Summary
Lignocellulosic biofuels represent a sustainable, renewable, and the only foreseeable alternative energy source to transportation fossil fuels. However, the recalcitrant nature of lignocellulose poses technical hurdles to an economically viable biorefinery. Low enzymatic hydrolysis efficiency and low productivity, yield, and titer of biofuels are among the top cost contributors. Protein engineering has been used to improve the performances of lignocellulose-degrading enzymes, as well as proteins involved in biofuel synthesis pathways. Unlike its great success seen in other industrial applications, protein engineering has achieved only modest results in improving the lignocellulose-to-biofuels efficiency. This review will discuss the unique challenges that protein engineering faces in the process of converting lignocellulose to biofuels and how they are addressed by recent advances in this field.
Introduction
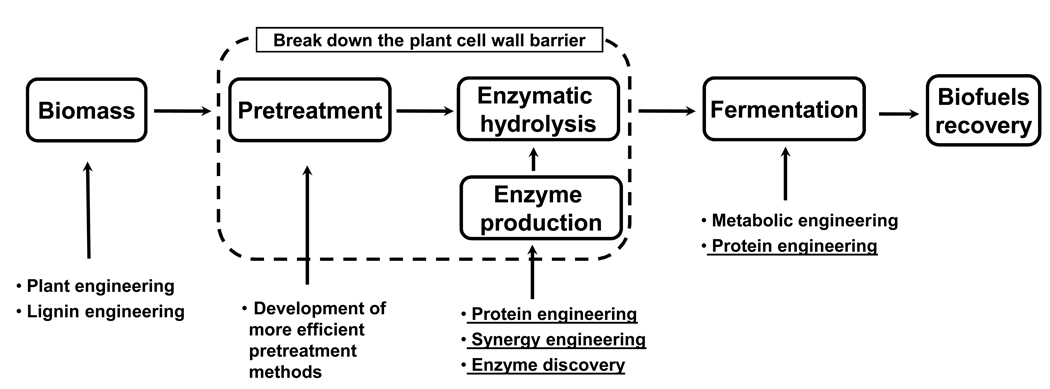
Modern society relies heavily on fossil fuels, which accounted for 88% of the global energy supply in 2007 [1]. Based on current fossil fuel reserves-to-production ratios, oil, natural gas, and coal could only last for approximately 40, 60, and 130 years, respectively [1]. To alleviate society’s dependence on fossil fuels and reduce greenhouse gas emissions, renewable energy sources have attracted intense political and academic attention. While other renewable energy sources, such as solar, wind, geothermal, and hydroelectric power, are more suitable for stationary power applications (electricity and heat), liquid fuels derived from biomass are the only foreseeable alternative to the petroleum products currently used in transportation [2••,3•,4••]. Although ethanol produced from corn or sugar cane currently dominates the biofuels market, it has limited agricultural growth potential and intrinsic physical drawbacks as a primary transportation fuel, such as high corrosivity, hygroscopicity, and low energy content [3•]. Therefore, it is highly desirable to produce alternative biofuels from a more sustainable resource, such as lignocellulose, which is derived from unusable portions of plant biomass in the form of agricultural, industrial, domestic, and forest residues. However, the recalcitrant crystalline structure of lignocellulosic biomass, which endows the plant cell wall with resistance to biodegradation, impedes its biological conversion to biofuels [2••]. The current lignocellulosic biofuel production process involves multiple costly and energy-intensive steps. Thus, significant technical advances in various fields are needed to lower the production cost to a level economically competitive with gasoline (Figure 1).
Figure 1.
A simplified overview of the traditional lignocellulose-to-biofuels process. This process involves multiple complex, costly, and energy-intensive steps, including pretreatment of plant biomass, enzyme production, enzymatic hydrolysis of pretreated biomass, and fermentation of the hydrolysate (monomeric sugars) to produce biofuels using engineered microorganisms. The most expensive processing steps, namely pretreatment, enzyme production, and enzymatic hydrolysis, are used to overcome the recalcitrance of biomass. Concerted effort from various fields is necessary to lower the production cost of lignocellulosic biofuels and the strategies covered in this review are underlined.
Enzymatic hydrolysis is one of the two most expensive processing steps (with the other, pretreatment, reviewed elsewhere [5]) in cellulosic biofuels production, which is mainly due to low enzyme catalytic efficiency. To achieve the same hydrolysis result, 40–100 times more enzyme is required to break down cellulose versus starch, although the enzyme production cost is not substantially different [6]. Therefore, engineering enzymes with improved catalytic efficiency is highly desirable for the commercialization of lignocellulosic biofuels. In addition, better enzymes might require less severe pretreatment conditions and thus reduce the formation of compounds inhibiting further hydrolysis and bioconversion of lignocellulose, resulting in a further reduction of production cost [5]. Another important processing step required for the economic success of lignocellulosic biofuels is microbial conversion of monomeric sugars to target biofuel molecules (Figure 1). Recent advances in metabolic engineering have enabled the production of various potential alternative biofuels in model microorganisms using monosaccharides as substrates (reviewed elsewhere [3•,7,8•]); however, the productivities and titers are too low to make them economically viable. This is due to the low activity of the pathway enzymes, as well as the low fuel tolerance and unbalanced redox state of the engineered microbes. In this review, we will discuss some of the most recent advances and applications of protein engineering in improving the performance of lignocellulose-degrading enzymes, as well as proteins involved in biofuel synthesis pathways, with an emphasis on how technical challenges could potentially be addressed by some of the new tools developed in the field.
Breaking down the plant cell wall barrier
The recalcitrant nature of the plant cell wall represents the biggest challenge in the development of lignocellulose-to-biofuels technologies. Its major structural component, cellulose, is protected by a matrix formed mainly by hemicellulose (the second most abundant component) and lignin, limiting the access of hydrolytic enzymes [2••]. In addition, cellulose forms a distinct crystalline structure, which cannot be penetrated by even small molecules such as water because of extremely tightly packing [9••]. The diverse architecture of plant cells themselves makes lignocellulose utilization more complicated, and different plant cell types might require completely different deconstruction methods [2••,9••]. While liberation of cellulose from the matrix is tackled by pretreatment [5] and lignin engineering [10], cellulose hydrolysis efficiency is the main focus of protein engineering. Efforts in this area include engineering of enzymes for improved specific activity, thermostability, and pH stability; and optimization of enzyme formulations for maximized synergy on different feedstock substrates.
Engineering cellulases
Cellulose is a linear homopolymer of glucose linked by β-1,4-glycosidic bonds. As the most abundant, yet the most recalcitrant constituent of plant cell wall, cellulose hydrolysis is a critical and challenging step, involving the action of three major types of cellulases: endoglucanases, exoglucanases (including cellodextrinases and cellobiohydrolases), and β-glucosidases. Microorganisms have evolved two strategies of utilizing their cellulases: discrete non-complexed cellulases that are typically secreted by aerobic bacteria and fungi, and complexed cellulases (cellulosome) that are typically expressed on the surface of anaerobic bacteria and fungi [9••]. While cellulosome engineering has mainly focused on optimizing the cellulosomal components (discussed in the section titled Engineering synergy), protein engineering has been applied to improve the performance of individual non-complexed cellulases. Despite continuing efforts to enhance non-complexed cellulase performances, the improvements obtained so far using protein engineering approaches [11] have been incremental, mainly due to the complexity of the insoluble substrates and the lack of high throughput screening/selection methods [12•].
Limited knowledge of the biochemical mechanisms involved in cellulose hydrolysis has limited the success achieved by rational and semi-rational design strategies in cellulase engineering, and no significant activity enhancement has been reported to date. Although cellulase activity on insoluble substrates is hard to predict, the stability of the cellulase itself could be very well modeled by the SCHEMA energy function [13]. Using a SCHEMA structure-guided recombination method, 15 highly diverse thermostable cellobiohydrolase hybrids (up to 7 °C higher than the most thermostable parent) were obtained by screening only a total of 73 variants. Considering the fact that protein stability enhances both mutational robustness and evolvability [14], this group of diverse cellobiohydrolases provides a better platform for improving their catalytic efficiency.
In an effort to adapt directed evolution to cellulase engineering, a high throughput selection method was recently developed based on chemical complementation to improve endoglucanase activity [15]. In this study, the authors elegantly designed an oligosaccharide surrogate by imbedding a cellotetraose between a methotrexate and a dexamethasone, which acted as a transcription inducer linking the hydrolysis activity of endoglucanases to the survival of a URA3-FOA counter-selection yeast strain. This method was of very high throughput and yielded two variants with improved catalytic efficiency (3.7- and 5.7-fold) from a family DNA shuffling library with a size of 108. However, since the selection was based on cleavage of a soluble substrate (methotrexate-cellotetraose-dexamethasone) by intracellular enzymes, it could not be used to engineer cellulase activity toward insoluble substrates. Given the fact that there is no clear correlation between enzyme activity on soluble substrates and that on insoluble substrates [12•], Chundawat and coworkers have geared their high throughput 96-well microplate technique toward more realistic solid substrates [16]. By using ultracentrifugal milling and a robotic multi-pipetting workstation, the issue of irreproducible solid substrate delivery (only crystalline cellulose (Avicel) and ammonia fiber expansion (AFEX) pretreated corn stover were tested in this study) was solved. Although no application of this system was reported, the integration of high throughput pretreatment [16], fermentation [17], and microplate format described here has the potential to enable high throughput engineering of the entire lignocellulose-to-biofuels process in a miniature biorefinery.
Engineering synergy
Due to the compositional complexity of the plant cell wall, synergistic action of a collection of enzymes with complementary activities is required for optimal degradation efficiency. In nature, synergy is best exemplified by a unique biomass-degrading machinery – the cellulosome. It exhibits a highly organized structure, in which many different types of carbohydrate-reactive enzymes, including cellulases, hemicellulases, and pectinases, are held together by a non-catalytic scaffoldin through high affinity, non-covalent interactions between enzyme-bound dockerins and cohesins in the scaffoldin [18]. Although the mechanism is not yet clearly defined, it is widely believed that, in addition to the enzyme-enzyme synergy (National Renewable Energy Lab, R&D 100 Award: www.nrel.gov/awards/2004hrvtd.html) that was also observed for non-complexed cellulases, the enzyme proximity [19], and enzyme-substrate [19] and enzyme-microbe [20] interactions are the key contributors to the enhanced synergy of a cellulosome [21]. Synergy engineering is still in its nascent stage. To advance significantly, better engineering tools or systems must be developed.
Inspired by the type-specific and species-specific interaction between cohesin-dockerin pairs, a designer cellulosome concept has been proposed to study and engineer a cellulosome for biotechnological applications [22]. By creating a chimeric scaffoldin consisting of divergent cohesins, a designer cellulosome allows incorporation of enzymes with different activities and origins in a composition- and spatially-defined manner. However, since all the cellulosomal components need to be purified and assembled in vitro, it is not economically feasible. This limitation might be overcome by combining the designer cellulosome concept with other recombinant expression strategies [22], such as (1) the intercellular complementation strategy [23•], which involves co-culturing several different recombinant strains each producing a cellulosomal component; or (2) cell surface display (Zhao, H., unpublished), which involves in vivo assembly of a cellulosome that is transported onto the cell surface via the secretion pathway. Pertinent to the second strategy, three types of cellulases have been displayed on yeast cell surface using α-agglutinin as the anchor protein, resulting in simultaneous saccharification and fermentation with a yield of 0.45 g of ethanol per 1 g of amorphous cellulose [24]. Recombinant organisms resulting from these engineering works have great potential in achieving consolidated bioprocessing (CBP), a highly compact and very promising process configuration that integrates enzyme production, hydrolysis, and fermentation in a single step [9••,25].
Pushing the limit of microbial biofuel production
Engineering biofuel biosynthetic enzymes
Doubts about the sustainability of ethanol as a liquid transportation fuel have sparked interest in engineering microbes for production of higher alcohols. Certain Clostridia have been known since the 1960s to produce 1-butanol, and heterologous expression of this pathway was recently demonstrated in E. coli [26,27] and S. cerevisiae [28]. The Liao group has since demonstrated that amino acid biosynthetic intermediates can be rerouted by the expression of heterologous enzymes to produce various branched and linear alcohols [29•,30–32]. While most of the work involved metabolic engineering, production of some alcohols required protein engineering to increase flux in the desired direction.
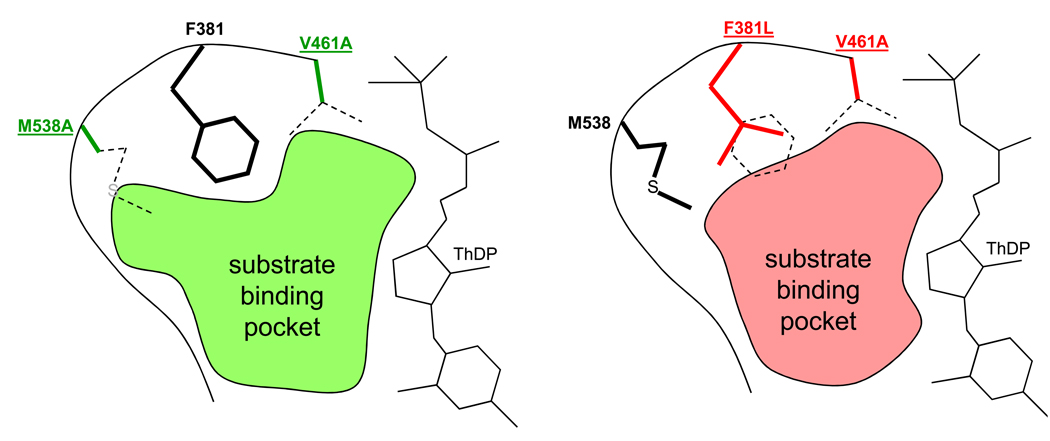
To produce 1-propanol and 1-butanol via the citramalate pathway, Atsumi and Liao performed directed evolution on citramalate synthase (CimA) from the thermophile Methanococcus jannaschii and selected for functional expression at moderate temperatures [31]. After six rounds of mutagenesis and selection under increasing selection pressure, they identified a mutant that was more active than the wild-type enzyme at moderate temperatures and was also insensitive to feedback inhibition. Using this variant, they were able to simultaneously produce high levels of 1-propanol and 1-butanol from glucose. In order to produce longer chain alcohols, Zhang and coworkers engineered the active site of both ketoisovalerate decarboxylase (KIVD) and 2-isopropylmalate synthase (IPMS or LeuA) to accept larger substrates by minimizing steric clashes that may arise when bound to larger substrates [30] (Figure 2). When expressed in a metabolically engineered E. coli, the two mutant enzymes could produce C5–C8 alcohols. In a final example for producing branched alcohols using engineered enzymes by the Liao group, they employed a previously described feedback insensitive mutant of IPMS that greatly increased the flux toward 3-methyl-1-butanol [32].
Figure 2.
A schematic stereoview of the active site of ketoisovalerate dehydrogenase (KIVD) [30]. In order to produce long-chain alcohols (C5–C8) from amino acid biosynthetic precursors, the active site of KIVD was modeled and altered to fit larger substrates. The shaded areas are representations of altered binding pockets and those of double mutants M538A/V461A (green) and F381L/V461A (red), which offer less steric hindrance to larger substrates like 2-keto-4-methylhexanoate. Dotted side chains are wild-type and highlight the changes in the substrate binding pocket as a result of mutations. ThDP: thiamine diphosphate, a co-substrate.
Engineering hemicellulose assimilation enzymes
Unlike cellulose, hemicellulose is a highly branched heterogeneous polymer of various pentoses, hexoses, and sugar acids. Among these, pentoses d-xylose and l-arabinose are the primary constituents, and as a result, significant effort has been made to engineer yeast to efficiently ferment these sugars into ethanol.
Protein engineering work for efficient fermentation of d-xylose has focused primarily on the fungal pentose assimilation pathway enzymes xylose reductase (XR) and xylitol dehydrogenase (XDH). The accumulation and secretion of intracellular xylitol, an intermediate in pentose assimilation, has led many to look into the cofactor preferences of the enzymes. XR generally prefers NADPH, whereas XDH prefers NAD+. The inability of yeast to regenerate these cofactors is thought to be a major bottleneck in this pathway. Several attempts have been made to close this loop by engineering one of the two enzymes such that both preferentially utilize either NAD+/NADH or NADP+/NADPH. Since XR can use both reduced cofactors, albeit with orders of magnitude difference in efficiency, decreasing its affinity for NADPH is a viable technique to force NADH use. Mutant XRs with disrupted electrostatic interactions with the 2’-phosphate of NADPH did indeed enhance ethanol production when expressed in yeast [33–36]. An alternative to engineering the cofactor preference of XR is to alter that of XDH. Unlike the more promiscuous XRs, all characterized XDHs have a strict preference for NAD+. Watanabe and coworkers first described the XDH cofactor preference reversal by structure-guided mutagenesis of residues in the cofactor binding site in proximity to the 2’-hydroxyl moiety of NAD+ [37]. They have since shown that this mutant does indeed enhance ethanol productivity from d-xylose when expressed in yeast [38–40]. Since then, there have been some other examples of similar engineering work [41], as well as observations of improved ethanol production [42].
l-Arabinose metabolism in recombinant yeast is also a difficult problem since l-arabinose assimilation is extremely slow. Cofactor preference is also an even more significant issue in this pathway since it uses an additional two oxidoreductases, l-arabinitol-4-dehydrogenase (LAD) and l-xylulose reductase (LXR), both of which create an even greater imbalance in the cofactor pool. Little work has been published to address this issue; although Zhao and coworkers have engineered an LAD with almost completely reversed cofactor preference from NAD-specific to NADP-specific using a combined structure-function guided and directed evolution method (Zhao, H., unpublished). No engineering work has been described for LXR, however. Another area of interest is engineering transporters for more efficient import of these “unnatural” sugars. Since yeast does not naturally metabolize these sugars, there are no transporters with high affinity toward them. Heterologous expression of pentose-specific transporters has demonstrated promising results [43–46], although engineering work may also be required to enhance the transport capacity and efficiency. Interest in this area ensures forthcoming results in the near future.
Engineering transcriptional factors
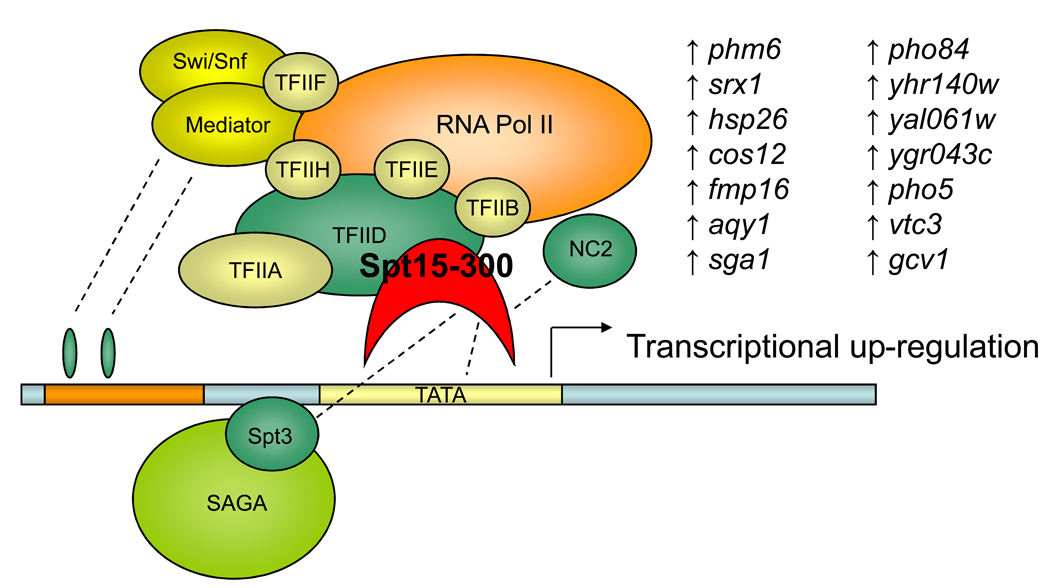
Most examples of protein engineering involved in biofuels production concentrate on increasing the catalytic efficiency of a single reaction. Engineering transcriptional factors creates a more global change in metabolism, and this strategy has been applied successfully in both E. coli and S. cerevisiae for improving biofuels production. The best known example is the process developed by the Stephanopoulos group known as global transcriptional machinery engineering (gTME) [47••] (Figure 3). Using error-prone PCR, they focused their mutagenesis on a transcription factor (Spt15p) and were able to increase ethanol tolerance of yeast. This work provided an alternative to adaptive evolution and demonstrated that complex phenotypic improvements were achievable by concentrating on a single cellular protein. The generality of this concept has been demonstrated with further examples in yeast for xylose fermentation [48], as well as in E. coli for enhanced alcohol tolerance, metabolite overproduction, and altering multiple phenotypes [49,50].
Figure 3.
A schematic representation of the global transcriptional machinery engineering gTME method developed by Stephanopoulos and coworkers [47••,49]. Complex phenotypic changes are possible via altered regulation of multiple genes. Such transformations can be achieved by concentrating mutagenesis on a single protein among the various components of yeast’s transcriptional machinery. Expression of the mutant TATA-binding transcriptional factor Spt15–300 (red) resulted in significant up-regulation of 14 genes. All these overexpressions act in a concerted manner to increase yeast’s tolerance to high concentrations of ethanol and glucose.
Apart from gTME, a previously recognized constitutively active mutant transcriptional factor in E. coli, crp* (cyclic AMP receptor protein), has been used to engineer E. coli for simultaneous utilization of glucose and d-xylose [51]. This mutant could be a useful tool for biofuels production from hemicellulosic sugars using E. coli as a platform organism, rather than yeast. Other opportunities for transcription factor engineering also exist within the zinc-finger family of proteins, an avenue as yet to be utilized for biofuels production.
Conclusions and future perspectives
Biofuels are of rapidly growing interest thanks to energy security, sustainability, and climate change. The first-generation biofuel technology has been used to produce ethanol from corn and sugar cane on a large scale in the United States and Brazil. However, the limited crop supply will not satisfy society’s growing energy demand; thus, the second-generation biofuel technology based on lignocellulose is under intense investigation. Several factors will influence the economic viability of lignocellulosic biorefinery (Figure 1). With the development of high throughput screening/selection methods, protein engineering will play an important role in producing new, more active enzymes for hydrolysis of biomass to sugars and subsequent microbial conversion of sugars to biofuel molecules, although the progresses reported to date have been incremental.
One possible reason for the limited success of protein engineering might be that the enzymes used as engineering templates so far were derived from a very limited sequence space – namely culturable microorganisms, which, on average, represent <1% of the genetic diversity found in nature [52]. To overcome this limitation imposed by traditional microbiological techniques, new strategies such as metagenomics [53] and single-cell genomics [54,55] were developed. A recent metagenomic study of a wood-degrading termite revealed hundreds of hitherto unknown glycoside hydrolase genes [56]. These novel cellulolytic proteins might expand the current plant-cell-wall-degrading enzyme paradigm and enable more fruitful protein engineering studies.
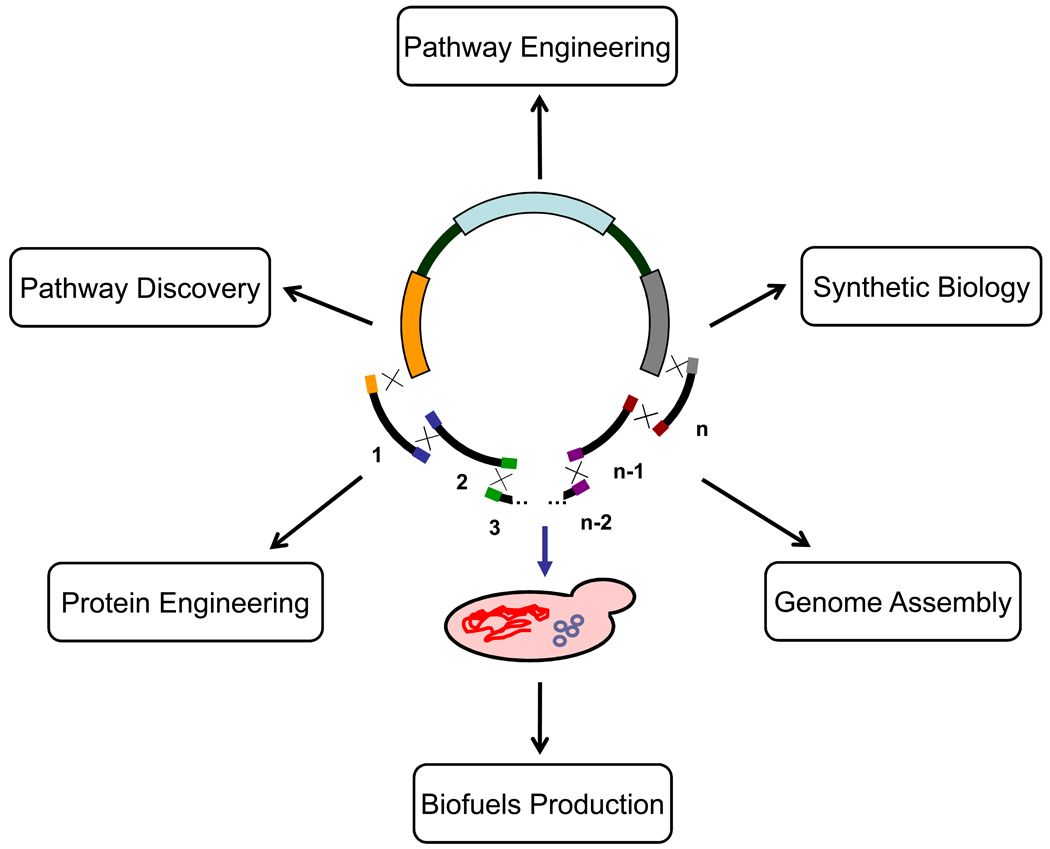
It is expected that metagenomics, single-cell genomics, and the genome sequencing projects of more than 40 cultivated cellulolytic microbes [57,58] will result in an exponential increase in the number of potential carbohydrate-reactive enzymes, as well as related biosynthesis and regulation pathways. Such massive genetic information requires the development of efficient gene cloning and expression tools to examine the putative protein functions in a context-dependent manner. The functional expression of putative genes in E. coli, S. cerevisiae, or other established industrial hosts might be difficult, especially those genes isolated from extreme environments [59]. Development of expression systems that include tRNA synthetase genes and/or stress-response elements might enable or improve the expression of such enzymes [59]. Of particular interest, S. cerevisiae has been recently shown to possess the ability of simultaneously taking up and correctly assembling DNA fragments into a large molecule in a single step [60,61• (Figure 4)]. This extraordinary ability will greatly accelerate and simplify the discovery, characterization, and engineering of individual genes and biochemical pathways applicable to biofuels production. It is especially useful to assemble large complex enzymatic pathways for consolidated bioprocessing.
Figure 4.
DNA assembler method developed by Zhao and coworkers [61•]. Multiple gene expression cassettes (encoding promoter-gene-terminator) are prepared by splicing PCR. Each fragment contains sequence homology to adjacent fragments at its termini enabling crossover events at both ends. All fragments are co-transformed and correctly assembled in S. cerevisiae with a linearized vector (or a helper fragment, not shown in the figure), yielding a plasmid encoding a functional pathway (or integration of a functional pathway into yeast chromosome). This method should significantly simplify the protein engineering of biochemical pathways and has various potential applications including biofuels production.
With the continuing development of new tools and scientific knowledge, significant advances will be made toward the development of next generation biofuels. Concerted efforts in protein engineering, metabolic engineering, plant engineering, chemical catalysis, and chemical process engineering will lead to an economically viable lignocellulosic biorefinery in the near future.
Acknowledgements
We gratefully acknowledge financial support from the British Petroleum Energy Biosciences Institute and National Institutes of Health (GM077596). N.N. also acknowledges Drickamer Fellowship support from the Department of Chemical and Biomolecular Engineering at the University of Illinois.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interests, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.BP Statistical Review of World Energy. 2008 June; www.bp.com/statisticalreview.
- ••2.Breaking the biological barriers to cellulosic ethanol: a joint research Agenda, DOE/SC-0095. U.S. Department of Energy Office of Science and Office of Energy Efficiency and Renewable Energy. 2006 www.doegenomestolife.org/biofuels/A roadmap from a workshop showcasing the benefits and great challenges of converting biomass to biofuels. The key barriers and suggested research strategies to address them are described.
- •3.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014.This paper summarizes the fossil fuel types and their biological counterparts. The strategies of engineering various metabolic pathways to produce some of the biofuels are discussed.
- ••4.Simonetti DA, Dumesic JA. Catalytic strategies for changing the energy content and achieving C--C coupling in biomass-derived oxygenated hydrocarbons. ChemSusChem. 2008;1:725–733. doi: 10.1002/cssc.200800105.While most research focuses on the biological conversion of biomass, this paper reviews chemical catalysis strategies.
- 5.Galbe M, Zacchi G. Pretreatment of lignocellulosic materials for efficient bioethanol production. Adv Biochem Eng Biotechnol. 2007;108:41–65. doi: 10.1007/10_2007_070. [DOI] [PubMed] [Google Scholar]
- 6.Merino ST, Cherry J. Progress and challenges in enzyme development for biomass utilization. Adv Biochem Eng Biotechnol. 2007;108:95–120. doi: 10.1007/10_2007_066. [DOI] [PubMed] [Google Scholar]
- 7.Fortman JL, Chhabra S, Mukhopadhyay A, Chou H, Lee TS, Steen E, Keasling JD. Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 2008;26:375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- •8.Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr Opin Biotechnol. 2008;19:414–419. doi: 10.1016/j.copbio.2008.08.008.This paper reviews metabolic engineering strategies in E. coli for the production of non-traditional biofuels.
- ••9.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002.A comprehensive review of the biochemical fundamentals of the cellulolytic enzymes and microorganisms system. The authors proposed two organism development strategies to achieve a consolidated bioprocessing.
- 10.Weng JK, Li X, Bonawitz ND, Chapple C. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol. 2008;19:166–172. doi: 10.1016/j.copbio.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Wen F, McLachlan M, Zhao H. Directed evolution: novel and improved enzymes. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc.; 2008. [Google Scholar]
- •12.Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24:452–481. doi: 10.1016/j.biotechadv.2006.03.003.This paper reviews the advantages and limitations of quantitative cellulase activity assays using soluble and insoluble substrates, which are the basis of protein engineering strategies.
- 13.Heinzelman P, Snow CD, Wu I, Nguyen C, Villalobos A, Govindarajan S, Minshull J, Arnold FH. A family of thermostable fungal cellulases created by structure-guided recombination. Proc Natl Acad Sci USA. 2009;106:5610–5615. doi: 10.1073/pnas.0901417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloom JD, Lu Z, Chen D, Raval A, Venturelli OS, Arnold FH. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 2007;5:29. doi: 10.1186/1741-7007-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peralta-Yahya P, Carter BT, Lin H, Tao H, Cornish VW. High-throughput selection for cellulase catalysts using chemical complementation. J Am Chem Soc. 2008;130:17446–17452. doi: 10.1021/ja8055744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chundawat SP, Balan V, Dale BE. High-throughput microplate technique for enzymatic hydrolysis of lignocellulosic biomass. Biotechnol Bioeng. 2008;99:1281–1294. doi: 10.1002/bit.21805. [DOI] [PubMed] [Google Scholar]
- 17.Isett K, George H, Herber W, Amanullah A. Twenty-four-well plate miniature bioreactor high-throughput system: assessment for microbial cultivations. Biotechnol Bioeng. 2007;98:1017–1028. doi: 10.1002/bit.21484. [DOI] [PubMed] [Google Scholar]
- 18.Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 19.Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Belaich A, Lamed R, Shoham Y, Belaich JP. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Zhang YH, Lynd LR. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci U S A. 2006;103:16165–16169. doi: 10.1073/pnas.0605381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding SY, Xu Q, Crowley M, Zeng Y, Nimlos M, Lamed R, Bayer EA, Himmel ME. A biophysical perspective on the cellulosome: new opportunities for biomass conversion. Curr Opin Biotechnol. 2008;19:218–227. doi: 10.1016/j.copbio.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Bayer EA, Lamed R, Himmel ME. The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol. 2007;18:237–245. doi: 10.1016/j.copbio.2007.04.004. [DOI] [PubMed] [Google Scholar]
- •23.Arai T, Matsuoka S, Cho HY, Yukawa H, Inui M, Wong SL, Doi RH. Synthesis of Clostridium cellulovorans minicellulosomes by intercellular complementation. Proc Natl Acad Sci U S A. 2007;104:1456–1460. doi: 10.1073/pnas.0610740104.A co-culturing strategy to produce cellulosomes is described, which might be useful in the development of consolidated bioprocessing.
- 24.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol. 2004;70:1207–1212. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, Toda H, Yamamoto S, Okino S, Suzuki N, Yukawa H. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol. 2008;77:1305–1316. doi: 10.1007/s00253-007-1257-5. [DOI] [PubMed] [Google Scholar]
- 28.Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7:36. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450.This paper demonstrates that the intermediates of the endogenous amino acid pathway in E. coli can be diverted to the production of higher alcohols through metabolic engineering.
- 30.Zhang K, Sawaya MR, Eisenberg DS, Liao JC. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc Natl Acad Sci U S A. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atsumi S, Liao JC. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl Environ Microbiol. 2008;74:7802–7808. doi: 10.1128/AEM.02046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connor MR, Liao JC. Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol. 2008;74:5769–5775. doi: 10.1128/AEM.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal B, Gorwa-Grauslund MF. The expression of a Pichia stipitis xylose reductase mutant with higher KM for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- 34.Petschacher B, Nidetzky B. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact. 2008;7:9. doi: 10.1186/1475-2859-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe S, Abu Saleh A, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153:3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe S, Pack SP, Saleh AA, Annaluru N, Kodaki T, Makino K. The positive effect of the decreased NADPH-preferring activity of xylose reductase from Pichia stipitis on ethanol production using xylose-fermenting recombinant Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2007;71:1365–1369. doi: 10.1271/bbb.70104. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe S, Kodaki T, Makino K. Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J Biol Chem. 2005;280:10340–10349. doi: 10.1074/jbc.M409443200. [DOI] [PubMed] [Google Scholar]
- 38.Matsushika A, Watanabe S, Kodaki T, Makino K, Inoue H, Murakami K, Takimura O, Sawayama S. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2008;81:243–255. doi: 10.1007/s00253-008-1649-1. [DOI] [PubMed] [Google Scholar]
- 39.Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S. Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase. J Biosci Bioeng. 2008;105:296–299. doi: 10.1263/jbb.105.296. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J Biotechnol. 2007;130:316–319. doi: 10.1016/j.jbiotec.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 41.Ehrensberger AH, Elling RA, Wilson DK. Structure-guided engineering of xylitol dehydrogenase cosubstrate specificity. Structure. 2006;14:567–575. doi: 10.1016/j.str.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Hou J, Shen Y, Li XP, Bao XM. Effect of the reversal of coenzyme specificity by expression of mutated Pichia stipitis xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Lett Appl Microbiol. 2007;45:184–189. doi: 10.1111/j.1472-765X.2007.02165.x. [DOI] [PubMed] [Google Scholar]
- 43.Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol. 2008;80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- 44.Leandro MJ, Spencer-Martins I, Goncalves P. The expression in Saccharomyces cerevisiae of a glucose/xylose symporter from Candida intermedia is affected by the presence of a glucose/xylose facilitator. Microbiology. 2008;154:1646–1655. doi: 10.1099/mic.0.2007/015511-0. [DOI] [PubMed] [Google Scholar]
- 45.Madhavan A, Tamalampudi S, Srivastava A, Fukuda H, Bisaria VS, Kondo A. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Appl Microbiol Biotechnol. 2009;82:1037–1047. doi: 10.1007/s00253-008-1818-2. [DOI] [PubMed] [Google Scholar]
- 46.Saloheimo A, Rauta J, Stasyk OV, Sibirny AA, Penttila M, Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol. 2007;74:1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- ••47.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969.This paper describes the use of gTME to engineer a recombinant S. cerevisiae strain with improved glucose/ethanol tolerance, a key trait essential for biofuels production but difficult to engineer by traditional methods.
- 48.Liu H, Yan M, Lai C, Xu L, Ouyang P. gTME for improved xylose fermentation of Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2008 doi: 10.1007/s12010-008-8431-9. [DOI] [PubMed] [Google Scholar]
- 49.Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Klein-Marcuschamer D, Santos CN, Yu H, Stephanopoulos G. Mutagenesis of the bacterial RNA polymerase alpha subunit for improving complex phenotypes. Appl Environ Microbiol. 2009;75:2705–2711. doi: 10.1128/AEM.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cirino PC, Chin JW, Ingram LO. Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol Bioeng. 2006;95:1167–1176. doi: 10.1002/bit.21082. [DOI] [PubMed] [Google Scholar]
- 52.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steele HL, Jaeger KE, Daniel R, Streit WR. Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol. 2009;16:25–37. doi: 10.1159/000142892. [DOI] [PubMed] [Google Scholar]
- 54.Hutchison CA, 3rd, Venter JC. Single-cell genomics. Nat Biotechnol. 2006;24:657–658. doi: 10.1038/nbt0606-657. [DOI] [PubMed] [Google Scholar]
- 55.Lasken RS. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 57.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19:210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 59.Hess M. Thermoacidophilic proteins for biofuel production. Trends Microbiol. 2008;16:414–419. doi: 10.1016/j.tim.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., 3rd One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •61.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991.This paper demonstrates the ease and efficiency of DNA assembler in constructing biochemical pathways.