Abstract
Assembly of an anastral spindle was modeled as a two-stage process: first, the aggregation of microtubule foci or asters around the chromosomes, and second, the elongation of cross-linked microtubules and onset of bipolarity. Several possibilities involving diffusion and transport were investigated for the first stage, and the most feasible was found to be binding of the asters to cytoskeletal filaments and directed transport toward the chromosomes. For the second stage, a differential-equation model was formulated and solved numerically; it involves cross-linking of microtubules with those aligned with the spindle axis and between microtubules bound to different chromosomes, and sliding of microtubules along the spindle axis to elongate the spindle. Ncd was postulated to perform both functions. The model shows that spindle formation begins with rapid cross-linking of microtubules, followed by elongation, which continues until the population of microtubules aligned with the spindle axis is depleted and microtubules cross-linking different chromosomes dominate. It also shows that when sliding is inhibited, short bipolar spindles still form, and if clustering is enhanced, normal-length spindles can form, although requiring longer assembly time. These findings are consistent with spindle assembly in live wild-type and ncd mutant Drosophila oocytes.
Introduction
Anastral spindles lack centrosomes at the poles and assemble by an unusual mechanism that differs from classical mitotic spindles, involving chromatin-mediated microtubule nucleation (1–4). This pathway of spindle assembly has been observed in oocytes of Drosophila, Xenopus, and the mouse, indicating that it may be widespread in evolution. The overall architecture of anastral spindles can also differ dramatically from classical mitotic spindles—studies of in vitro-assembled Xenopus extract spindles show that the spindles consist of randomly interspersed long and short microtubules with minus ends distributed throughout the spindle (5,6). Recent work has further demonstrated that microtubules grow both from the chromosomes toward the poles and from the poles toward the chromosomes in anastral spindles of live Drosophila oocytes (7). Remarkably, this implies that microtubules in the oocyte spindles are of mixed polarity, rather than oriented predominantly with minus ends at the poles, as in classical mitotic spindles. Microtubule dynamics was found to be similar at the spindle poles and equator, differing from mitotic spindles of syncytial blastoderm embryos, but consistent with the implied mixed polarity of the meiotic spindle microtubules (7).
Assembly of a bipolar anastral spindle is thought to involve microtubule cross-linking and sliding, mediated by motors and other microtubule-associated proteins. Available models take into account the ability of motors to form microtubule asters or organize microtubules into arrays resembling spindles (8,9), as well as nucleation near chromatin and microtubule sliding. For example, the slide-and-cluster model proposes that microtubules, nucleated near chromosomes, slide toward the poles with their minus ends leading, mediated by a plus-end-directed motor, and are clustered by a minus-end motor, and that microtubule loss is stochastic (10). Spindle length in this model is affected by microtubule sliding velocity, which decreases with distance from the chromosomes, with poles forming at the positions where the average sliding velocity is zero. The slide-and-cluster model addresses several properties of oocyte spindles, but it is based on microtubule and spindle dynamics in Xenopus extract spindles—these spindles undergo assembly in vitro and their dynamics could differ substantially from live oocytes. The model does not address initial steps in the formation of the spindle, nor does it account for the recently reported mixed polarity of microtubules in the spindle based on live analysis of Drosophila oocytes, caused by microtubule growth from the poles as well as the chromosomes (7), which may also be true of meiotic spindles of other species.
Here we derive a mathematical model for anastral spindle assembly and maintenance that takes into account observations in live oocytes during assembly and steady-state spindle maintenance. Our model incorporates the motor-driven microtubule sliding of the slide-and-cluster model, but differs from this model in several important respects. These include that only one motor is required to mediate both microtubule sliding and clustering, and that microtubules in the spindle are oriented with mixed polarity. The spindle in our model is formed by cross-linking microtubules associated with individual chromosomes, as observed for the Drosophila oocyte meiosis I spindle (3). We also address a key event of the initial microtubule nucleation during anastral spindle assembly—association of motor-bound foci or asters with the chromosomes—and show that the asters require directed transport to move toward the chromosomes.
Results and Discussion
Aggregation of microtubules at the chromosomes
Anastral spindle assembly and maintenance was modeled in two stages using the Drosophila oocyte meiosis I spindle as a model. The first stage is the aggregation of asters of microtubules with the condensed meiotic chromosomes or karyosome. The asters, bound to the Ncd kinesin-14 motor fused to GFP, have been observed as spots of bright fluorescence that originate at random locations in the germinal vesicle away from the karyosome. There are two candidate mechanisms for movement of the asters to the karyosome: diffusion and directed transport.
Diffusion and aster aggregation with the karyosome
Diffusion is likely to play a role, given the Brownian motion of the oocyte cytoplasm, but diffusion alone is unlikely to be the mechanism because of the high probability of the asters escaping to places elsewhere in the cell. The germinal vesicle is spherical and ∼32 ± 2 μm in diameter just before breakdown (n = 5); the karyosome, which is much smaller, ∼4.4 ± 0.3 μm in diameter (n = 5), lies close to one edge, within ∼1 μm of the germinal vesicle membrane (11). The nuclear envelope is breaking down at the time of aster formation, so an aster could easily diffuse out of the germinal vesicle. Given a particle in Brownian motion at a distance R from the center of a sphere of radius r (with R > r), the probability P(R) of the particle contacting the sphere is r/R (12). It is now possible to calculate the fraction of asters in the germinal vesicle that will be captured by the karyosome in the absence of directed transport. We can approximate the karyosome by a sphere of radius rk and the germinal vesicle by a sphere of radius rgv (see Table S1 in the Supporting Material for symbols). Consider a spherical coordinate system with the origin at the center of the karyosome and the center of the germinal vesicle at ρ = S, θ = 0. By the law of cosines, this gives ρ = rk for the karyosome membrane or boundary and
| (1) |
for the germinal vesicle boundary (Fig. 1). The fraction of asters captured is expected to be the average of the aster capture probability over the germinal vesicle, which is the integral of capture probabilities over the germinal vesicle (excluding the karyosome), divided by the volume of the germinal vesicle (excluding the karyosome),
| (2) |
since P(ρ) is rk/ρ in the absence of directed transport, regardless of the diffusion coefficient. This fraction is 16% for rk = 2.2 μm, rgv = 16 μm, and S = 12.8 μm, using the values for rk and rgv that we measured from live images of the germinal vesicle and karyosome during early oocyte spindle assembly (3) and the observation that the karyosome is ∼1 μm from the edge of the germinal vesicle (11). The calculated fraction of asters captured ranges from 15 to 20% over the possible values of S, and even doubling the karyosome size while holding the germinal vesicle size constant yields only 33% capture. Thus, most of the asters will never reach the karyosome by diffusion alone, in contrast to our observations based on analysis of time-lapse sequences, where an average of 90 ± 3% of asters migrating in the plane of focus were captured (n = 64, total = 71 asters in nine oocytes). This means that the asters are unlikely to reach the karyosome by diffusion alone and are probably being transported to the karyosome.
Figure 1.
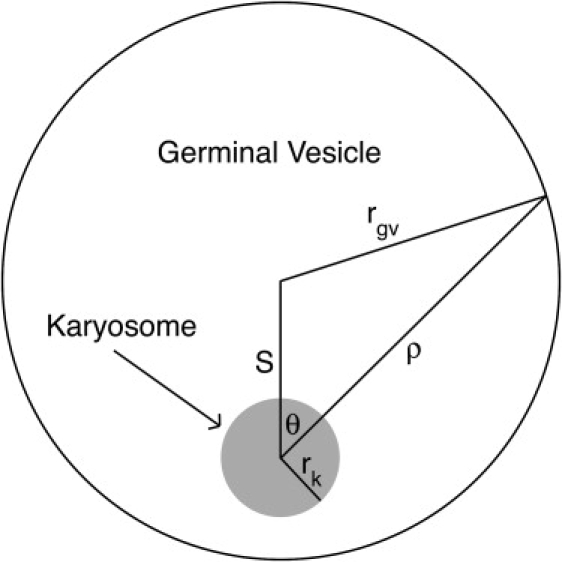
Geometry of the germinal vesicle and karyosome, showing the relationship between ρ and θ (in the spherical coordinate system) for a given point on the membrane or boundary of the germinal vesicle. In live oocytes, rk and rgv are ∼2.2 μm and 16 μm, respectively; placing the karyosome 1 μm from the edge of the germinal vesicle yields S = rgv–rk−1 μm = 12.8 μm.
Transport of asters to the karyosome
Two types of transport can be considered. The first is that the asters diffuse in a drifting medium and the second is that they bind cytoskeletal structures, e.g., microtubules or actin filaments, that direct them toward the karyosome. We consider the first possibility unlikely because it requires drift from a large area and concentration at the relatively small karyosome. Thus, the microtubule asters must bind to fibers or tracks along which they are transported toward the karyosome. The finding of a kinesin-14 microtubule motor, Ncd, associated with the asters (3) does not necessarily provide information regarding the postulated cytoskeletal tracks—the tracks along which transport is proposed to occur have not been visualized in oocytes labeled either with rhodamine-tubulin or Ncd-GFP. Moreover, we and others have observed that the Ncd motor does not bind to cytoplasmic microtubules; instead, the motor binds specifically to the microtubule asters that migrate toward the karyosome and the spindle microtubules (2,3). The Ncd motor could cross-link and bundle microtubules to form the asters, but transport of the asters along cytoskeletal tracks to the karyosome most likely involves a different motor.
Capture of asters by the karyosome
The high fraction of asters that is captured implies that nearly all of them eventually bind to cytoskeletal filaments that direct them toward the karyosome, although not all may be bound at a given time. We derived a model to show that not all of the asters are necessarily bound at a given time. Consider a karyosome with a given number of tracks or fibers emerging from it; the spatial density of fibers is proportional to 1/ρ2, thus the pseudo-first order association rate constant for asters binding the fibers, k∗on, is proportional to 1/ρ2. If we let the dissociation rate constant, koff, be constant and assume that binding is at equilibrium with a fraction Ceq of asters bound, then
| (3) |
where the first equality comes from setting the time derivative of aster density equal to zero and
| (4) |
is a constant that measures the strength of binding (stable, complete binding is Kb = 0 and no binding is Kb = ∞). Now the asters obey a Fokker-Planck equation,
| (5) |
where f is a function of space and time representing the number of asters per unit volume, D = the diffusion coefficient, v = the transport velocity, and Feq = 1-Ceq is the fraction of asters not bound at a given point in space. P(R) in the case of directed transport can be derived from Eq. 5 using the same method used by Berg (12) for the no-transport case. First, we derived a steady-state solution for f. Setting the time derivative equal to zero in Eq. 5 and solving for f yields
| (6) |
where the constants C1 and C2 can be derived from the boundary conditions. The constant Q can be defined as
| (7) |
It quantifies the importance of transport relative to diffusion. We derived a steady-state solution to Eq. 5 that is continuous but not smooth at ρ = R (we can arbitrarily normalize by setting f(R) = 1), but that satisfies f(rk) = 0 because of capture of asters that arrive at the karyosome and f(∞) = 0:
| (8) |
We can calculate the fraction captured, P(R), from the flux of asters toward the karyosome (Jin) and the flux outward (Jout) at ρ = R. The flux of asters along the radial dimension is
| (9) |
To calculate Jout, we calculate Jρ at ρ = R using the equation for f on ρ ≥ R; to calculate Jin, we calculate −Jρ at ρ = R using the equation for f on ρ ≤ R. This yields
| (10) |
Note that in the limit of no transport (Q→0), this reduces to P(R) = rk/R as in the pure-diffusion case given above—this is revealed by expanding the exponentials in the last equation in Eq. 10 as a Taylor series up to first order. Likewise, in the limit of highly prevalent transport (large Q), P(R) = 1: all asters are captured—when Q/R > 10, 1-P(R) < 0.00005.
We can integrate P(R) and divide by volume, as in Eq. 2, to derive the total captured fraction of asters in the germinal vesicle when transport is involved. To derive an analytical solution, it is necessary to switch the order of the integrals in ρ and θ, but Eq. 1 is still used to define the germinal vesicle boundary. The fraction captured is given by
| (11) |
The fraction of asters initially bound, Cinit, may be expressed in terms of Kb; we simply average Ceq (given by Eq. 3) over the germinal vesicle excluding the karyosome:
| (12) |
Expanded forms of Eqs. 11 and 12 are given in the Supporting Material.
Aster capture by the karyosome in live oocytes
We measured the asters in time-lapse sequences to estimate a diffusion coefficient; they averaged 1.81 ± 0.08 μm (measured along the longest dimension; mean ± SE, n = 42 asters in eight oocytes). This yields D = 0.008–0.01 μm2/s, modeling the aster as 2–3 microtubules of the same length as the aster, and assuming that Ncd-GFP is bound, on average, to one in four tubulin subunits; we also assumed Mr = 27 kDa and D = 15.0 μm2/s for GFP (13), D ∝ M−1/3, and a ∼50-fold effect of nucleoplasm viscosity on D compared to aqueous solution (14). We estimated the velocity of the asters from time-lapse sequences of spindle assembly in live oocytes (3), based on the germinal vesicle radius and aggregation time, by dividing the diameter of the germinal vesicle by the time from nuclear envelope breakdown to the completion of aggregation. Assuming that the karyosome lies to one side of the germinal vesicle and that the asters originate randomly in the germinal vesicle, some of the asters will be near the far side of the germinal vesicle from the karyosome. These asters will traverse the diameter of the germinal vesicle over the time from nuclear envelope breakdown to completion of aggregation. Asters that begin nearer to the karyosome will tend to be captured earlier. This strategy gave a velocity estimate of 3.3 ± 0.5 μm/min, or 0.055 μm/s (n = 5). This is 4–5 times slower than the velocity of Ncd in vitro (15), which is roughly consistent with the slowing of kinesin-transported microtubules in viscous media observed by Hunt et al.; this indicates that the transport rate is reasonable for Ncd or a motor of similar speed (16). Using the MATLAB function fzero, an estimate of D = 0.01 μm2/s and the same geometric parameters as in the diffusion section above, we found a value for Kb = 0.14 μm−2 that matched the observed 90% capture rate (this corresponds to Cinit = 4.4%, with binding reaching 90% within a micron or so of the karyosome).
Thus, the roughly estimated parameters of rgv = 16 μm, rk = 2.2 μm, S = 12.8 μm, D = 0.01 μm2/s, v = 0.055 μm/s, and Kb = 0.14 μm−2 give a predicted 90% aster capture rate, consistent with our observations in live oocytes. Small deviations in these parameters produce similar capture rates. For example, D = 0.008 μm2/s yields 94% capture, and capture ranges from 89% to 96% if the karyosome is moved from the edge of the germinal vesicle (S = 13.8 μm) to the middle (S = 0). As noted above, deviations in v, D, or Kb only have an effect if Q changes, so, for example, an increase in v can be offset by a proportionate increase in D or Kb.
The mechanism of aster capture in Drosophila oocytes is not certain based on current data; however, addition of cytochalasin D and monastrol in mouse oocytes did not appear to disrupt the process (4), indicating that actin and kinesin-5, respectively, are not involved. Thus, one or more microtubule-based motors other than kinesin-5 are likely to be responsible.
Spindle elongation and the onset of bipolarity
Migration of the microtubule asters to the karyosome is followed by rapid growth of new microtubules outward in random directions from the chromosomes, resulting in a large number of poorly organized microtubules associated with the chromosomes at the beginning of spindle assembly that rapidly become organized around individual chromosomes (3). The second phase of our model describes spindle elongation and the onset of bipolarity (Fig. 2). It assumes that initially a few microtubules attached to the chromosomes are aligned with the presumptive spindle axis and that motor activity on these aligned microtubules results in net elongation rather than contraction. The model also assumes that motor activity leads to cross-linking and alignment of microtubules relative to the spindle axis and cross-linking of microtubules attached to different chromosomes. Ncd has recently been shown to cross-link microtubules aligned in parallel with one another and to slide antiparallel microtubules along one another in vitro (17), and is proposed here to perform both of these functions in the oocyte meiosis I spindle. The model further assumes that microtubule polymerization and depolymerization are at equilibrium, so that there is no net change in the total amount of microtubules.
Figure 2.
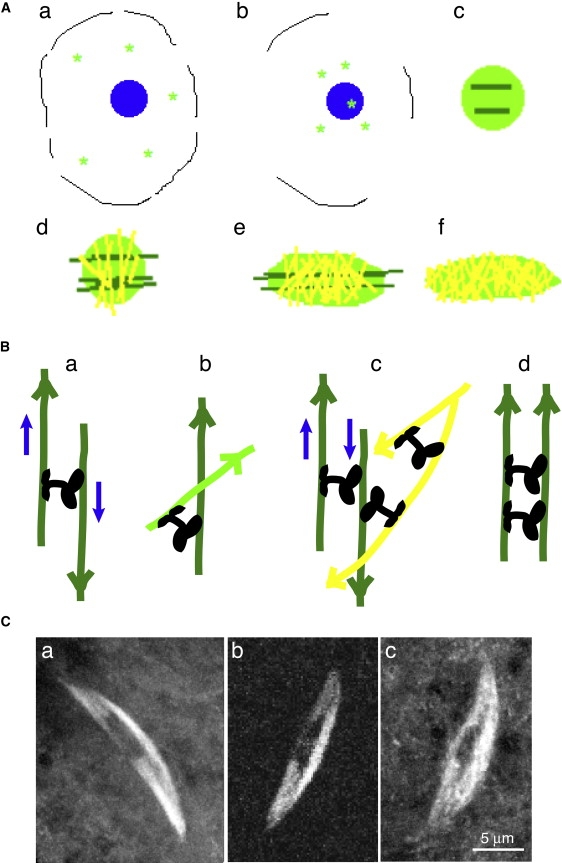
(A) Model for meiosis I spindle assembly. (a) Asters (bright green) in the germinal vesicle at nuclear envelope breakdown are (b) transported toward the karyosome (blue), which they bind, (c) creating a mass of randomly aligned microtubules (bright green) around the chromosomes, with a few aligned with the chromosomes or nascent spindle axis (dark green). The aligned microtubules are nucleated by asters bound to the chromosomes and grow parallel to their surface. (d) Some microtubules (yellow) are cross-linked between different chromosomes, and others (dark green) are cross-linked in alignment with the spindle axis, (e) allowing elongation of the spindle. The spindle elongates, but this process spreads out the microtubules aligned with the spindle axis, dispersing them, and elongation ceases, yielding (f) a bipolar spindle with randomly oriented microtubules. Figure not to scale. (B) A detailed view of (a) sliding, (b) cross-linking, (c) sliding of cross-linked microtubules, and (d) cross-linked microtubules parallel to the spindle axis. Microtubule colors the same as in panel A; arrowheads denote the plus end. Ncd motors, black; blue arrows, microtubule sliding direction. (C) Live Drosophila oocyte meiosis I spindles labeled with (a) GFP-α-tubulin, (b) Ncd-GFP, and (c) EB1-GFP, a microtubule plus-end-binding protein. Bar, 5 μm.
We define a spindle axis, associated with a coordinate x, the distance along the axis from a given point to the chromosomes. We let the density of unaligned microtubules be ρ1 and the density of spindle axis-aligned microtubules be ρ2. At the beginning of spindle elongation, an independent set of microtubules is likely to be aggregated around each chromosome; however, over time Ncd can create a population of microtubules cross-linking different chromosomes. We let the density of these cross-linking microtubules be ρ3. All three densities, ρ1, ρ2, and ρ3, depend on x and time. At time 0, randomly aligned microtubules predominate, having a region of constant density at low x (near the chromosomes) and decaying exponentially with x outside of that region. We also assume a much smaller initial population (<10%) of aligned and thus elongation-capable microtubules that is concentrated around the chromosomes and falls off exponentially with x. The interchromosomal cross-linked microtubule population is initially zero.
All three populations are subject to two kinds of net changes: conversion from one population to another, and poleward movement. Conversion takes place by cross-linking of randomly aligned microtubules or dissociation of cross-linked or aligned microtubules; rates of conversion may be approximated based on existing microtubule populations. First, randomly aligned microtubules can be cross-linked to microtubules aligned parallel to the spindle axis; we take this rate to be kaρ1ρ2, assuming first-order kinetics in each microtubule pool. However, for entropic reasons, aligned microtubules will tend to lose alignment over time; we let its rate be kdρ2, assuming this is also a first-order process. Likewise, randomly aligned microtubules attached to different chromosomes can be cross-linked together, and the rate of this may be taken as kcρ12, assuming first-order kinetics in the microtubule pools on each side of the potential cross-link. However, interchromosomal and intrachromosomal cross-links compete, so these linkages may be broken, with a rate taken as kdcρ1ρ3.
Net poleward microtubule movement is assumed only to occur due to transport or elongation of the microtubules aligned parallel to the spindle axis (i.e., due to changes in ρ2). Other mechanisms, such as poleward flux, are not considered here, for simplicity and because they have not been demonstrated to play a role in the Drosophila oocyte spindle, which is the basis of this model. At steady state, other microtubules may stochastically move toward the poles if there is greater depolymerization there, but the population as a whole will not show net movement. A given microtubule slides at a velocity proportional to the amount of aligned microtubules between itself and the equator. Since Ncd can bind two different microtubules, one by its head and one by its tail, its motion need only be minus-end-directed on the head-bound microtubule. The other microtubule can be oriented in either direction and movement by Ncd thus can still cause elongation, as with any motor. The assumption is based on this, along with the limitation of microtubule motion by Stokes drag, causing the force propelling a microtubule (which is proportional to the amount of motor—and thus roughly to the amount of aligned microtubules—between the given microtubule and the equator) to be proportional to its velocity. We let this velocity at a distance x from the equator be
| (13) |
where ke is a constant dependent on motor concentrations. This gives a continuity equation for ρ2, which provides its time evolution:
| (14) |
It also gives the time evolution of the total spindle length L:
| (15) |
Unaligned microtubules of either population are likely to be pulled by this sliding, but at a slower speed,
| (16) |
where kr is dependent on ke and on the extent of cross-linking between spindle axis-aligned and nonaligned microtubules, kr < ke. This also yields a continuity equation, providing the time evolution of ρ1 and ρ3:
| (17) |
The mechanism for this sliding most likely involves motors walking along the spindle-aligned and heavily cross-linked microtubules, whose minus ends are oriented toward the poles; microtubules that are bound to the tail of the moving motor would thus be propelled toward the poles, creating the sliding motion. Because the microtubules are attached to fixed structures in the middle of the spindle—the chromosomes—that receive equal force from both directions (and thus no net force), this system allows a single motor to bring about spindle expansion to a steady state, differing from the case of two asters that either fuse or dissociate completely when subjected to force from a single motor (8).
The model reaches equilibrium as ρ2 goes to zero, since this allows all time derivatives to go to zero. Qualitatively, the basis of steady state in this model is that the population of aligned microtubules driving the elongation is depleted by being spread out and cross-linked in other directions, including to microtubules associated with other chromosomes. At steady state, there would be no more microtubules aligned with the spindle axis than with any other direction, so that elongation is not feasible. The equilibrium length will, however, be dependent on initial amounts of microtubules as well as sliding and cross-linking rates. At equilibrium,
| (18) |
everywhere. Note that since motor concentrations and microtubule assembly/disassembly rates may vary over time, spindle elongation and contractions may occur stochastically, even at steady state, e.g., Zou et al. (18).
It is important to note that this equilibrium is unlikely to last throughout meiosis; instead, changes in cross-linking are necessary after spindle assembly to segregate the chromosomes. This is supported by observations in mouse oocyte spindles of bipolar spindle formation preceding biorientation of chromosomes and oscillation of chromosomes on the metaphase plate, indicating that the microtubule organization at the completion of spindle assembly does not support chromosome segregation or other directional motion of the chromosomes (4). This reorganization would correspond to changes in motor activity, analogous to the changes in mitotic spindles that cause the chromosomes to be first aligned on the metaphase plate and then segregated. The microtubules involved in oocyte meiotic chromosome segregation, however, differ from the spindle axis-aligned microtubules described above that are depleted at the end of spindle assembly (the ρ2 population), as these would disperse the chromosomes throughout the pole regions instead of segregating them into groups. In other words, the new microtubules must retain the cross-linking at the poles (characteristic of the ρ3 population, which is dominant at the end of spindle assembly).
Diffusional effects on spindle elongation
An additional factor that may contribute to spindle elongation is longitudinal diffusion; microtubules have been observed to exhibit this random motion during inhibition of motor ATPase activity or with truncated motors (19,20), and a localized distribution of particles subjected to diffusion will spread out over time, so a population of microtubules aligned to the spindle axis is expected to spread out over time with an effect similar to motor-induced sliding, but much weaker and slower. This effect is thus unlikely to be significant unless the motor ATPase is inhibited or truncated motors are present.
Overall features of the model
This model resembles the slide-and-cluster model proposed previously for anastral spindle assembly (10), insofar as it incorporates both sliding and clustering, and takes the rates of these processes to be responsible for the shape of the meiotic spindle. However, it differs from this model in that it starts with a population of poorly aligned microtubules growing out in all directions from asters associated with the chromosomes, as observed in live Drosophila and mouse oocytes at the initial stages of spindle assembly (3,4), rather than only on nucleation at the chromosomes. It is also novel in postulating a single motor, Ncd, to perform both sliding and clustering, although recognizing a potential minor role for diffusion, or possibly other motors working alongside or opposing Ncd. In this, and in its deterministic nature, it also differs from the model proposed by Schaffner and José (9); stochastic simulations could be performed in this model, but deterministic solutions are likely to be simpler and more informative, and thus have been used in this study.
Predictions of the model
The model agrees with the key confirmed predictions in Burbank et al. (10):
-
1.
Spindles reach a steady state based on intrinsic factors—in this case, the kinetic coefficients, which are dependent primarily on motor concentrations, and initial conditions consisting of a disorganized microtubule population associated with the chromosomes.
-
2.
Some minus ends will be found throughout the spindle, due to the random alignment of the microtubules.
-
3.
Microtubules slide toward the poles with the leading end being primarily the end that is clustered (i.e., cross-linked) the most at the poles. Assuming that this cross-linking is due to Ncd, a minus-end motor, the minus ends would be leading. This would be similar to Fig. 2 B, a and c, but with the two microtubules attached to the Ncd motor parallel to one another instead of antiparallel.
-
4.
Microtubules will slide against each other more slowly near the poles because of the smaller population of spindle axis-aligned microtubules at the poles.
-
5.
The pole position is dependent on global microtubule sliding rates, as expressed by ke and kr, rather than on the position of a nucleating and organizing center. Positional variation of sliding velocities, which is determined by the initial conditions and some position-independent constants, can also affect pole position.
-
6.
The microtubule velocity gradient is produced by initial conditions and cross-linking; perturbations in either of these could inhibit elongation and pole formation. In particular, inhibition of cross-linking between different populations of microtubules will likely lead to multipolar or multiple spindles.
Our model also produces predictions not made by previous models:
-
1.
Spindle assembly, including both sliding and clustering, should be capable of proceeding with only Ncd as the motor. The disruptive effects of ncd null and loss-of-function mutants on meiotic spindle assembly provides evidence of the central role that Ncd plays (2,3). It is possible that other motors are involved and may affect spindle length, shape, or assembly time, but spindles should assemble without them. A test of this prediction would be to analyze mutants of other candidate motors.
-
2.
Many microtubules are not oriented parallel to the spindle axis; instead, they are diagonal or even perpendicular to the axis, in the case of shorter microtubules. This is especially true at steady state. This could be tested by EB1 particle tracking.
-
3.
Cross-linking between microtubules associated with individual chromosomes begins early, but at first is mainly localized near the equator. This matches the observation that during early spindle assembly, chromosomes do not drift apart even though focused poles have not yet formed.
-
4.
Because of the cross-linking between chromosomes by Ncd, minus ends are likely to be concentrated to a large degree at the poles and in a band that runs between the poles down the center of the spindle, rather than in the peripheral parts of the spindle. Due to the presence of the chromosomes, it may be somewhat difficult to distinguish this configuration from polar localization. This conclusion is consistent with studies reporting enhanced minus-end populations at the poles, but a significant number of minus ends elsewhere.
-
5.
Since Ncd is responsible for both sliding and clustering, inhibition or loss of Ncd function will cause loss of bipolarity and loss of adherence between chromosomes. This has been observed in spindles of ncd null and loss-of-function oocytes (2,3). If only sliding activity is inhibited, ke and kr will decrease markedly, although they will probably not reach zero because of longitudinal diffusion. As described in Perturbations below, this will slow down spindle assembly, and higher cross-linking rates will become necessary to attain the same equilibrium spindle length.
Example “normal” simulation
Simulations were performed in arbitrary units. Time points from a “normal” simulation are shown in Fig. 3. In each graph, the density of unaligned microtubules, ρ1, is in bright green; the density of spindle axis-aligned microtubules, ρ2, in dark green; and the density of microtubules cross-linking different chromosomes, ρ3, in yellow. For the initial conditions, ρ1 = 1 for 10 units of space around the equator and then falls off exponentially with relative rate = 0.03, while ρ2 = 0.1 at the equator and immediately falls off exponentially at the same rate (Fig. 3 A). Concentrations (ρ) versus distance from equator (x) are shown in the graphs.
Figure 3.
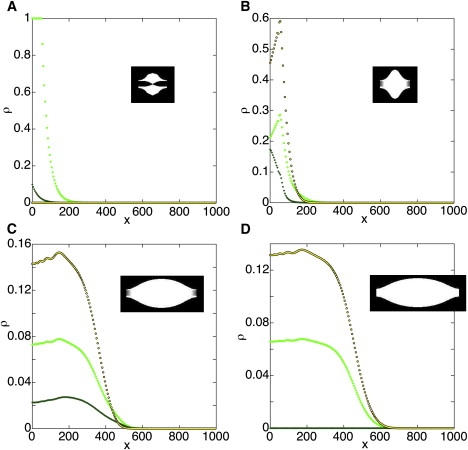
A simulation representing normal behavior. ka = 1, kd = 0.1, kc = 4, kdc = 2, kr = 0.3, and ke = 0.6. ρ1 (randomly aligned microtubules (MTs), bright green), ρ2 (spindle axis-aligned MTs, dark green), and ρ3 (interchromosomal cross-linking, yellow with black outlines). The simulation was performed in arbitrary units. (A) Initial conditions. (B) The simulation begins with rapid cross-linking (t = 5 time units from start); then (C) elongation is observed (t = 50). (D) As the spindle reaches steady state, ρ2 is depleted (t = 500). A depiction of the spindle is shown at each time point.
A set of parameters that appeared fairly realistic was ka = 1, kd = 0.1, kc = 4, kdc = 2, kr = 0.3, and ke = 0.6. The simulation begins with rapid cross-linking (Fig. 3 B), and then elongation is observed (Fig. 3 C). As ρ2 is depleted, the spindle reaches steady state (Fig. 3 D).
This steady state is consistent with the results reported by Liang et al. for microtubule dynamics in live oocyte spindles (7). The spindle comprises microtubules with minus-ends oriented in either direction, toward the chromosomes at the equator or toward the poles, and polymerization and depolymerization occur in both directions because of dynamic instability. Liang et al. observed depletion of fluorescence from either the poles or the equator of a fluorescently-labeled spindle when the equator or a pole, respectively, was bleached repeatedly in fluorescence loss in photobleaching (FLIP) assays. The rate of depletion depended primarily on the amount of fluorescent protein that was bleached, not where the protein was located. Bleaching the equator, which is thicker and consists of more microtubules, and thus has more fluorescently-labeled protein to bleach, resulted in a slightly higher rate constant for fluorescence loss or kFLIP at the poles, than bleaching a pole had on kFLIP at the equator. Similarly, bleaching half the equator markedly lowered kFLIP at the poles. These dynamics are likely to be similar throughout the spindle, consistent with the fluorescence-recovery-in-photobleaching (FRAP) data reported by Liang et al. (7), and the conclusion from this study that microtubule dynamics are similar at the poles and equator of the oocyte meiosis I spindle.
Perturbations
Some perturbations were also tried; as in the model of Burbank et al. (10), spindle assembly was capable of completing under a variety of different conditions. For example, ka = 1, kd = 0.1, kc = 0.4, kdc = 0.2, kr = 0.03, and ke = 0.06 produced a spindle in a similar timescale, albeit ∼20% shorter and with a much larger ρ2 reached during assembly and then disappearing as steady-state ensued. The spindle produced by the normal parameter set above, except with sliding slowed by 10-fold, also formed successfully, although the spindle was reduced in length by nearly half and the assembly time was approximately doubled relative to the first parameter set (Fig. 4 A).
Figure 4.
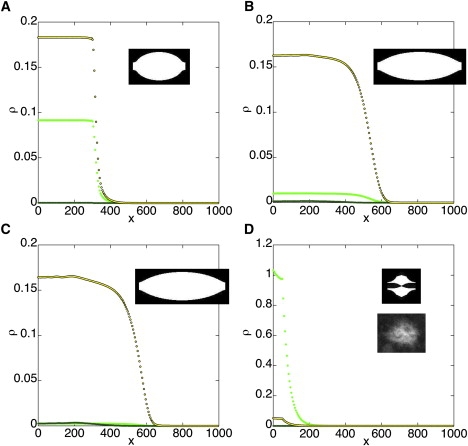
Perturbed spindles, shown at the end of the assembly time. (A) Inhibited sliding: ka = 1, kd = 0.1, kc = 4, kdc = 2, kr = 0.03, ke = 0.06, and t = 700. (B) Inhibited sliding and enhanced cross-linking: ka = 2, kd = 0.025, kc = 8, kdc = 0.5, kr = 0.02, ke = 0.04, and t = 1200. (C) More strongly inhibited sliding and enhanced cross-linking: ka = 4, kd = 0.0125, kc = 16, kdc = 0.25, kr = 0.006, ke = 0.012, and t = 4000. (D) Inhibition of both sliding and cross-linking: ka = 0.025, kd = 0.1, kc = 0.1, kdc = 2, kr = 0.01, ke = 0.02, and t = 500. (Bottom inset) A Drosophila oocyte spindle mutant for ncd2, thought to be defective in ATPase activity and thus to exhibit inhibition of both sliding and cross-linking. Colors and units as in Fig. 3.
The N600K mutation in Ncd severely inhibits any Ncd-dependent motility, including sliding; however, it increases tightness of motor binding to microtubules and thus of cross-linking (21). As shown above, inhibition of sliding in our model increases spindle assembly time and decreases equilibrium spindle length; however, spindle length can be normal or even aberrantly long if cross-linking is strengthened. A simulation was performed (Fig. 4 B) in which sliding was reduced 15-fold relative to the normal spindle above (Fig. 3), but cross-linking was strengthened. This strengthening allowed normal spindle length to be recovered over a longer assembly process. Note that given enough time and cross-linking, a spindle with very inhibited sliding can still reach normal length, as shown in Fig. 4 C with 50-fold inhibited sliding relative to the original.
If ke = kr = 0 (i.e., there is no sliding), then growth does not occur. However, as noted above, even in the absence of motor there is likely to be some diffusional sliding. If cross-linking and sliding are both strongly inhibited, the spindle does not elongate and the chromosomes do not become cross-linked together (Fig. 4 D). The spindle essentially arrests at early stages of assembly, consistent with behavior observed for ncd2, a severe loss-of-function mutant thought to be defective in ATPase activity (3).
A bidirectional Ncd has been shown to produce mostly normal meiosis I spindles (3)—this is consistent with the model since the mutant motor is expected to have both sliding and cross-linking capabilities, with any inhibition likely to be related to sliding due to a tendency to slide in both directions.
Estimation of constants
To estimate the values of the kinetic constants during the process of spindle elongation, the spindle lengths and assembly times from the normal simulation were compared to values observed in live oocytes. The length of the mature meiosis I spindle is ∼15 μm (18), corresponding to 600 arbitrary distance units, and it assembles in ∼40 min (3), corresponding to 500 arbitrary time units. This yields an estimated value of 0.025 μm for the distance unit of the simulation and 0.08 min for the time unit. The steady-state tubulin concentration in the oocyte meiosis I spindle was roughly estimated at ∼30 μM, assuming a cytoplasmic concentration of 1–2 μM Ncd-GFP in oocytes, as in embryos (22), and that Ncd-GFP is bound on average to one in four tubulin subunits in the spindle. This corresponds to ∼0.2 arbitrary density units in the simulation and gives 150 μM for its density unit. Consequently, the “normal” parameter values may be estimated to be ka = 0.08 min−1 μM−1, kd = 1 min−1, ke = 0.05 min−1 μM−1, kr = 0.03 min−1 μM−1, kc = 0.3 min−1 μM−1, and kdc = 0.2 min−1 μM−1.
Summary
In our model, anastral spindle assembly is initiated by the aggregation of microtubule asters around the chromosomes; this process is mediated by binding to cytoskeletal filaments and directed transport, together with diffusion—diffusion alone would result in a smaller fraction of asters reaching the chromosomes than observed. Elongation and establishment of the bipolar spindle then occurs by sliding and cross-linking of microtubules, which we assign to three populations: those with random alignment; those aligned with the spindle axis; and those cross-linking different chromosomes. This model results in the assembly of normal spindles (Fig. 3), whereas perturbations cause abnormal spindles to form (Fig. 4). The kinesin-14 Ncd is posited to perform both sliding and cross-linking; other motors may be involved but are not predicted to be essential. The effects of perturbations on spindle assembly are consistent with observations from Ncd mutant studies. The model produces several testable predictions, including the role of other motors, as well as the presence of many microtubules perpendicular to or diagonal to the spindle axis, especially at steady state.
Supporting Material
One table and two equations are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01362-9.
Supporting Material
Acknowledgments
This study was supported by grants to S.A.E. from the National Institutes of Health (GM046225) and March of Dimes (1-FY07-443). M.A.H. is a 2008 Goldwater scholar.
References
- 1.Heald R., Tournebize R., Blank T., Sandalzopoulos R., Becker P. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 2.Matthies H.J.G., McDonald H.B., Goldstein L.S.B., Theurkauf W.E. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sköld H.N., Komma D.J., Endow S.A. Assembly pathway of the anastral Drosophila meiosis I oocyte spindle. J. Cell Sci. 2005;118:1745–1755. doi: 10.1242/jcs.02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuh M., Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Yang G., Houghtaling B.R., Gaetz J., Liu J.Z., Danuser G. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat. Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
- 6.Burbank K.S., Groen A.C., Perlman Z.E., Fisher D.S., Mitchison T.J. A new method reveals microtubule minus ends throughout the meiotic spindle. J. Cell Biol. 2006;175:369–375. doi: 10.1083/jcb.200511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Z.-Y., Hallen M.A., Endow S.A. Mature Drosophila meiosis I spindles comprise microtubules of mixed polarity. Curr. Biol. 2009;19:163–168. doi: 10.1016/j.cub.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nédélec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J. Cell Biol. 2002;158:1005–1015. doi: 10.1083/jcb.200202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaffner S.C., José J.V. Biophysical model of self-organized spindle formation patterns without centrosomes and kinetochores. Proc. Natl. Acad. Sci. USA. 2006;103:166–171. doi: 10.1073/pnas.0604721103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burbank K.S., Mitchison T.J., Fisher D.S. Slide-and-cluster models for spindle assembly. Curr. Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 11.Lancaster O.M., Cullen C.F., Ohkura H. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J. Cell Biol. 2007;179:817–824. doi: 10.1083/jcb.200706067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg H.C. Princeton University Press; Princeton, NJ: 1983. Random Walks in Biology. [Google Scholar]
- 13.Sprague B.L., Pego R.L., Stavreva D.A., McNally J.G. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachsmuth M., Waldeck W., Langowski J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 2000;298:677–689. doi: 10.1006/jmbi.2000.3692. [DOI] [PubMed] [Google Scholar]
- 15.Endow S.A., Higuchi H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature. 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- 16.Hunt A.J., Gittes F., Howard J. The force exerted by a single kinesin molecule against a viscous load. Biophys. J. 1994;67:766–781. doi: 10.1016/S0006-3495(94)80537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fink G., Hajdo L., Skowronek K.J., Reuther C., Kasprzak A.A. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat. Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 18.Zou J., Hallen M.A., Yankel C.D., Endow S.A. A microtubule-destabilizing kinesin motor regulates spindle length and anchoring in oocytes. J. Cell Biol. 2008;180:459–466. doi: 10.1083/jcb.200711031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vale R.D., Soll D.R., Gibbons I.R. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- 20.Chandra R., Endow S.A., Salmon E.D. An N-terminal truncation of the ncd motor protein supports diffusional movement of microtubules in motility assays. J. Cell Sci. 1993;104:899–906. doi: 10.1242/jcs.104.3.899. [DOI] [PubMed] [Google Scholar]
- 21.Song H., Endow S.A. Decoupling of nucleotide- and microtubule-binding in a kinesin mutant. Nature. 1998;396:587–590. doi: 10.1038/25153. [DOI] [PubMed] [Google Scholar]
- 22.Hallen M.A., Ho J., Yankel C.D., Endow S.A. Fluorescence recovery kinetic analysis of γ-tubulin binding to the mitotic spindle. Biophys. J. 2008;95:3048–3058. doi: 10.1529/biophysj.108.134593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


