. Furthermore, higher NT5C3 gene expression correlated with lower intracellular gemcitabine and AraC phosphorylated metabolite concentrations. To define the nature and extent of common genetic variation in NT5C3 and the possible functional effect of that variation on NT5C3 function, we have now performed a resequencing study of NT5C3 using 240 ethnically defined DNA samples, followed by functional characterization of coding region SNPs and SNPs associated with NT5C3 gene expression in the lymphoblastoid cell lines from which DNA used to resequenced the gene was obtained. Results of this study provide information with regard to common sequence variation in NT5C3, as well as the functional consequences of that sequence variation.
Materials and Methods
DNA samples
DNA samples from 60 Caucasian-American (CA), 60 African-American (AA), 60 Han Chinese-American (HCA) and 60 Mexican-American (MA) unrelated subjects (sample sets HD100CAU, HD100AA, HD100CHI, HD100MEX) were obtained from the Coriell Cell Repository (Camden, NJ). All of these DNA samples had been collected and anonymized by the National Institute of General Medical Sciences before deposit, and all subjects had provided written consent for the use of their DNA for experimental purposes. The present study was reviewed and approved by the Mayo Clinic Institutional Review Board.
NT5C3 gene resequencing
Each of the 240 DNA samples studied was used to perform PCR amplifications of all NT5C3 exons, splice junctions and a portion of the 5’-FR. Primer sequences and PCR amplification conditions are listed in the Supplementary Material (Table 1). Amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility with an ABI 3700 DNA sequencer using BigDye™ dye terminator sequencing chemistry (Perkin-Elmer, Boston, MA). To exclude PCR-related artifacts, independent amplifications were performed for any SNP observed in only a single DNA sample or for any sample with an ambiguous chromatogram. The sequencing chromatograms were analyzed using Mutation Surveyor version 2.2 [5]. GenBank accession numbers for the NT5C3 reference sequences used in these experiments were NT_007819.16 and NM_001002009.1. All sequence data were deposited in PharmGKB with accession number PA31802.
NT5C3 expression constructs and transient expression
The wild type (WT) cDNA open reading frame (ORF) for four different NT5C3 isoforms were cloned into the eukaryotic expression vector pcDNA3.1 D/V5-His-TOPO vector (Invitrogen, Carlsbad, CA). The WT construct encoding 297 amino acid protein was then used to perform site-directed mutagenesis with the QuikChange kit (Stratagene, La Jolla, CA). The sequences of primers used to perform site-directed mutagenesis are also listed in the Supplementary Material (Table 1). The DNA sequences of all inserts were confirmed by sequencing. COS-7 cells were then transfected with expression constructs encoding WT and variant NT5C3 allozymes, as well as “empty” vector that lacked an insert as a control, using the TransFast reagent (Promega, Madison, WI) at a charge ratio of 1:1. After 48 h, the cells were harvested in a buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2 and 1 mM dithiothreitol. The cells were homogenized with a Polytron homogenizer (Brinkmann Instruments, Westbury, NY), followed by centrifugation at 100,000 × g for 1 h, and supernatant preparations were stored at - 80°C for use in functional genomic studies.
NT5C3 enzyme assays and substrate kinetic studies
Nucleotidase activity was measured with an HPLC-based assay. Specifically, 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2 and 1 mM DTT were mixed with the substrate, CMP, 0.004-2.5 mM, and recombinant NT5C3 in a final volume of 500 μl. The reaction mixture was incubated at 37°C for 60 min, and the reaction was stopped by adding 100 μl of the assay mixture to 50 μl of ice-cold 1.2M HClO4, followed by incubation for 10 min at 0°C. This mixture was then centrifuged for 1 min, and 130 μl of the supernatant was collected and neutralized with 35μl 1M K2CO3, followed by centrifugation. The supernant was injected onto an HPLC column to measure the reaction product, cytidine. The HPLC was equipped with a 150 mm × 4.6 mm C18-reverse phase column and was eluted with 50 mM potassium phosphate buffer, pH 5.0, at a flow rate of 1 ml/min, as described previously [6].
Western blot analysis
COS-7 cell cytosol was subjected to electrophoresis on 10 % Tris-HCl acrylamide gels loaded on the basis of cotransfected β-galactosidase activity measured with the Promega β-Galactosidase Enzyme Assay System (Promega, Madison, WI). Proteins were then transferred to PVDF membranes (BioRad), and the membranes were incubated with rabbit polyclonal anti-NT5C3 antibody (GenWay Biotech., San Diego, CA) or anti-HA antibody (Sigma-Aldrich, Inc., St. Louis, MO), followed by secondary antibody. Immunoreactive proteins were detected using the ECL Western Blotting System (Amersham Pharmacia, Piscataway, NJ). The IPLab Gel H (Biosystemetica, Plymouth, UK) system and the NIH image program (http//rsb.info.nih.gov/nih-image) were used to quantify immunoreactive proteins, and the data were expressed as a percentage of the WT protein intensity on the same gel.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed with the LightShift™ Chemiluminescent EMSA Kit (Pierce, Rockford, IL). Nucleotide sequences of biotin-labeled oligonucleotides for the I4(-114) SNP were 5’-AAAGTATTGGG/ATTGGTGCAAA-3’ and were for the I6(9) SNP 5’-GGTAAGTGTA/GTATTAGGCA-3’. Specifically, each reaction (20 μl), contained 100 fmol of 3’-end labeled probe, 10 μg of nuclear extract prepared from COS-7, HepG2 or Su86 cells, 1 μg poly(dI-dC), 2.5% glycerol, 0.05% NP-40, 5 mM MgCl2 and 1 × binding buffer. A 100-fold molar excess of unlabeled probe was used to perform competition experiments. The mixtures were incubated for 45 min at room temperature. Samples were separated on a non-denaturing 5% polyacrylamide gel at 4°C and were then transferred to a nylon membrane (Pierce, Rockford, IL). DNA-protein interactions were detected by use of the Chemiluminescant Nucleic Acid Detection Module Kit (Pierce, Rockford, IL) and were visualized by autoradiography.
NT5C3 expression and exon array analysis
Expression array analyses were performed using Affymetrix U133 Plus 2.0 GeneChips as described previously [4]. Exon array analyses were performed with total RNA isolated from 8 randomly selected lymphoblastoid cells. Biotin-labeled cDNA was fragmented and hybridized to Affymetrix Human Exon 1.0 ST Array chips. The exon array data were normalized by GCRMA using the Partek® Genomics Suite (http://www.partek.com/software) [7].
Human NT5C3 homology model
Three unpublished NT5C3 crystal structures are currently publicly available (PDB accession numbers 2CN1, 2JGA, and 2VKQ). We chose to use 2VKQ because it had the highest resolution of the 3 structures. The amino acid substitution for the single nonsynonymous cSNP observed was modeled using molecular figures prepared using Molscript [8] and Raster3D [9].
Data analysis
Values for π, θ and Tajima’s D were calculated as described by Tajima [10]. D’ values, a measure of linkage disequilibrium that is independent of allele frequency, were calculated as described by Hartl and Clark [11] and Hedrick [12]. Haplotype analysis was performed as described by Schaid et al. [13] using the E-M (expectation-maximization) algorithm. The association analysis between genetic polymorphisms and NT5C3 mRNA expression array data was performed with PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) [14]. Least square regression was used to perform the association in PLINK. Mean protein and Km values were compared using students t-test and ANOVA (GraphPad Software Inc., La Jolla, CA).
Results
NT5C3 gene resequencing
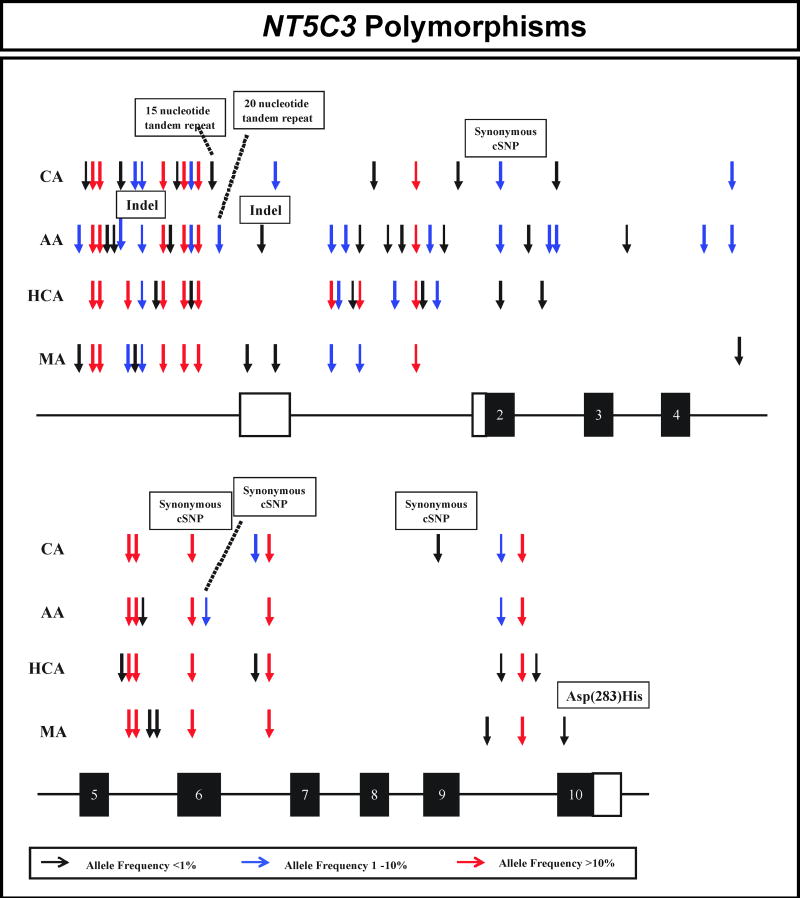
The areas of NT5C3 resequenced included exons, splice junctions, and a portion of the 5’-FR using 240 DNA samples from 4 ethnic groups. The polymorphisms observed in NT5C3 are listed in Table 1 and are displayed graphically in Figure 1. Sixty-one polymorphisms, including 57 SNPs, 2 indels and 2 tandem repeats were observed. One nonsynonymous (G847C, Asp283His) and 4 synonymous cSNPs, T9C, C276T, T306C and G759A, were identified (Table 1). The allele encoding His283 was observed in only a single MA sample. Three of the four synonymous SNPs had an allele frequency greater than 1.0% in at least one ethnic group (Table 1 and Figure 1). Forty-eight of the 61 NT5C3 polymorphisms were not present in public databases (Table 1). All polymorphisms identified were deposited in the NIH PharmGKB database.
Table 1.
Human NT5C3 polymorphisms. Locations in the gene, nucleotide sequence alterations, amino acid sequence alterations, and minor allele frequencies of polymorphisms in the four populations studied are listed. If a polymorphism had already been deposited in a public database (dbSNP or HapMap), an rs number is indicated. All other polymorphisms listed are unique to this study. Polymorphisms within exons have been boxed. IVS is intervening sequence. The numbering scheme for nucleotide positions is based on assignment of the (+1) position to the “A” in the translation initiation codon, with nucleotides 5’ to that position assigned negative, and those 3’ within the cDNA assigned positive numbers. Nucleotide positions in introns are numbered from the nearest splice site, with distances from 3’ splice junction assigned positive and distances from 5’ splice junctions assigned negative numbers. AA, African-American; CA, Caucasian-American; HCA, Han Chinese-American; MA, Mexican-American; FR, flanking region; UTR, untranslated region.
| NT5C3 Polymorphism | ||||||||
|---|---|---|---|---|---|---|---|---|
| Frequency of Variant Allele |
||||||||
| Gene Location | Nucleotide | Nucleotide Change | Amino Acid Change | AA | CA | HCA | MA | HapMap and/or dbSNP |
| 5’-FR | -1273 | A→C | 0.100 | 0.000 | 0.000 | 0.008 | ||
| 5’-FR | -1270 | T→C | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5’-FR | -1145 | T→G | 0.200 | 0.358 | 0.258 | 0.442 | rs34307182 | |
| 5’-FR | -1143 | G→A | 0.283 | 0.292 | 0.317 | 0.133 | rs6976843 | |
| 5’-FR | -1018 | T→C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 5’-FR | -944 | T→C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 5’-FR | -884 | G insertion | 0.033 | 0.008 | 0.000 | 0.000 | ||
| 5’-FR | -693 | C→G | 0.000 | 0.000 | 0.142 | 0.017 | ||
| 5’-FR | -656 | C→T | 0.000 | 0.017 | 0.000 | 0.008 | ||
| 5’-FR | -654 | G→A | 0.017 | 0.042 | 0.092 | 0.083 | ||
| 5’-FR | -653 | T→C | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 5’-FR | -552 | G→C | 0.333 | 0.342 | 0.417 | 0.250 | rs10262141 | |
| 5’-FR | -496 | C→T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 5’-FR | -373 | C→T | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5’-FR | -354 | T→C | 0.225 | 0.342 | 0.275 | 0.225 | rs13228827 | |
| 5’-FR | -340 | T→G | 0.042 | 0.017 | 0.008 | 0.000 | ||
| 5’-FR | -302 | A→G | 0.333 | 0.342 | 0.417 | 0.250 | rs13228639 | |
| 5’-FR | -267 | 15 nucleotide tandem repeat | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5’-FR | -258 | 20 nucleotide tandem repeat | 0.025 | 0.000 | 0.000 | 0.000 | ||
| 5’-UTR | -194 | T→C | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 5’-UTR | -134 | deletion of GGTGGG | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 5’-UTR | -67 | T→C | 0.000 | 0.033 | 0.000 | 0.008 | ||
| IVS 1 | -5780 | C→G | 0.025 | 0.000 | 0.133 | 0.017 | ||
| IVS 1 | -5767 | A→T | 0.000 | 0.000 | 0.025 | 0.000 | ||
| IVS 1 | -5757 | A→T | 0.017 | 0.000 | 0.000 | 0.000 | ||
| IVS 1 | -5756 | T→A | 0.000 | 0.000 | 0.008 | 0.000 | ||
| IVS 1 | -5714 | A→G | 0.008 | 0.000 | 0.133 | 0.017 | ||
| IVS 1 | -5705 | C→A | 0.000 | 0.008 | 0.000 | 0.000 | ||
| IVS 1 | -5654 | A→G | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 1 | -5635 | G→A | 0.000 | 0.000 | 0.017 | 0.000 | ||
| IVS 1 | -5633 | A→G | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 1 | -5418 | G→A | 0.225 | 0.342 | 0.283 | 0.225 | rs10230500 | |
| IVS 1 | -5125 | T→G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| IVS 1 | -5059 | G→A | 0.050 | 0.000 | 0.000 | 0.000 | ||
| IVS 1 | -4900 | G→A | 0.000 | 0.000 | 0.058 | 0.000 | ||
| IVS 1 | -4645 | G→T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 1 | -19 | A→C | 0.000 | 0.009 | 0.000 | 0.000 | ||
| Exon 2 | 9 | T→C | 0.033 | 0.017 | 0.008 | 0.000 | rs17170223 | |
| IVS 2 | 139 | A→G | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 2 | -307 | T→A | 0.000 | 0.000 | 0.008 | 0.000 | ||
| IVS 2 | -237 | G→A | 0.017 | 0.000 | 0.000 | 0.000 | ||
| IVS 2 | -4 | T→G | 0.033 | 0.008 | 0.000 | 0.000 | ||
| IVS 3 | 103 | C→T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 4 | 35 | C→T | 0.050 | 0.000 | 0.000 | 0.000 | ||
| IVS 4 | -114 | G→A | 0.025 | 0.017 | 0.008 | 0.000 | rs11974256 | |
| IVS 5 | 77 | C→G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| IVS 5 | 266 | G→A | 0.250 | 0.342 | 0.417 | 0.242 | rs3750119 | |
| IVS 5 | 430 | G→A | 0.250 | 0.342 | 0.417 | 0.242 | rs3750118 | |
| IVS 5 | 533 | T→G | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 5 | 548 | A→T | 0.000 | 0.000 | 0.000 | 0.008 | ||
| IVS 5 | 647 | G→T | 0.000 | 0.000 | 0.000 | 0.008 | ||
| Exon 6 | 276 | C→T | 0.250 | 0.342 | 0.417 | 0.242 | rs3750117 | |
| Exon 6 | 306 | T→C | 0.100 | 0.000 | 0.000 | 0.000 | ||
| IVS 6 | 9 | A→G | 0.000 | 0.042 | 0.000 | 0.008 | ||
| IVS 6 | -71 | C→T | 0.117 | 0.325 | 0.275 | 0.225 | rs2392209 | |
| Exon 9 | 759 | G→A | 0.000 | 0.008 | 0.000 | 0.000 | ||
| IVS 9 | 52 | T→A | 0.000 | 0.000 | 0.000 | 0.008 | ||
| IVS 9 | 61 | A→G | 0.025 | 0.017 | 0.008 | 0.000 | ||
| IVS 9 | -267 | A→G | 0.225 | 0.342 | 0.283 | 0.225 | rs2893457 | |
| IVS 9 | -38 | C→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| Exon 10 | 847 | G→C | Asp (283) His | 0.000 | 0.000 | 0.000 | 0.008 | |
Figure 1.
Human NT5C3 genetic polymorphisms. Arrows indicate the locations of polymorphisms, with different colors indicating minor allele frequencies. Black rectangles represent portions of exons encoding the open-reading frame (ORF). Open rectangles represent portions of exons encoding 5’ and 3’-UTRs. AA is African-American, CA is Caucasian-American, HCA is Han Chinese-American, MA is Mexican-American, and indel is an insertion/deletion event.
We also determined “nucleotide diversity”, a quantitative measure of genetic variation, adjusted for allele frequency, in all four ethnic groups by calculating θ, a population mutation measure that is theoretically equal to the neutral mutation variable, and π, average heterozygosity per site [5]. Values for Tajima’s D, a test of the neutral mutation hypothesis, were also estimated (Table 2). Although negative values for Tajima D indicate a departure from neutrality, none of the values listed in Table 2 was statistically significant.
Table 2.
NT5C3 genetic diversity and neutrality test values. Estimates of two measures of nucleotide diversity, π and θ, are listed, as well as Tajima’s D, a test of the “neutral” mutation hypothesis. The p values refer to Tajima’s D.
| Population | π × 104 | θ × 104 | Tajima’s D | P value |
|---|---|---|---|---|
| African-American | 8.62 ± 4.5 | 11.15 ± 3.2 | -0.691 | 0.51 |
| Caucasian-American | 8.97 ± 4.8 | 8.39 ± 2.5 | 0.251 | 0.81 |
| Han Chinese-American | 9.88 ± 5.2 | 8.41 ± 2.5 | 0.517 | 0.62 |
| Mexican-American | 7.06 ± 3.9 | 7.26 ± 2.2 | -0.079 | 0.94 |
Haplotype and linkage disequilibrium analysis
We also performed population-specific linkage disequilibrium and haplotype analysis for NT5C3. A total of 22 haplotypes had a frequency of over 1% (Supplementary Material Table 2). Haplotype designations were based on the encoded amino acid sequence of the allozyme, with the WT sequence designated as *1. The *2 haplotype contained the SNP that encoded His283. Letter designations were then added based on decreasing frequencies, using AA samples as the “base”. The haplotype containing the nonsynonymous cSNP had a frequency of less than 1%, but was included in the table because of the variant amino acid sequence (Supplementary Material Table 2). Haplotype frequencies differed greatly among the four ethnic groups. For example, 11 haplotypes with an allele frequency over 1% were observed in AA subjects while 8, 9 and 6 were observed in CA, HCA and MA subjects, respectively (Supplementary Material Table 2). *1B and *1C were observed with high frequency in all four populations, while many haplotypes were observed in only a single ethnic group (Supplementary Material Table 2). Graphical representations of population-specific D’ values across the NT5C3 gene for the four populations studied are shown in Supplementary Figure 1.
Functional genomic studies
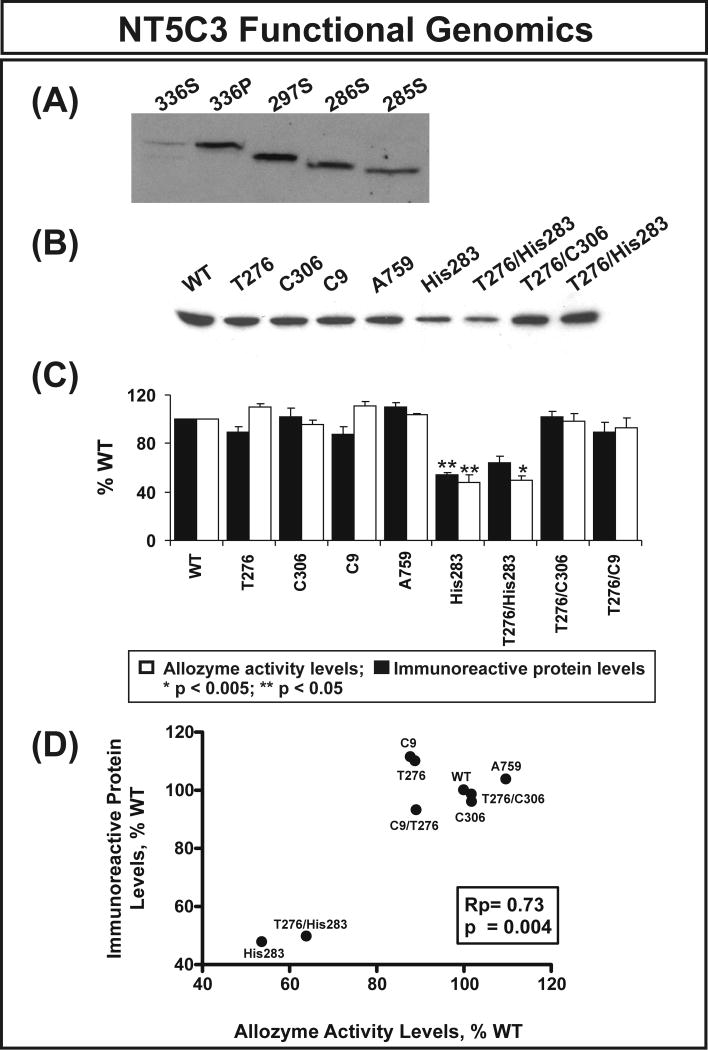
Expression constructs for the NT5C3 WT and the His283 variant allozyme were created to determine the effect of the nonsynonymous SNP on levels of protein, enzyme activity and substrate kinetics. However, before performing these studies, we had to decide which constructs to create since several alternatively spliced NT5C3 transcripts have been described. Specifically, four different NT5C3 isoforms can be expressed. Isoforms 1 (297 amino acids) and 3 (286 amino acids) [15] are expressed in reticulocytes and lymphocytes. Isoform 4 (285 amino acids) [16] is expressed only in reticulocytes, and isoform 2 (336 amino acids) [17] is induced in Raji cells by interferon alpha in lupus inclusions (http://beta.uniprot.org/uniprot/Q9H0P0). In order to determine whether there might be differences among isoforms in terms of catalytic ability, we created expression constructs for isoforms consisting of 285, 286, 297 and 336 amino acids. To determine whether these constructs encoded the expected protein, supernatants from cells transfected with 4 isoforms were used to perform Western blot analysis using anti-HA antibody. Since the 336 isoform is membrane-bound, we also included cell pellet for the 336 isoform in the Western blot analysis. As showed in Figure 2A, all four constructs encode proteins with the expected molecular weight and the majority of the 336 isoform was in the cell pellet, as anticipated. Apparent Km values for the 297, 286 and 285 amino acid residue proteins were 145±5.6, 413±37 and 321±24 μM, respectively, with CMP as a substrate (Table 3A). The 336 amino acid isoform had no activity with CMP as substrate, and none of the isoforms had detectable enzyme activity with IMP as a substrate, consistent with previous reports that NT5C3 only has activity with pyrimidine analogs as substrate [18, 19]. Since the DNA used to resequence the NT5C3 gene had been isolated from lymphoblastoid cell lines, we performed exon array analysis using RNA from 8 pooled lymphoblastoid cell lines to determine which isoform was most highly expressed in those cells. We found that the 297 amino acid isoform (NM_001002009.1) was highly expressed. Therefore, the expression constructs that we studied in the next series of experiments were created based on the 297 amino acid isoform.
Figure 2.
NT5C3 allozyme functional genomics. (A) Western blot analysis with anti-HA antibody for four different NT5C3 isoforms. Expression constructs encoding 4 different transcripts were transfected into COS-1 cells. Cells were then lyzed and cell supernatant (S) and pellet (P) were used for the analysis as indicated. (B) Representative Western blot analysis for the nonsynonymous and synonymous cSNPs studied. (C) Levels of NT5C3 allozyme activity and immunoreactive protein as a percentage of WT. Each bar represents the average of 6 independent transfections (mean±SE). All values were corrected for transfection efficiency. (D) Correlation between average levels of NT5C3 immunoreactive protein and enzyme activity.
Table 3.
(A) 5’-Nucleotidase enzyme activity was measured in COS-7 cell lines transfected with different NT5C3 isoforms. Activity was measured by HPLC with CMP as a substrate. No activity was detected IMP as substrate. ND is nondetectable. * p <0.005 when compared with both isoform 3 and isoform 4. (B) Recombinant NT5C3 (297 amino acid isoform) allozyme substrate kinetics. CMP, Ara-CMP and GemMP substrate kinetics for WT and variant NT5C3 allozymes were determined. Values listed represent mean ± SEM (n ≥ 6).
| (A) | ||
|---|---|---|
| NT5C3Isoforms | Km (μM) | |
| Isoform 1 (336 amino acids) NM_001002010 | ND | |
| Isoform 2 (297 amino acids) NM_001002009 | 145 ± 5.6* | |
| Isoform 3 (286 amino acids) | 413 ± 37 | |
| Isoform 4 (285 amino acids) | 321 ± 24 | |
| (B) | ||
| Apparent KmValues for NT5C3 (297 amino acids) Allozymes (μM) | ||
| Substrates | WT | His283 |
| CMP | 145 ± 5.6 | 133 ± 11 |
| Ara-CMP | 100 ± 5.0 | 91 ± 3.1 |
| Gem-MP | 88 ± 6.1 | 78 ± 5.0 |
In addition to the nonsynonymous cSNP that we had observed in a single MA sample, we also created expression constructs for the 4 common synonymous SNPs, since synonymous SNPs have also been shown to influence mRNA levels [20]. Our haplotype analysis showed tight linkage disequilibrium among several variant NT5C3 alleles, including T276/C9, T276/C306 and T276/His283. Therefore, we also created expression constructs that contained these haplotypes, and all of these NT5C3 expression constructs were transfected into COS-7 cells. The cells were also co-transfected with β-galactosidase to make it possible to correct for possible variation in transfection efficiency. Immunoreactive protein levels were then measured by quantitative Western blot analysis and levels of enzyme activity were measured in the same samples. Levels of NT5C3 immunoreactive protein, as well as the enzyme activity for these constructs are shown graphically in Figure 2. The His283 and T276/His283 variants showed a decrease in protein levels of approximately 50% when compared with WT (Figure 2B). Similar results were observed when enzyme activity was assayed using cytidine monophosphate (CMP) as a substrate (Figure 2C). Previous finding suggested that the most common mechanism by which nonsynonymous cSNP effect function is due to decreased protein level [21]. In order to determine whether NT5C3 nonsynonymous cSNPs had the same effects, we correlated the protein level and enzyme activity for each allozyme and the results showed significant correlation between these two, consistent with previous finding (Rp = 0.73, p = 0.004) (Figure 2D) [22, 23].
Because of the possibility that part of the difference in level of enzyme activity that we had observed for the variant allozyme might be due to an alterations in substrate kinetics, substrate kinetic studies were also performed. Cytidine monophosphate, cytosine arabinoside monophosphate and gemcitabine monophosphate were used as substrates for recombinant allozymes. Ten different substrate concentrations ranging from 0.004 to 2.5 mM were assayed. Apparent Km values observed for each substrate (CMP, Ara-CMP and Gem-MP) are listed in Table 3B. The apparent Km value for the 297 amino acid isoform that we observed was consistent with that found by Pagli and Valentine with CMP as substrate [19]. Although the Km values did not differ significantly among allozymes, it was clear that gemcitabine-MP, CMP and Ara-CMP were good substrates for NT5C3, consistent with our previous observations during an association study [4].
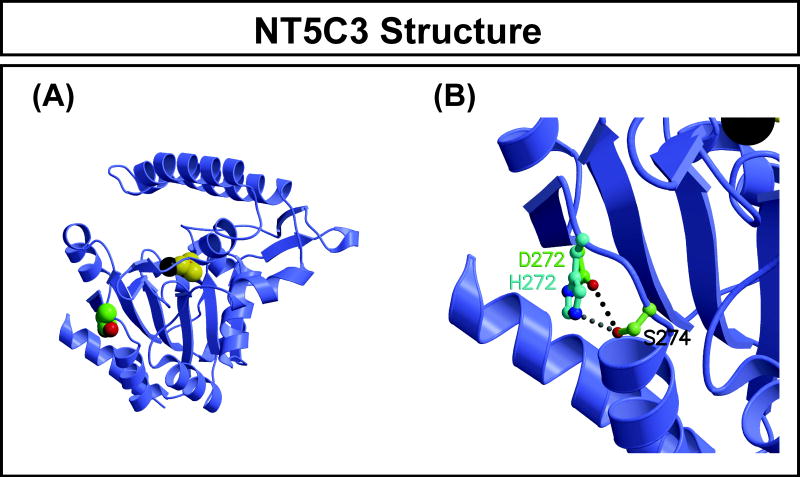
NT5C3 structural model
As mentioned previously, a major mechanism by which nonsynonymous cSNPs affect function is due to their effect on protein quantity [21, 24]. Therefore, we wanted to determine whether the Asp283His SNP might alter protein structure and, as a result, stability. The x-ray crystal structure of NT5C3 has been solved at 2.50 Å (PDB identifier 2VKQ). Therefore, we were able to map the variant amino acid encoded by the nonsynonymous cSNP in NT5C3 onto this structure (Figure 3) and computationally substitute the variant residue to determine whether the variant amino acid was compatible with the WT protein structure. This NT5C3 structure was for the 286-residue isoform with magnesium and beryllium trifluoride ligands bound in the active site, so residue Asp272 in the crystallized isoform corresponds to Asp283 in the 297-residue isoform. The Asp272 side chain is solvent exposed and directed away from the active site. The His272 substitution could be structurally accommodated easily with a simple adjustment of the side chain torsion angles. Since the N-terminal residues are flexible and solvent-accessible, the additional residues in the 297-residue isoform are unlikely to affect the local environment of Asp272/Asp283, although we modeled the variant amino acid on a static structure without considering the dynamic effect of the structure or the cellular environment to which the protein might be exposed.
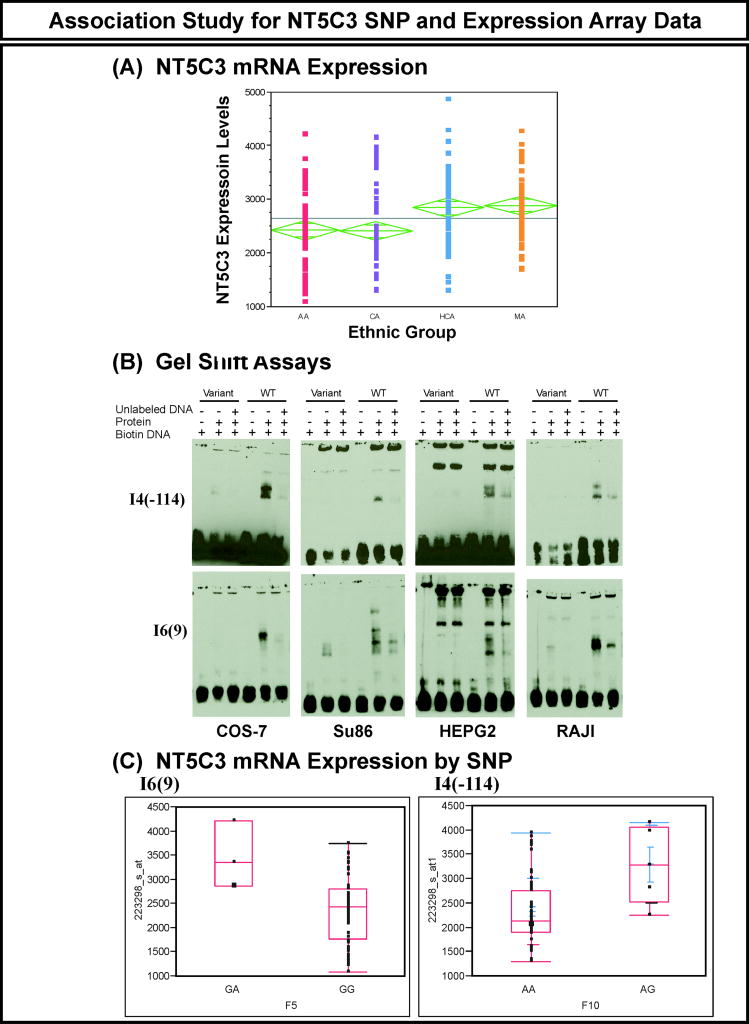
Figure 3.
(A) NT5C3 expression levels in the 240 lymphoblastoid cells studied. Each dot represents one individual sample. Expression level was plotted against each ethnic group. ANOVA analysis was performed to compare the expression level among the 4 ethnic groups. (B) EMSA experiments showing “shifts” observed with I4(-114) and I6(9) WT and variant nucleotides in the presence of nuclear extract from the four cell lines indicated. (C) Quantile boxplot of NT5C3 expression based on genotype at the I6(9) and I4(-114) SNPs.
NT5C3 genotype-phenotype association study
SNPs in regulatory regions can also play an important role in variation in function as a result of their impact on transcription. In order to identify SNPs that might contribute to variation in NT5C3 expression, we performed an association study using NT5C3 SNPs and NT5C3 expression array data for the 240 lymphoblastoid cell lines from which the DNA used to resequence the gene had been obtained. We observed approximately 4-fold variation in basal NT5C3 expression levels in these cell lines. The level of expression was significantly different among ethnic groups, with HCA and MA appearing to have higher expression in these cell lines (Figure 4A). Although none of the SNPs was significant after Bonferroni correction, we functionally characterized the top 3 SNPs, I6(9), I9(61), I4(-114) with uncorrected p values = 0.001, 0.0013 and 0.0013, respectively, as well as one SNP in exon 2 with a p value = 0.03. The translation initiation codon for the 297 amino acid isoform is located in exon 2. We also performed the analysis within each ethnic group I6(9) was the top SNP, with an unadjusted p value= 0.007 in CA samples and I9(61) and I4(-144) were top two candidate SNPs with unadjusted p values =0.01 in AA samples. To determine whether there might be transcription factors binding to regions containing any of these SNPs, we performed EMSAs using oligonucleotides containing either the WT or variant sequences. We used nuclear extracts from four different cell lines, COS-7, HepG2, a pancreatic cancer cell line, SU86, and RAJI cells, to perform the gel shift assays. We observed different protein binding patterns for the WT and variant sequences for two SNPs, I4(-114) and I6(9) (Figure 4B). No shift was observed with either WT or variant sequences for I9(61) or E2(9) (data not shown). We then used the AliBaba (ttp://www.gene-regulation.com/pub/programs.html) and Transfact (http://www.gene-regulation.com/pub/databases.html) programs to search for possible transcription factors that might bind to SNPs that displayed a shift on EMSA. Since the NT5C3 I4(-114) and I6(9) SNPs showed different protein binding patterns, we also performed genotype-phenotype associations for these two SNPs, and the wild type I4(-114)GG and I6(9)AA genotypes were both associated with lower expression levels when compared with heterozygous sequences (Figure 4C). These two SNPs showed strong linkage disequilibrium, with r2 = 0.736. However, these SNPs were not associated with gemcitabine or AraC cytotoxicity (IC50 values) in these cell lines, which might be due to the lack of homozygous variants in these cell lines.
Figure 4.
Human NT5C3 homology model. Three NT5C3 crystal structures are available (PDB accession numbers 2CN1, 2JGA, and 2VKQ). We used 2VKQ because it has the highest resolution of the 3 structures available. This structure contains residues 14-286 (isoform 3), and has BeF3 and Mg ions in the active site. We studied the 297 amino acid isoform (isoform 2) since our lymphoblastoid cell lines express that isoform. Therefore, our nonsynonymous SNP is at position 283 in the 297 amino acid isoform but is located at position 272 in the 286 amino acid isoform. (A) The NT5C3 molecule is shown as a blue ribbon structure. Bound in the active site are magnesium (dark gray) and BeF3- (the pink beryllium is barely visible) with yellow fluorine ions, drawn as space-filling spheres. The side chain of Asp272 is also drawn as space-filling spheres, with green carbon atoms. (B) Closeup of Asp272 and His272 superposition to show that the surface solvent-exposed location of Asp272 provides adequate space for His272 such that it can also form a hydrogen bond with Ser274, although from a slightly different direction.
Discussion
The 5’-nucleotidases are a family of enzymes that catalyzes the dephosphorylation of nucleoside monophosphates, including nucleoside analogs that are used to treat a variety of diseases [3, 25]. This family of enzymes includes 5 cytosolic enzymes as well as one mitochondrial and one membrane-bound nucleotidase [25]. NT5C3 is a cytosolic enzyme that catalyzes the dephosphorylation of pyrimidine analogues [25]. Therefore, it plays an important role in both endogenous nucleoside and nucleotide pool balance as well as response to pyrimidine analogues such as gemcitabine and AraC. In a previous study, we showed that NT5C3 expression is highly associated with gemcitabine and AraC sensitivity using a model system consisting of a large number of lymphoblastoid cell lines [4]. The present study represents an attempt to determine common genetic variation in NT5C3 and its effect on NT5C3 function. Specifically, we performed a comprehensive NT5C3 gene resequencing study using 240 ethnically defined DNA samples, followed by functional genomic characterization.
We observed a total of 61 polymorphisms in this gene, 48 of which were novel. NT5C3 deficiency has been associated with hemolytic anemia [18, 26, 27], and a series of mutations were identified in patients with NT5C3 deficiency [15, 28, 29]. However, none of those mutations was observed in our samples. Since NT5C3 has 4 possible isoforms, we created expression constructs for isoform 1 (336 amino acids), isoform 2 (297 amino acids), isoform 3 (286 amino acids) and isoform 4 (285 amino acids), followed by substrate kinetic studies. We did not detect any activity with the longest isoform that included 336 amino acids. Our results showed that Km values for the 285 and 286 amino acid isoforms differed significantly from that of the 297 amino acid isoform (p<0.005) (Table 3). However, since the 297 residue isoform is highly expressed in the lymphoblastoid cells, our functional studies were performed with this isoform.
Among the SNPs identified during our resequencing study was one nonsynonymous cSNP that changed the encoded amino acid from Asp to His at position 283, as well as 4 common synonymous SNPs within the ORF. Therefore, we created expression constructs for all 5 of these coding SNPs as well as 3 haplotypes that contained two variants each (C9/T276, T276/C306, T276/His283). After transient expression of these constructs in COS-7 cells together with β-galactosidase as a control for transfection efficiency, we performed quantitative Western blot analysis to determine the effect of these SNPs on protein levels. We observed a significant decrease in protein level and enzyme activity for both the His283 and T276/His283 variants, but no difference in apparent Km values (Figure 2, Table 3). This observation was consistent with previous findings that one of the most common mechanisms by which nonsynonymous SNPs affect function is due to decreased protein levels [21, 30]. We did not observe any effect of the synonymous cSNPs, although synonymous cSNPs can affect mRNA secondary structure and stability [20]. A homology model of NT5C3 showed that the His283 variant amino acid is located far from the active site (Figure 3). Therefore, this variant allozyme might not be expected to have a significant impact on substrate kinetics – consistent with our experimental results (see Table 3).
We also determined apparent Km values for two cytidine analog monophosphates, gemcitabine monophosphate and AraC monophosphate. Our previous association study demonstrated an important role for NT5C3 in determining sensitivity to gemcitabine and AraC in lymphoblastoid cell lines [4]. The results of the current study showed that NT5C3 is able to catalyze the dephosphorylation of these two cytidine analog monophosphates with Km values even lower than that for CMP. Not only can SNPs within the coding region have a significant impact on protein function, but also SNPs in regulatory regions can influence function through their effect on transcription. We took advantage of the fact that the DNA samples used for NT5C3 resequencing were isolated from 240 lymphoblastoid cell lines for which we also had expression array data. Therefore, we were able to perform a SNP-NT5C3 expression association study to determine whether any of the SNPs that we had observed might be associated with mRNA levels. Although none of the SNPs showed significant p values after correction for multiple comparisons, we performed EMSAs for 4 SNPs, including the 3 top SNPs on the basis of uncorrected p values and one SNP in exon 2. Gel shift assays demonstrated that two SNPs, I6(9) in intron 6 and I4(-114) in intron 4, showed different DNA-protein binding patterns for variant when compared with WT sequence (Figure 4B). The genotype-phenotype association studies for I6(9) and I4(-114) showed decreased NT5C3 expression with the wild type sequences for these two linked SNPs when compared with heterozygous samples (Figure 4C). Although these two SNP did not show a significant association with gemcitabine and AraC IC50 which could be due to the lack of homozygosity in these lymphoblastoid cell lines, these results indicate that these SNPs should be included in future NT5C3 genotyping studies.
In summary, we have performed a comprehensive series of studies of the pharmacogenomics of a gene encoding an important cytosolic nucleotidase – NT5C3. These experiments resulted in the identification of a large number of novel SNPs and haplotypes that were not represented in the HapMap or other public databases. Functional characterization of the nonsynonymous cSNP encoding Asp283His showed decreased enzyme activity and decreased protein level for the His283 allozyme. Two intron SNPs showed different nuclear protein binding patterns for variant nucleotide sequences, compatible with the results of genotype-phenotype correlation studies between SNPs and NT5C3 expression in 240 lymphoblastoid cell lines. However these two SNPs did not associate with gemcitabine and AraC IC50 for these cell lines, which might be due to the lack of homozygous variant in these cell lines. These results significantly increase our knowledge of common genetic variation in NT5C3 and their effect on function. They may also help to shed light on future translational pharmacogenomic studies of nucleoside analog drugs.
Supplementary Material
Acknowledgments
This study was supported in part by National Institutes of Health (NIH) grants K22 CA130828 (L.W.), R01 CA138461 (L.W.), R01 GM28157 (R.M.W.), R01 GM35720 (R.M.W.), R01 CA132780 (R.M.W.), U01 GM61388 (The Pharmacogenetics Research Network) (R.M.W.) and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award (R.M.W.).
References
- 1.Chiarelli LR, Fermo E, Abrusci P, Bianchi P, Dellacasa CM, Galizzi A, et al. Two new mutations of the P5’N-1 gene found in Italian patients with hereditary hemolytic anemia: the molecular basis of the red cell enzyme disorder. Haematologica. 2006;91:1244–1247. [PubMed] [Google Scholar]
- 2.Donadelli M, Costanzo C, Beghelli S, Scupoli MT, Dandrea M, Bonora A, et al. Synergistic inhibition of pancreatic adenocarcinoma cell growth by trichostatin A and gemcitabine. Biochim Biophys Acta. 2007;1773:1095–1106. doi: 10.1016/j.bbamcr.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maring JG, Groen HJ, Wachters FM, Uges DR, de Vries EG. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5:226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- 6.Amici A, Emanuelli M, Raffaelli N, Ruggieri S, Magni G. One-minute high-performance liquid chromatography assay for 5’-nucleotidase using a 20-mm reverse-phase column. Anal Biochem. 1994;216:171–175. doi: 10.1006/abio.1994.1022. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–8. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- 8.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 9.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 10.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl DL, Clark AG. Organization of genetic variation, in Principles of Population Genetics. Chapter 3. Sinauer Associates, Inc.; Sunderland, MA: 2000. pp. 95–107. [Google Scholar]
- 12.Hedrick PW. Genetics of Populations. 3. Boston, MA: Jones and Bartlett Publishers; 2005. [Google Scholar]
- 13.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marinaki AM, Escuredo E, Duley JA, Simmonds HA, Amici A, Naponelli V, et al. Genetic basis of hemolytic anemia caused by pyrimidine 5’ nucleotidase deficiency. Blood. 2001;97:3327–3332. doi: 10.1182/blood.v97.11.3327. [DOI] [PubMed] [Google Scholar]
- 16.Kanno H, Takizawa T, M S, Fujii H. Molecular basis of Japanese variants of pyrimidine 5’-nucleotidase deficiency. Br J Haematol. 2004;126:265–271. doi: 10.1111/j.1365-2141.2004.05029.x. [DOI] [PubMed] [Google Scholar]
- 17.Rich SA, Bose M, Tempst P, Rudofsky UH. Purification, microsequencing, and immunolocalization of p36, a new interferon-alpha-induced protein that is associated with human lupus inclusions. J Biol Chem. 1996;271:1118–1126. doi: 10.1074/jbc.271.2.1118. [DOI] [PubMed] [Google Scholar]
- 18.Amici A, Magni G. Human erythrocyte pyrimidine 5’-nucleotidase, PN-I. Arch Biochem Biophys. 2002;397:184–190. doi: 10.1006/abbi.2001.2676. [DOI] [PubMed] [Google Scholar]
- 19.Paglia DE, Valentine WN. Characteristics of a pyrimidine-specific 5’-nucleotidase in human erythrocytes. J Biol Chem. 1975;250:7973–7979. [PubMed] [Google Scholar]
- 20.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 21.Weinshilboum R, Wang L. Pharmacogenetics: inherited variation in amino acid sequence and altered protein quantity. Clin Pharmacol Ther. 2004;75:253–258. doi: 10.1016/j.clpt.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Moyer AM, Salavaggione OE, Wu TY, Moon I, Eckloff BW, Hildebrandt MA, et al. Glutathione S-transferase P1: gene sequence variation and functional genomic studies. Cancer Res. 2008;68:4791–4801. doi: 10.1158/0008-5472.CAN-07-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyer AM, Salavaggione OE, Hebbring SJ, Moon I, Hildebrandt MA, Eckloff BW, et al. Glutathione S-transferase T1 and M1: gene sequence variation and functional genomics. Clin Cancer Res. 2007;13:7207–16. doi: 10.1158/1078-0432.CCR-07-0635. [DOI] [PubMed] [Google Scholar]
- 24.Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- 25.Hunsucker SA, Mitchell BS, Spychala J. The 5’-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther. 2005;107:1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Valentine WN, Anderson HM, Paglia DE, Jaffé ER, Konrad PN, Harris SR. Studies on human erythrocyte nucleotide metabolism. II. Nonspherocytic hemolytic anemia, high red cell ATP, and ribosephosphate pyrophosphokinase (RPK, E.C.2.7.6.1) deficiency. Blood. 1972;39:674–684. [PubMed] [Google Scholar]
- 27.Valentine WN, Bennett JM, Krivit W, Konrad PN, Lowman JT, Paglia DE, et al. Nonspherocytic haemolytic anaemia with increased red cell adenine nucleotides, glutathione and basophilic stippling and ribosephosphate pyrophosphokinase (RPK) deficiency: studies on two new kindreds. Br J Haematol. 1973;24:157–167. doi: 10.1111/j.1365-2141.1973.tb05736.x. [DOI] [PubMed] [Google Scholar]
- 28.Chiarelli LR, Fermo E, Zanella A, Valentini G. Hereditary erythrocyte pyrimidine 5’-nucleotidase deficiency: a biochemical, genetic and clinical overview. Hematology. 2006;11:67–72. doi: 10.1080/10245330500276667. [DOI] [PubMed] [Google Scholar]
- 29.Chiarelli LR, Morera SM, Galizzi A, Fermo E, Zanella A, Valentini G. Molecular basis of pyrimidine 5’-nucleotidase deficiency caused by 3 newly identified missense mutations (c.187T>C, c.469G>C and c.740T>C and a tabulation of known mutations. Blood Cells Mol Dis. 2008;40:295–301. doi: 10.1016/j.bcmd.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.