Abstract
The food-borne pathogen Campylobacter jejuni is dependent on a functional flagellum for motility and the export of virulence proteins that promote maximal host cell invasion. Both the flagellar and non-flagellar proteins exported via the flagellar type III secretion system contain a sequence within the amino-terminus that directs their export from the bacterial cell. Accordingly, we developed a genetic screen to identify C. jejuni genes that encode a type III secretion amino-terminal sequence that utilizes the flagellar type III secretion system of Yersinia enterocolitica and a phospholipase reporter (yplA). We screened a library of 321 C. jejuni genes and identified proteins with putative type III secretion amino-terminal sequences. One gene identified by the screen was Cj1242. We generated a mutation in Cj1242, and performed growth rate, motility, secretion and INT 407 cell adherence and internalization assays. The C. jejuni Cj1242 mutant was not altered in growth rate or motility when compared with the wild-type strain, but displayed an altered secretion profile and a reduction in host cell internalization. Based on the phenotype of the C. jejuni Cj1242 mutant, we designated the protein Campylobacter invasion antigen C (CiaC). Collectively, our findings indicate that CiaC is a potentially important virulence factor.
Introduction
Campylobacter jejuni, a Gram-negative pathogen, is one of the leading bacterial causes of gastroenteritis worldwide (Allos, 2001; Westrell et al., 2009). The clinical presentation of C. jejuni-mediated disease varies from one individual to another, where some individuals have watery diarrhoea and others experience diarrhoea with blood (Blaser et al., 1983; Friedman et al., 2004). The reason for the variation in clinical presentation is not known. We speculate that both the unique virulence factors of the infecting strain as well as the host innate immune response influence the presentation and severity of disease (Larson et al., 2008). The most severe form of campylobacteriosis, which is characterized by fever, severe abdominal cramps and diarrhoea containing blood and leucocytes, likely results from C. jejuni invasion of the intestinal epithelium. Indeed, intracellular bacteria have been observed by electron microscopy examination of samples from C. jejuni-infected individuals with acute infectious colitis characterized by diarrhoea with blood (van Spreeuwel et al., 1985).
Campylobacter jejuni must be metabolically active and secrete proteins from the flagellar type III secretion system (T3SS) for maximal invasion of host epithelial cells (Konkel and Cieplak, 1992; Konkel et al., 1993; 2004). The proteins synthesized and secreted by C. jejuni upon cocultivation with epithelial cells are termed Campylobacter invasion antigens (Cia) (Konkel et al., 1999). The importance of the Cia proteins in C. jejuni pathogenesis has been demonstrated with a ciaB mutant, which is deficient in Cia protein secretion. The severity and time of onset of disease in piglets inoculated with a C. jejuni ciaB null mutant is significantly attenuated when compared with a C. jejuni wild-type isolate. The piglets inoculated with the C. jejuni ciaB null mutant did not develop diarrhoea until 3 days post inoculation whereas all piglets inoculated with a C. jejuni wild-type isolate developed diarrhoea within 24 h (Raphael et al., 2005).
Gram-negative bacteria have evolved distinct secretion systems to actively transport proteins across their membranes (Thanassi and Hultgren, 2000; Pallen et al., 2003; Kostakioti et al., 2005). The T3SS is characterized by the export of proteins across both membranes of the bacterium, which normally occurs upon bacteria–host cell contact (Cornelis, 2006; Galan and Wolf-Watz, 2006). In C. jejuni, the only T3SS is the flagellar apparatus (Parkhill et al., 2000). Previous work has demonstrated that the secretion of C. jejuni Cia and other virulence proteins is dependent on a functional flagellar T3SS (Konkel et al., 2004; Poly et al., 2007). Precedence for the secretion of a virulence factor from the flagellum was first demonstrated with Yersinia enterocolitica (Schmiel et al., 1998; 2000; Young et al., 1999), which utilizes the flagellar T3SS to export a phospholipase termed YplA.
The majority of the proteins secreted from C. jejuni, including the Cia virulence proteins, have not yet been identified due in part to low levels of protein secretion under in vitro conditions. The aim of this study was to identify a virulence protein that is secreted from the C. jejuni flagellar T3SS. As a first step in the identification of putative Campylobacter-secreted proteins (Csp), we tested if CiaB would be recognized and secreted from the well-characterized flagellar T3SS of Y. enterocolitica (Warren and Young, 2005). Based on the finding that CiaB was secreted from Y. enterocolitica, we developed a screen that utilized Y. enterocolitica and the YplA effector protein to identify C. jejuni genes that encode amino-terminal residues that facilitate protein secretion in a T3SS-dependant manner (i.e. T3S amino-terminal sequences) (Schmiel et al., 2000; Berring et al., 2004; Warren and Young, 2005). We demonstrated that the screen had the potential to identify putative Csp with T3S amino-terminal sequences using known C. jejuni flagellar secreted proteins. We report the identification of 42 C. jejuni proteins with amino-terminal sequences that promote secretion from the Y. enterocolitica flagellar T3SS. From this list, one gene (Cj1242) encoding a hypothetical protein was selected for additional study. We generated a mutation in Cj1242, and examined the growth rate, motility, secretion profile and adherence and invasion properties of the C. jejuni Cj1242 mutant relative to the wild-type isolate. The C. jejuni Cj1242 mutant displayed an altered secretion profile and reduced host cell invasion, demonstrating that Cj1242 is a virulence protein.
Results
The CiaB protein is secreted via the Y. enterocolitica flagellar T3SS
Based on the finding that CiaB is secreted via the flagellar T3SS of C. jejuni (Konkel et al., 1999), we reasoned that CiaB should be recognized and secreted in a T3SS-dependent manner in a heterologous system. To test this possibility, the full-length ciaB gene was cloned into the pMMB207 plasmid and conjugated into the Y. enterocolitica JB580v wild-type strain and Y. enterocolitica GY4492, a mutant lacking any functional T3SS (pYV8081-ΔflhDC ysaT). These bacterial strains and plasmids are described in Table 1. Whole-cell lysate and supernatants were collected from the Y. enterocolitica strains cultured under conditions to induce the secretion of the flagellar outer proteins (Fops) (i.e. 2 h at 26°C in TYE broth medium). The Fops represent a set of at least 12 proteins secreted from the flagellar T3SS, including the flagellar filament proteins FleABC. As expected, the Y. enterocolitica JB580v wild-type strain secreted the Fops, whereas the Y. enterocolitica pYV8081-ΔflhDC ysaT mutant did not secrete the Fops (Fig. 1A). The supernatants were also probed with the mouse monoclonal flagellin-specific antibody 15D8 for the detection of the Y. enterocolitica FleABC flagellar filament proteins (38–40 kDa) (Kapatral and Minnich, 1995). The FleABC proteins were detected in the supernatants of Y. enterocolitica JB580v wild-type strain, demonstrating that the flagellar T3SS was functional, whereas the FleABC proteins were not detected from supernatants of the Y. enterocolitica pYV8081-ΔflhDC ysaT T3SS mutant (Fig. 1B). Importantly, the CiaB protein (73 kDa) was detected in the supernatant of the Y. enterocolitica JB580v wild-type strain, but not the Y. enterocolitica flagellar mutant, as judged by immunoblot analysis with a rabbit polyclonal CiaB-specific antibody (Fig. 1C). The detection of CiaB protein in the supernatant was not due to bacterial cell lysis, as the cytoplasmic protein sigma 70 (σ70) was not detected in the supernatants (Fig. 1D). As an additional control, we found that CiaB was synthesized and could be detected in the whole-cell lysate of the Y. enterocolitica pYV8081-ΔflhDC ysaT T3SS mutant (Fig. 1E). As expected, the Y. enterocolitica cytoplasmic protein σ70 was detected in the whole-cell lysates prepared from each of the bacterial strains (Fig. 1F). Collectively, these results indicate that CiaB is recognized as a flagellar T3 protein secreted by Y. enterocolitica.
Table 1.
Strains and plasmids.
| Genotype | Source or reference | |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| NCTC 11168 | Wild-type genome sequenced strain | Parkhill et al. (2000) |
| F38011 | Clinical isolate | Rivera-Amill and Konkel (1999) |
| F38011 flgB | Insertion disruption of flgB, non-motile and Cia protein secretion deficient | Konkel et al. (2004) |
| Y. enterocolitica | ||
| JB580v | Serogroup O:8, Nal yenR (r− m+) | Kinder et al. (1993) |
| GY4478 | JB580v, pYV8081− | Young and Young (2002) |
| GY4757 | JB580v ΔyplAB, pYV8081− | Warren and Young (2005) |
| GY4492 | JB580v ΔflhDC ysaT::TnMod-RKm, pYV8081− | Young and Young (2002) |
| Escherichia coli | ||
| S17-1 lambda pir | recA thi pro hsdR− M+ RP4::2-Tc::Mu::Km Tn7 pir | Simon et al. (1983) |
| Inv-alpha F′ | F′endA1 recA1 hsdR17 (r−, m+) supE44 thi-1 gyrA96 relA1 f80lacZDM15 D(lacZYA-argF) U169l- | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′proAB lacIZΔM15 Tn10) | Stratagene |
| Plasmids | ||
| pMMB207 | mob+, low copy vector containing an inducible tac promoter (Ptac), Cm | Morales et al. (1991) |
| pMEK250 | pMMB207 harbouring the full-length 1.9 kb ciaB gene driven by Ptac | This study |
| pTM100 | mob+, derivative of pACYC184, Cm Tet | Michiels and Cornelis (1991) |
| pCSP50 | Pcat upstream of NdeI and BglII sites for directional cloning of fusions with 5′-truncated yplA (lacking nucleotides 4–150) and complete yplB locus cloned into pTM100 EcoRI site, Tet | This study |
| pCSP50-yplA 1–108 | Nucleotides 1–108 of yplA fused to truncated yplA in pCSP50 | This study |
| pCSP50-flaA 1–108 | Nucleotides 1–108 of flaA (Cj1339c) fused to truncated yplA in pCSP50 | This study |
| pCSP50-flaC 1–108 | Nucleotides 1–108 of flaC (Cj0720c) fused to truncated yplA in pCSP50 | This study |
| pCSP50-ciaB 1–108 | Nucleotides 1–108 of ciaB fused to truncated yplA in pCSP50 | This study |
| pCSP50-cysM 1–108 | Nucleotides 1–108 of cysM fused to truncated yplA in pCSP50 | This study |
| pBluescript II SK+ | Phagemid cloning vector | Stratagene |
| pMW10 | C. jejuni–E. coli shuttle vector, Kan | Wosten et al. (1998) |
| pBSK-Kan2 | pBluescript II SK+ with original ampicillin cassette replaced by the native promoter and apha3 gene from pMW10, Kan | This study |
| pBSK-Kan2:delCj1242 | pBSK-Kan2 with Cj1242 internal deletion and disrupted with tetO from pUOA3, Kan Tet | This study |
| pRY111 | C. jejuni–E. coli shuttle vector, pWKS29 MCS, Cm | Yao et al. (1993) |
| pRY111:Cj1242 | pRY111 with a 1.7 kb fragment encompassing Cj1242, Cm | This study |
Fig. 1.
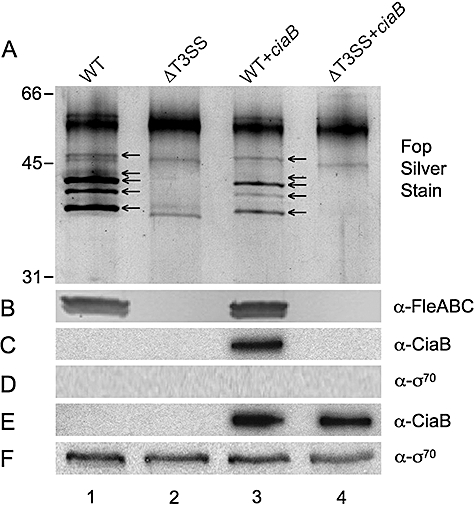
The C. jejuni CiaB protein is secreted via the Y. enterocolitica flagellar T3SS. Supernatants (A–D) and whole-cell lysates (E and F) were analysed by SDS-PAGE coupled with silver staining or immunoblot analysis. A. Silver stain showing the flagellar outer proteins (Fops) and FleABC. B. Immunoblot probed with the flagellin antibody (FleABC, 38–40 kDa). C. Immunoblot probed with the CiaB antibody (CiaB, 73 kDa). D. Immunoblot probed with the RNA polymerase σ70 antibody. E. Immunoblot probed with the CiaB antibody. F. Immunoblot probed with the σ70 antibody. Lanes: 1, Y. enterocolitica wild-type harbouring the empty pMMB207 vector (WT); 2, Y. enterocolitica pYV8081–ΔflhDC ysaT flagellar mutant harbouring the empty pMMB207 vector (ΔT3SS); 3, Y. enterocolitica wild-type harbouring the pMMB207 vector containing the C. jejuni ciaB gene (WT + ciaB); and 4, Y. enterocolitica pYV8081–ΔflhDC ysaT flagellar mutant harbouring the pMMB207 vector containing the C. jejuni ciaB gene (ΔT3SS + ciaB).
C. jejuni T3S amino-terminal sequences promote secretion from the Y. enterocolitica flagellar T3SS
All proteins exported via a T3SS contain an amino-terminal sequence to direct their export from the bacterial cell. Moreover, previous work has shown that a T3SS protein can be: (i) recognized and secreted by more than one T3SS in the same bacterium and (ii) recognized and secreted from bacteria that belong to other genera (Young and Young, 2002; Lee and Galan, 2004; Badea et al., 2009). As CiaB was secreted via the C. jejuni and Y. enterocolitica flagellar T3SS, we hypothesized that the CiaB amino-terminus would direct the export of a fusion protein from Y. enterocolitica in a T3SS-dependent manner. In addition, we hypothesized that the amino-termini of two other C. jejuni flagellar-secreted proteins, FlaA and FlaC, would also promote secretion of a fusion protein. To test this hypothesis, we generated the pCSP50 shuttle vector encoding the Y. enterocolitica yplA phospholipase gene as a reporter (Fig. 2). The Y. enterocolitica YplA enzyme is an A2 phospholipase and is secreted under flagellar T3SS inducing conditions in vitro (Schmiel et al., 1998; Berring et al., 2004). Warren and Young (2005) determined that the YplA enzyme's T3S amino-terminal sequence is localized within the first 20 residues. The pCSP50 shuttle vector incorporates a constitutive promoter (cat) upstream of NdeI and BglII cloning sites, a 5′-truncated yplA gene (eliminating the first 50 amino acids including the T3S amino-terminal sequence), and the yplB chaperone gene. The amino-terminal deletion of the Y. enterocolitica YplA protein abolished its secretion, but not its enzymatic (phospholipase) activity (not shown) (Hatic et al., 2002).
Fig. 2.
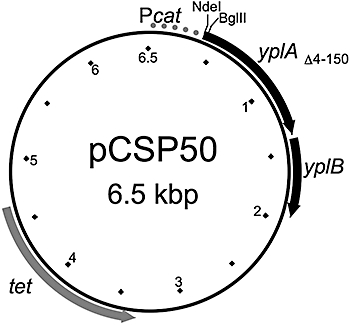
The pCSP50 shuttle vector. The NdeI and BglII sites flank the 5′ end of a truncated yplA gene and facilitate directional cloning to generate fusions with 108 bp amino-terminal sequences from C. jejuni genes.
The Y. enterocolitica JB580v wild-type strain secretes YplA under flagellar T3SS-inducing conditions and the enzymatic activity can be detected on phospholipase agar (PLA) plates (not shown). The hydrolysis of Tween 80 in PLA plates results in a fatty acid precipitate that forms a halo surrounding the YplA secretion-competent colonies. In contrast, the Y. enterocolitica yplAB strain GY4757, generated for use in conjunction with a YplA reporter, showed no detectable phospholipase activity. Similarly, the Y. enterocolitica yplAB strain harbouring the native pCSP50 vector was secretion negative (Fig. 3A). However, when the first 108 nucleotides of yplA (1–36 AA encoding sequence) was fused to the truncated yplA gene, the YplA fusion protein was secreted and detected on PLA plates.
Fig. 3.
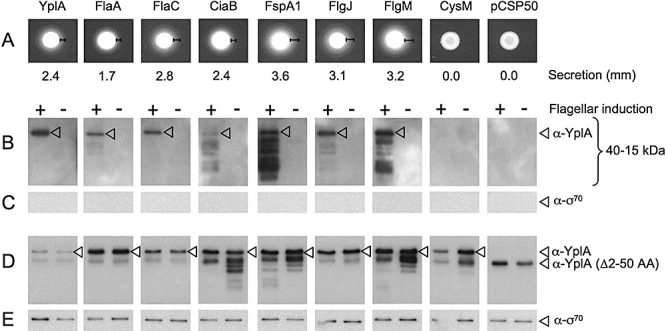
Secretion of YplA fusion proteins from Y. enterocolitica under flagellar T3SS-inducing conditions. The first 36 amino acids of each indicated protein was fused to YplA encoded on vector pCSP50. The YplA secretion zone widths (mm) were measured from the edge of the bacterial growth to the outer edge of the fatty acid precipitate. Detection of the YplA fusion protein by immunoblot analysis was done with cultures grown under flagellar-inducing conditions (lanes marked ‘+’) and non-inducing conditions (lanes marked ‘−’). A. A representative PLA assay is shown indicating YplA fusion proteins and secretion zone widths. B. Immunoblot analysis of supernatants probed with the YplA antibody. C. Immunoblot analysis of supernatants probed with the RNA polymerase σ70 antibody. D. Immunoblot analysis of whole-cell lysates probed with the YplA antibody. E. Immunoblot analysis of whole-cell lysates probed with the σ70 antibody.
To provide proof of concept for the screen for C. jejuni genes harbouring T3S amino-terminal sequences, we generated yplA fusions with the first 108 nucleotides of three genes encoding proteins known to be secreted via the C. jejuni flagellar T3SS (FlaA, FlaC, CiaB) (Fig. 3A). As predicted, all three fusions with YplA were secreted and detected on PLA plates. In contrast, a fusion of the first 108 nucleotides of the C. jejuni gene for CysM was generated to serve as a T3SS negative control, and no secretion was observed. CysM is a 32.4 kDa cytoplasmic protein (O-acetylserine sulfhydrylase B) involved in cysteine biosynthesis (Garvis et al., 1997).
Identification of C. jejuni genes harbouring putative T3S amino-terminal sequences
The results from the native CiaB secretion assay and the YplA reporter assay demonstrated that the Y. enterocolitica flagellar system could be utilized to identify a C. jejuni protein with a T3S amino-terminal sequence. Thus, a total of 359 genes from the 1654 identified ORFs from the C. jejuni NCTC 11168 sequence were selected to test via the YplA reporter assay (Parkhill et al., 2000). These genes/ORFs were chosen for analysis as the deduced amino acid sequences lack predicted membrane-spanning domains, periplasmic domains, Sec-dependent signals or Tat-dependent signals. No genes were found to encode type I Sec-independent motifs. Primers were designed to amplify the first 108 encoding bases of all 359 ORFs and facilitate directional cloning into the shuttle vector pCSP50 to generate translational fusions with the truncated YplA reporter. The first 108 bp for 341 of the 359 ORFs were successfully cloned and sequence confirmed in the Escherichia coli S17-1 λ-pir donor strain. From this fusion library, 321 vectors were successfully conjugated into the Y. enterocolitica yplAB host strain and characterized for YplA secretion on PLA plates (Table S1). Table 2 lists the 42 C. jejuni genes that harbour amino-terminal sequences that resulted in YplA secretion zone widths greater than or equal to that obtained with the CiaB amino-terminus from the Y. enterocolitica yplAB strain after 12 h incubation on PLA plates.
Table 2.
C. jejuni genes encoding a putative T3S amino-terminal sequence.
| Genea | Locus | Product description |
|---|---|---|
| flgMb,c | Cj1464 | Anti-sigma 28 factor |
| fspA1b,e | Cj0859c | Flagellar secreted protein, virulence factor |
| rrcb | Cj0012c | Non-haem iron protein, rubrerythrin |
| Cj0036 | Cj0036 | Hypothetical protein |
| Cj1242d | Cj1242 | Hypothetical protein |
| flgJb,d,f | Cj1463 | Flagellar rod protein |
| Cj0073cd | Cj0073c | Conserved hypothetical protein |
| Cj0122 | Cj0122 | Hypothetical protein |
| Cj0125c | Cj0125c | Hypothetical protein |
| Cj0140 | Cj0140 | Hypothetical protein |
| Cj0239c | Cj0239c | NifU protein homologue |
| Cj0251c | Cj0251c | Conserved hypothetical protein |
| Cj0787d | Cj0787 | Conserved hypothetical protein |
| Cj0788d | Cj0788 | Hypothetical protein |
| Cj0015c | Cj0015c | Hypothetical protein |
| Cj0021cb | Cj0021c | Putative fumarylacetoacetate hydrolase family protein |
| Cj0030 | Cj0030 | Hypothetical protein |
| hemN | Cj0363c | Putative oxygen-independent coproporphyrinogen III oxidase |
| Cj0416 | Cj0416 | Hypothetical protein |
| Cj0449c | Cj0449c | Conserved hypothetical protein |
| Cj0849c | Cj0849c | Conserved hypothetical protein |
| Cj1300b | Cj1300 | Putative SAM domain containing methyltransferase |
| Cj1543b | Cj1543 | Putative allophanate hydrolase subunit 2 |
| Cj0188cd | Cj0188c | Putative kinase |
| Cj0254 | Cj0254 | Hypothetical protein |
| Cj0391c | Cj0391c | Hypothetical protein |
| Cj0717b | Cj0717 | Putative ArsC family protein |
| Cj0973 | Cj0973 | Hypothetical protein |
| Cj1006cb,d | Cj1006c | Putative MiaB-like tRNA modifying enzyme |
| Cj1057cb | Cj1057c | Putative coiled-coil protein |
| Cj1089c | Cj1089c | Hypothetical protein |
| Cj1310c | Cj1310c | Hypothetical protein (617 family) |
| Cj1348cb | Cj1348c | Putative coiled-coil protein |
| Cj1497c | Cj1497c | Hypothetical protein |
| Cj0069 | Cj0069 | Hypothetical protein |
| Cj0668b | Cj0668 | Putative ATP/GTP-binding protein |
| Cj0681 | Cj0681 | Hypothetical protein |
| Cj0706d | Cj0706 | Conserved hypothetical protein |
| Cj0916c | Cj0916c | Conserved hypothetical protein |
| Cj1162cb | Cj1162c | Putative heavy-metal-associated domain protein |
| Cj1232 | Cj1232 | Hypothetical protein |
| Cj1505cb | Cj1505c | Putative two-component response regulator (SirA-like protein) |
C. jejuni gene yplA fusions with a secretion zone width greater than or equal to the zone obtained for the ciaB : yplA fusion strain. Listed in descending order of secretion zone width; ascending locus number for equivalent zones.
Not annotated in original NCTC 11168 sequence analysis (Parkhill et al., 2000).
Upregulated when grown in the presence of DOC (Malik-Kale et al., 2008).
C. jejuni–YplA fusion proteins are secreted to the culture supernatant by the Y. enterocolitica flagellar T3SS
To confirm that the YplA fusion enzyme activity measured by the PLA plate assay was the result of secretion through the Y. enterocolitica flagellar T3SS, we tested several strains by immunoblot analysis. Y. enterocolitica strains harbouring the pCSP50 vector with C. jejuni amino-terminal sequences were grown in broth culture under conditions that induced or repressed synthesis of the flagellar system. Importantly, the C. jejuni amino-terminal sequences fused to YplA were only detected in the supernatants of strains cultured under flagellar-inducing conditions (Fig. 3B). The amount of protein secreted into the supernatant, as judged by immunoblot analysis was roughly proportional to that measured by the PLA plate assay and varied according to the C. jejuni amino-terminal sequence. As predicted from the PLA plate assays, there were no reactive bands detected from the supernatants for the strains harbouring the CysM : YplA fusion or the pCSP50-truncated YplA. To evaluate the supernatants for bacterial lysis, which would result in the release of cytoplasmic proteins, the blots were probed using a mouse monoclonal antibody to the cytoplasmic protein σ70. No reactive band was detected for σ70 in any supernatants (Fig. 3C). Immunoblot analysis of the whole-cell lysates with a rabbit polyclonal YplA-specific antibody confirmed that the YplA fusion proteins (33.0–33.4 kDa, depending on amino-terminal sequence) were being synthesized under both flagellar and non-flagellar conditions (Fig. 3D). In addition, a band of consistent intensity was detected for both growth conditions corresponding to σ70 in the whole-cell lysate samples (Fig. 3E). Cumulatively, these data indicate that the YplA fusion proteins were secreted from the flagellar T3SS.
Functional classification of C. jejuni proteins harbouring putative T3S amino-terminal sequences
The functional classifications of the 42 proteins harbouring putative T3S amino-terminal sequences were obtained from the Sanger Institute website (http://www.sanger.ac.uk/) (Gundogdu et al., 2007). The majority of the C. jejuni NCTC 11168 proteins were classified as either conserved hypothetical proteins (16 proteins) or proteins of unknown function (14 proteins). Noteworthy is that two flagellar-related proteins (FlgM, FlgJ) and a pathogenicity-related protein (FspA) were identified among the proteins harbouring a T3S amino-terminal sequence, which had not been characterized when this study commenced.
Cj1242 is secreted from C. jejuni
To confirm that one of the proteins identified using the phospholipase indicator agar assay was secreted from the flagellar T3SS of C. jejuni, we generated a Cj1242 deletion mutant. Cj1242 was chosen because the Cj1242–YplA fusion protein was highly secreted from Y. enterocolitica (Table 2), the gene is predicted to be monocistronic and is upregulated when C. jejuni is cultured under conditions that induce virulence genes (Malik-Kale et al., 2008). The Cj1242 gene is capable of encoding a protein with a Mr 12 164. The growth rate and motility of the Cj1242 deletion mutant were indistinguishable from that of the C. jejuni wild-type strain (not shown). We then performed secretion assays to determine if the C. jejuni Cj1242 mutant was capable of Cia protein export.
The profile of Cia proteins detected from the C. jejuni F38011 wild-type strain was similar to that observed in previous work (Konkel et al., 2004). In contrast with the wild-type strain, the secretion profile of the C. jejuni Cj1242 mutant lacked one band of 12.2 kDa (Fig. 4A). The 12.2 kDa band was restored in the C. jejuni Cj1242 complemented strain, which was transformed with a plasmid harbouring a wild-type copy of Cj1242 in trans. Secreted proteins were not detected for the C. jejuni wild-type strain when fetal bovine serum (FBS) was omitted from the labelling medium, which is consistent with previous work indicating that components within serum are sufficient to induce Cia protein secretion (Konkel et al., 1999; Rivera-Amill et al., 2001). In addition, secreted proteins were not detected for the C. jejuni flgB mutant incubated with FBS, which is consistent with previous work indicating that Cia protein secretion is dependent on a functional flagellar secretion apparatus (Konkel et al., 2004). The presence of the Cia proteins in the supernatants from the wild-type strain, Cj1242 mutant and Cj1242-complemented strain was not due to bacterial lysis, because a 32.4 kDa band was not detected in supernatants probed with the CysM antibody (Fig. 4B). Coomassie brilliant blue (CBB R-250) staining of cell lysates from the secretion assay confirmed that equivalent quantities of protein were loaded (not shown), and an autoradiograph of the dried gel demonstrated equivalent labelling of cellular proteins with [35S]-methionine (Fig. 4C). A 32.4 kDa band was detected in the whole-cell lysates with the CysM antibody (Fig. 4D). Cumulatively, these data indicate that Cj1242 (CiaC) was secreted from the flagellar T3S system.
Fig. 4.
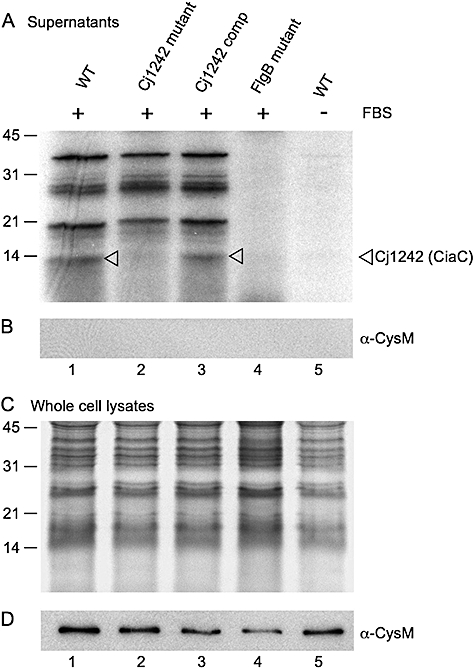
Secretion of Cj1242 (CiaC) from the C. jejuni flagellar T3SS. Isolates were incubated in medium containing [35S]-methionine and supplemented with 1% FBS or without FBS as described in Experimental procedures. Supernatants (A and B) and whole-cell lysates (C and D) were analysed by SDS-PAGE coupled with autoradiography and immunoblot analysis. A. Autoradiograph of supernatant samples; CiaC (12.2 kDa) protein is indicated by an arrowhead. B. Immunoblot of supernatant samples probed with the CysM antibody (32.4 kDa). C. Autoradiograph of whole-cell lysates. D. Immunoblot of whole-cell lysates probed with the CysM antibody. Lanes: (1) C. jejuni F38011 wild-type with 1% FBS; (2) C. jejuni F38011 Cj1242 mutant with 1% FBS; (3) C. jejuni F38011 Cj1242 mutant complemented with pRY111 : Cj1242 with 1% FBS; (4) C. jejuni F38011 flgB mutant with 1% FBS; (5) C. jejuni F38011 wild type without FBS.
Cj1242 (CiaC) is required for maximal C. jejuni invasion of host cells
Possible differences in bacterial adhesion and invasion between the C. jejuni wild-type strain and Cj1242 mutant were explored by the inoculation of human INT 407 epithelial cells. Quantification of adherent (i.e. cell-associated) and intracellular bacteria by the gentamicin-protection assay revealed that the adherence of the C. jejuni wild-type and the Cj1242 mutant to the INT 407 cells was indistinguishable from one another, but that the C. jejuni Cj1242 mutant was reduced in host cell invasion when compared with the wild-type isolate (P < 0.01) (Table 3). Based on the deficiency in host cell internalization, we designated the protein encoded by Cj1242 as Campylobacter invasion antigen C (CiaC).
Table 3.
Adherence and internalization of the C. jejuni wild-type strain and isogenic mutants.
| Numbers of viable bacteria |
|||
|---|---|---|---|
| Bacterial strain | Adherent | Internalized | I/Aa |
| C. jejuni wild-type | (7.1 ± 0.6) × 105 | (3.3 ± 0.5) × 104 | 4.6 |
| C. jejuni Cj1242 | (7.6 ± 1.2) × 105 | (6.2 ± 1.9) × 103 | 0.82b |
| C. jejuni ciaB | (7.2 ± 1.2) × 105 | (4.0 ± 0.6) × 103 | 0.56b |
| E. coli XL1-Blue | (1.8 ± 0.4) × 105 | (1.7 ± 1.2) × 102 | 0.09 |
Per cent of internalized bacteria relative to adherent bacteria.
Internalization of the C. jejuni Cj1242 and C. jejuni ciaB mutants was significantly different from the wild-type strain (P < 0.01) as judged by analysis using unpaired Student's t-tests.
Discussion
The goal of this study was to identify a C. jejuni secreted protein. To accomplish this goal, we developed a screen using Y. enterocolitica to identify genes from C. jejuni that contained a T3S amino-terminal sequence. As a first step, we showed that the full-length CiaB protein from C. jejuni was synthesized by Y. enterocolitica and exported via the flagellar T3SS. We then demonstrated that the amino-terminal sequences of the C. jejuni CiaB, FlaA and FlaC proteins were sufficient to drive secretion of a YplA fusion protein from Y. enterocolitica. FlaA, FlaC and CiaB proteins are known to be secreted from the C. jejuni flagellum (Konkel et al., 2004; Song et al., 2004). Collectively, these data demonstrate proof of concept for screening C. jejuni proteins for T3S amino-terminal sequences using the Y. enterocolitica PLA plate assay. We then utilized the assay to test for the presence of T3S amino-terminal sequences in 321 genes from C. jejuni. Using the criteria outlined, a total of 42 C. jejuni genes were identified that encode amino-terminal sequences that promoted YplA fusion secretion from Y. enterocolitica at levels equal to or higher than the CiaB : YplA fusion protein. One of the 42 genes identified was Cj1242, which we demonstrate is a potentially important virulence determinant.
While the study was in progress, information on three of the 42 C. jejuni proteins identified in the YplA screen was published by other research groups. These studies identified two flagellar-related proteins (FlgM, FlgJ) and a pathogenicity-related protein (FspA1). FlgM (Cj1464) is an anti-sigma factor involved in blocking the promoter binding activity of σ28 and the cytoplasmic levels can be controlled by secretion through the flagellar T3SS (Hendrixson and DiRita, 2003; Wosten et al., 2004). Although the precise role of FlgJ (Cj1463) in C. jejuni is unknown, FlgJ of Salmonella enterica is a two-domain protein consisting of an N-terminal domain (including the T3S amino-terminal sequence) involved in flagellar rod formation and a C-terminal region involved in flagellar L ring and hook formation (Nambu et al., 1999; Hirano et al., 2001). Interestingly, the C. jejuni FlgJ protein contains the corresponding N-terminal region as found in other ε-proteobacteria (including H. pylori), but it lacks the C-terminal acetylmuramidase region found in most β- and γ-proteobacteria (Pallen et al., 2005; Nambu et al., 2006). FspA is a 15.5 kDa protein that is secreted from C. jejuni via the flagellum (Poly et al., 2007). Two variant forms of FspA (A1 and A2) have been identified among C. jejuni strains. FspA2 was found to associate with the host cell monolayer and induce apoptosis when added to cell culture in purified form. Validation of the YplA screen described herein lies in the finding that the amino-termini of FlaA, FlaC, CiaB, FlgM, FlgJ and FspA all drive YplA export from Yersinia via the flagellar T3SS, whereas fusion of the amino-terminus of a known cytoplasmic protein (CysM) to YplA did not. Importantly, FlaA, FlaC, CiaB, FlgM and FspA all contribute to C. jejuni pathogenesis.
Previous work in our laboratory has demonstrated that culturing C. jejuni with physiological concentrations of the bile acid deoxycholate (DOC) results in the upregulation of 150 genes (Malik-Kale et al., 2008). DOC is also known to induce the synthesis of the Campylobacter invasion antigens (Cia) that are secreted via the flagellar T3SS (Konkel and Cieplak, 1992; Konkel et al., 1993; 1999; 2004; Rivera-Amill et al., 2001). We found that eight of the genes induced by DOC also harbour T3S amino-terminal sequences as judged by PLA plate assay. These genes are of interest because C. jejuni cultured in the presence of DOC stimulates this bacterium's pathogenic activity, which is evidenced by an increase in the kinetics of C. jejuni-host cell invasion (Malik-Kale et al., 2008).
The ultimate goal of this study was to identify a C. jejuni Cia virulence protein. The first Cia protein (CiaB) was identified in 1999 (Konkel et al., 1999), but the remaining Cia proteins have proven difficult to identify using traditional proteomic approaches, due in part to low levels of protein secretion under in vitro conditions. We selected Cj1242 for further characterization because the Cj1242–YplA fusion protein resulted in a high level of secretion and the gene is upregulated in C. jejuni cultured with DOC. We generated a Cj1242 mutant and then performed growth rate, motility, protein secretion and cell adherence/internalization assays. The C. jejuni Cj1242 mutant growth rate in Mueller–Hinton (MH) broth and its motility on 0.4% agar were indistinguishable from the C. jejuni wild-type strain (not shown). The profile of secreted proteins from the C. jejuni Cj1242 mutant lacked one band of the mass predicted for the Cj1242 protein (12.2 kDa). We performed adherence and internalization assays with the Cj1242 mutant and INT 407 cells, and found that there was no significant difference in the adherence of this mutant to INT 407 cells relative to the C. jejuni wild-type strain. However, the gentamicin-protection assay revealed the internalization of the C. jejuni Cj1242 mutant was significantly reduced when compared with the wild-type strain (P < 0.01). Based on the deficiency in host cell internalization, we designated the protein encoded by Cj1242 as Campylobacter invasion antigen C (CiaC).
We consider a C. jejuni strain yielding a per cent I/A of greater than 1 as both invasive and pathogenic, as inoculation of piglets with these strains results in clinical symptoms that resemble those of human campylobacteriosis, including diarrhoea with blood in the stool (Raphael et al., 2005). Inoculation of newborn piglets with C. jejuni wild-type strain (secretion-positive isolates) results in more severe disease when compared with a C. jejuni ciaB isogenic mutant (i.e. deficient in secretion of all Cia proteins). Noteworthy is that the I/A ratio (i.e. the per cent of adherent bacteria that invade epithelial cells) for the C. jejuni ciaC mutant is less than 1 (I/A = 0.82%), which is similar with the C. jejuni ciaB mutant (I/A = 0.56%). Based on this invasion ratio, we hypothesize that the C. jejuni ciaC mutant (i.e. deficient in secretion of one Cia protein) would also cause less severe disease than a wild-type strain. Our findings indicate that CiaC is required for C. jejuni to efficiently invade epithelial cells, and invasion is a virulence attribute of strains known to cause severe campylobacteriosis.
Analysis of the deduced amino acid sequences of the C. jejuni proteins found to harbour a putative T3S amino-terminal sequence revealed some additional information. We utilized two recently developed programs for prediction of T3S proteins (Arnold et al., 2009; Löwer and Schneider, 2009) to analyse the C. jejuni proteins for T3 amino-terminal sequences and compare the results with our YplA fusion data. While the results were slightly different for each program, at most only 10.6% of the 321 C. jejuni proteins tested via the YplA reporter assay are predicted to be secreted. In contrast, 14 of the 42 (33.3%) C. jejuni proteins listed in Table 2 are predicted to be secreted by one or both of the prediction programs. Of interest, both algorithms predicted CiaC to be secreted, but neither predicted FlaA and CiaB to be secreted. The failure of these programs to identify known C. jejuni flagellar-secreted proteins, including FlaA and CiaB, highlight the need for experimental validation of prediction algorithms.
The deduced amino sequences of two of the 42 proteins contain domains that suggest that they could be localized to the cytoplasm. Cj0012c is annotated as ruberythrin, a protein that protects against oxidative stress (Sztukowska et al., 2002; Mydel et al., 2006). The amino-terminus (i.e. 36 amino acids) of Cj0012c contains a small non-haeme iron domain found in the desulforedoxin and desulfoferrodoxin proteins of some methanogens and sulphate/sulphur reducers (Marchler-Bauer et al., 2007). Cj0363c is annotated as a putative oxidoreductase by inclusion in the cluster of an orthologous group (COG0635) for oxygen-independent coproporphyrinogen III oxidase (hemN). Noteworthy is that Cj0363c is distantly related to other proteins in COG0635C (Cj0363c, Cj0580c and Cj0992c) and it does not reside in the vicinity of other hem cluster genes on the C. jejuni chromosome. Moreover, the predicted products of Cj0363c, Cj0580c and Cj0992c contain a radical S-adenosylmethionine (SAM) domain. Radical SAM proteins catalyse diverse reactions, including methylation, isomerization, sulphur insertion, ring formation, anaerobic oxidation and protein radical formation. Evidence exists that these proteins generate radical species by reductive cleavage of SAM through an unusual iron-sulphur centre (Sofia et al., 2001). Although there is no experimental evidence indicating the cellular localization of either Cj0012c or Cj0363c in C. jejuni, these two examples highlight the need to analyse each putative T3S protein identified in our screen. It is possible that some of the genes identified using the PLA plate assay may not possess functional T3S amino-terminal sequences recognized in C. jejuni, or the amino-terminal region may be folded and/or inaccessible in the native protein. However, recent work also indicates that some bacteria secrete virulence proteins that were previously believed to be located solely in the cytosol (Boel et al., 2005).
Campylobacter jejuni harbours only one T3SS, the flagellum. As a first step in the identification of a C. jejuni virulence protein, we sought to identify genes from C. jejuni that harbour T3S amino-terminal sequences that direct their export from the flagellum. We report 42 C. jejuni proteins with putative T3S amino-terminal sequences. Moreover, we demonstrated that a mutation in one previously uncharacterized C. jejuni gene, Cj1242, resulted in an isolate with an altered secretion profile and reduced host cell invasion. We have also demonstrated that the secretion of CiaC is dependent upon a functional flagellar apparatus, which serves to further highlight the importance of the flagellar secretion system in the export of C. jejuni virulence proteins. We are currently investigating whether the other proteins identified in this study are secreted from C. jejuni and contribute to pathogenesis. The phospholipase reporter assay described herein demonstrates that there is a remarkable level of conservation in T3SS protein recognition among the proteobacteria; C. jejuni is a member of the delta-epsilon subdivision of proteobacteria and Y. enterocolitica is a member of the gamma subdivision. Based on this finding, we submit that the phospholipase reporter system can be used to identify genes harbouring T3S amino-terminal sequences from a variety of bacteria that possess less well-characterized T3SS.
Experimental procedures
Bacterial strains, plasmids and media
The bacterial strains and plasmids are described in Table 1. All Y. enterocolitica strains used in this study were derived from strain JB580v (Kinder et al., 1993). C. jejuni strains were cultured with MH broth or agar supplemented with 5% citrated bovine blood and incubated at 37°C under microaerobic conditions (5% O2, 10% CO2, 85% N2) with chloramphenicol (Cm, 8 μg ml−1), kanamycin (Kan, 50 μg ml−1) or tetracycline (Tet, 2 μg ml−1). E. coli strains were cultured at 37°C with Luria–Bertani (LB) broth or agar with Cm (15 μg ml−1), Kan (50 μg ml−1) or Tet (15 μg ml−1). Y. enterocolitica strains were incubated at 26°C in LB broth or agar supplemented with Cm (10 μg ml−1), nalidixic acid (Nal, 20 μg ml−1) or Tet (10 μg ml−1).
C. jejuni gene selection for T3S amino-terminal sequence screen
We selected genes to screen for T3S amino-terminal sequences from the original annotation of the C. jejuni NCTC 11168 sequence (Parkhill et al., 2000). Of 1654 ORFs, 359 were chosen for analysis following the elimination of genes encoding proteins with known functions or containing membrane-spanning domains, periplasmic domains, Sec-dependent signals or Tat-dependent signals. No genes were identified with known type I Sec-independent motifs.
Recombinant DNA procedures with the pMMB207 and pCSP vectors
Vector pMMB207, harbouring a 1.9 kb fragment encompassing the full-length ciaB gene, was PCR-amplified from C. jejuni NCTC11168 chromosomal DNA using primers CiaB-F1 (5′-GGA TCC AAA GTT AAA AAG GAG AAT AAA AGT ATG) and CiaB-R1 (5′-TTA TTT TTT CTT ATA TCT TTC AAA TTC TC). Correct orientation of the ciaB gene was determined by inducing expression from the Ptac promoter with 5 mM isopropyl β-D-1-thiogalactopyranoside. Constructs were confirmed by DNA sequencing and conjugated into the Y. enterocolitica wild-type and mutant strains.
To facilitate the identification of C. jejuni genes that harbour T3S amino-terminal sequences, the pCSP50 shuttle vector was generated. The pCSP50 vector includes a tet cassette, a constitutive promoter (cat), a 5′-truncated yplA gene (lacking 150 nucleotides encoding the native T3S amino-terminal sequence) and the yplB gene (cognate chaperone). The NdeI and BglII sites facilitated directional cloning of C. jejuni sequences as fusions with the truncated yplA. The first 108 bp of the amino-terminal regions of 328 C. jejuni genes were PCR-amplified with primers containing restriction sites for directional cloning into pCSP50. The amplicons and pCSP50 vector were digested with NdeI and BglII, DNA fragments ligated and E. coli S17-1 λ-pir was transformed with Tet selection. Cloned C. jejuni gene fragments were confirmed by PCR fragment size and sequence analysis. Vectors were conjugated into Y. enterocolitica strains and confirmed by agarose gel electrophoresis of restriction digested plasmid preparations.
Phospholipase indicator agar assay and analysis
Medium for detecting secretion of the YplA phospholipase and YplA fusion proteins from Y. enterocolitica was prepared as described previously (Young and Young, 2002). Y. enterocolitica strains were incubated overnight in LB broth with shaking at 26°C. Fop secretion was induced by spotting 1.5 μl of culture on TYE PLA medium (1% tryptone, 0.5% yeast extract, 1.5% agar, 1% Tween 80 and 1 mM CaCl2), and incubation at 26°C. Each isolate was tested for secretion at least three times from at least two independent PLA plate assays to ensure reproducible results. The conjugates were tested on PLA plates in groups of 16 in addition to a Y. enterocolitica strain expressing wild-type YplA as a positive control. All plates were scanned at 300 dpi resolution (12, 24 and 48 h) to create a digital archive of the secretion results. The secretion zone widths were measured manually from digital images using select tools in Adobe Photoshop CS2 version 9.0.2 (Adobe Systems Incorporated, USA). The 24 h secretion zone widths for the positive controls were consistent for all PLA plates (n = 22, average = 3.3 mm, standard deviation = 0.12 mm).
Rabbit antibodies to YplA and CysM
Polyclonal antibodies against recombinant YplA and recombinant CysM were produced in female New Zealand White rabbits by subcutaneous injection of 100 μg of the immunogens in TiterMax Gold (Sigma). Subsequent booster injections of 50 μg of the immunogens in Freund's incomplete adjuvant were administered 2 and 4 weeks after the primary immunizations. Blood was collected from the rabbits by terminal bleeds. The sera were processed and stored at −80°C. Antibody generation in the New Zealand White rabbits was performed using a protocol approved by the Institutional Animal Care and Use Committee (IACUC protocol #2433) at Washington State University.
Determination of Fop and YplA fusion protein secretion by immunoblot
Yersinia enterocolitica strains were incubated overnight in LB broth with shaking at 26°C. Fop secretion was induced by inoculation of TYE broth (1% tryptone, 0.5% yeast extract) with 1× TYE broth-washed Y. enterocolitica cultures and incubation with shaking at 26°C for 4–6 h. The OD540 of all cultures was determined, the cells washed 1× with TYE to remove secreted proteins and suspended in fresh TYE at an OD540 of 0.5 for the 0 h time point of the secretion assay. After 2 h of shaking at 26°C, OD540 were determined for normalization of whole-cell lysate samples, and 1 ml of each supernatant harvested by filtration through 0.22 μm sterile filters. Secreted proteins were precipitated by addition of 111 μl of 6.1 N trichloroacetic acid (10% vol vol−1 TCA final), minimum of 1 h incubation at −20°C and centrifugation with two acetone washes. Precipitated proteins were dissolved in 50 μl of single-strength electrophoresis sample buffer and heated to 95°C for 5 min. Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (12% polyacrylamide) SDS-PAGE with the discontinuous buffer system described by Laemmli (1970). The proteins were electrophoretically transferred to polyvinylidene fluoride membranes (Immobilon P; Millipore Corp., Bedford, MA) for immunoblot analysis. Bound antibodies were detected with peroxidase-conjugated goat anti-rabbit immunoglobulin G or peroxidase-conjugated goat anti-mouse immunoglobulin G. Immunoblot development was done by chemiluminescence (Western Lightning, PerkinElmer Life Sciences) and film exposure (Biomax MR film, Kodak).
Generation of the C. jejuni Cj1242 deletion mutant and complement strain
The Cj1242 gene was disrupted by homologous recombination between the disrupted Cj1242 gene on a suicide vector and the Cj1242 gene in the chromosome. The Cj1242 gene on the suicide vector had been disrupted by insertion of a TetO cassette as outlined below. A 900-base-pair fragment upstream of the C. jejuni F38011 Cj1242 gene was amplified using the primers Cj1242F1SstI (5′-TTG AGC TCG CTC TAG CTA TAA TGG TCA CAG) and Cj1242R1SstII (5′-AAC CGC GGC ATT TGA TGT TTT TTG AGT ATT ATC) and cloned into the pCR2.1 cloning vector (TA cloning system; Invitrogen) as outline by the supplier. An 806-base-pair fragment downstream of the Cj1242 gene was amplified using the primers Cj1242F2SstII (5′-TTC CGC GGA CTT CGG CAG ATG AAT TTC AAG) and Cj1242R2XhoI (5′-AAC TCG AGG TAA GCT TTA AGG CAT CAT AGA C) and cloned into a separate pCR2.1 cloning vector. The upstream fragment was then restriction-digested with SstI and SstII, gel-purified and ligated into the pCR2.1 cloning vector harbouring the downstream fragment. A 2.4 kb TetO cassette was amplified from pUOA3 (Taylor et al., 1987) with primers containing SstII sites and cloned into the SstII site of the pCR2.1 Cj1242 construct. The resultant 4.1 kb insert was then excised by SstI and XhoI restriction digest, gel-purified and ligated into pBSK-Kan2. The resultant suicide vector was sequence-confirmed and electroporated into the C. jejuni F38011 isolate. Transformants were selected on MH blood agar containing Tet 2 μg ml−1. Tet-resistant isolates were screened for Kan sensitivity, indicating a double-crossover homologous recombination event and loss of the suicide vector. Tet cassette integration into the C. jejuni Cj1242 gene was confirmed by PCR.
Construction of a complementation vector for the Cj1242 gene was accomplished by cloning a PCR product obtained with primers Cj1242F1SstI and Cj1242R2XhoI. The 1.7 kb amplicon encompassing Cj1242 was digested with SstI and XhoI, gel-purified and ligated into shuttle vector pRY111. The resultant pRY111 : Cj1242 complementation vector was sequence-confirmed and electroporated into the C. jejuni F38011 wild-type strain. Transformants were selected on MH blood agar containing Cm 8 μg ml−1 and presence of the vector encoded copy of Cj1242 was confirmed by PCR.
INT 407 cell adherence and internalization assays
A stock culture of INT 407 cells (human embryonic intestine, ATCC CCL 6) was obtained from the American Type Culture Collection. The cells were cultured in MEM supplemented with 10% FBS at 37°C in a humidified, 5% CO2 incubator. The day prior to an assay, each well of a 24-well tissue culture tray was seeded with 1.5 × 105 cells and incubated for 18 h at 37°C in a humidified, 5% CO2 incubator. The following day, the cells were rinsed with MEM-1% FBS and inoculated with approximately 5 × 107 cfu of a bacterial suspension. The tissue culture trays were centrifuged at 600 g for 5 min to promote bacteria–host cell contact, and incubated at 37°C in a humidified, 5% CO2 incubator. For the adherence assays, the plates were incubated for 30 min. The cell monolayers were then rinsed three times with PBS, epithelial cells lysed with a solution of 0.1% (vol vol−1) Triton X-100 (Calbiochem, La Jollo, CA) and bacterial suspensions were serially diluted and spread onto MH blood plates. The number of viable, adherent bacteria was determined by counting the resultant colonies. To assess bacterial internalization, the inoculated cell monolayers were incubated for 3 h, rinsed three times with MEM-1% FBS, and incubated for an additional 3 h in MEM-1% FBS containing a bactericidal concentration of gentamicin (250 μg ml−1). The number of internalized bacteria was then determined as outlined above for the adherence assays. The reported values represent the mean counts ± standard deviations derived from triplicate wells. All assays in this study were repeated a minimum of three times to ensure reproducibility and performed at a multiplicity of infection between 50 and 500. Regardless of the multiplicity of infection, the phenotype of the C. jejuni Cj1242 mutant relative to the wild-type strain was always the same.
C. jejuni secretion assay
The C. jejuni F38011 strain and isogenic Cj1242 mutant was metabolically labelled with [35S]-methionine as described elsewhere (Konkel and Cieplak, 1992). Briefly, isolates were harvested from biphasic culture on MH agar supplemented with 0.1% DOC and resuspended in MEM lacking methionine supplemented with or without dialysed albumin depleted FBS to an OD540 = 0.3. [35S]-methionine was then added and inocula were incubated at 37°C for 3 h under microaerophilic conditions. After incubation, supernatant fluids were concentrated 10-fold by precipitation with 4 vols of ice-cold 1 mM HCl-acetone. The pellets were air-dried and dissolved in an equal amount of water and double-strength sample buffer. Equal volumes of the concentrated samples were subjected to 12% SDS-PAGE. The gel was dried, exposed to film for 5 days, and developed to acquire the auroradiograph.
Bioinformatics
Operon and regulon prediction was performed by query of the MicrobesOnline site (Alm et al., 2005). In silico T3S protein prediction was performed using ‘EffectiveT3’ (http://www.chlamydiadb.org; Arnold et al., 2009) and ‘Modlab’ software (http://gecco.org.chemie.uni-frankfurt.de/index.html; Löwer and Schneider, 2009).
Acknowledgments
We thank Dr Glenn M. Young (University of California-Davis) for providing the Y. enterocolitica strains, Dr Scott A. Minnich (University of Idaho-Moscow) for providing the mouse monoclonal flagellin specific antibody 15D8, Dr William G. Miller (US Department of Agriculture, Albany, CA) for sequence analysis of the C. jejuni NCTC 11168 genome, Rebecca C. Flanagan for performing INT 407 cell adherence and internalization assays and Dr Daelynn Buelow for performing C. jejuni secretion assays. We also thank Dr Buelow, Charles L. Larson, Jason M. Neal-McKinney for critical review of the manuscript. This work was supported from funds awarded to M.E.K. from the National Institute of Health, Department of Health and Human Services under contract number NO1-AI-30055.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline Website for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, et al. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5:e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea L, Beatson SA, Kaparakis M, Ferrero RL, Hartland EL. Secretion of flagellin by the LEE-encoded type III secretion system of enteropathogenic Escherichia coli. BMC Microbiol. 2009;9:30. doi: 10.1186/1471-2180-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berring E, Brancato S, Grant K, Schaper E, Kadavil S, Smagin H, et al. Destabilization of phospholipid model membranes by YplA, a phospholipase A2 secreted by Yersinia enterocolitica. Chem Phys Lipids. 2004;131:135–149. doi: 10.1016/j.chemphyslip.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Wells JG, Feldman RA, Pollard RA, Allen JR. Campylobacter enteritis in the United States. A multicenter study. Ann Intern Med. 1983;98:360–365. doi: 10.7326/0003-4819-98-3-360. [DOI] [PubMed] [Google Scholar]
- Boel G, Jin H, Pancholi V. Inhibition of cell surface export of group A streptococcal anchorless surface dehydrogenase affects bacterial adherence and antiphagocytic properties. Infect Immun. 2005;73:6237–6248. doi: 10.1128/IAI.73.10.6237-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, et al. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis. 2004;38(Suppl. 3):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Garvis SG, Tipton SL, Konkel ME. Identification of a functional homolog of the Escherichia coli and Salmonella typhimurium cysM gene encoding O-acetylserine sulfhydrylase B in Campylobacter jejuni. Gene. 1997;185:63–67. doi: 10.1016/s0378-1119(96)00631-2. [DOI] [PubMed] [Google Scholar]
- Gundogdu O, Bentley SD, Holden MT, Parkhill J, Dorrell N, Wren BW. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics. 2007;8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatic SO, Picking WL, Young BM, Young GM, Picking WD. Purification and characterization of two active derivatives of recombinant YplA, a secreted phospholipase from Yersinia enterocolitica. Biochem Biophys Res Commun. 2002;292:463–467. doi: 10.1006/bbrc.2002.6690. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50:687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- Hirano T, Minamino T, Macnab RM. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol. 2001;312:359–369. doi: 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Minnich SA. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Cieplak W., Jr Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect Immun. 1992;60:4945–4949. doi: 10.1128/iai.60.11.4945-4949.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Mead DJ, Cieplak W., Jr Kinetic and antigenic characterization of altered protein synthesis by Campylobacter jejuni during cultivation with human epithelial cells. J Infect Dis. 1993;168:948–954. doi: 10.1093/infdis/168.4.948. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. Mechanisms of protein export across the bacterial outer membrane. J Bacteriol. 2005;187:4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson CL, Christensen JE, Pacheco SA, Minnich SA, Konkel ME. Campylobacter jejuni secretes proteins via the flagellar type III secretion system that contribute to host cell invasion and gastroenteritis. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3rd edn. Washington, D.C.: American Society for Microbiology; 2008. pp. 315–332. [Google Scholar]
- Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- Löwer M, Schneider G. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One. 2009;4:e5917. doi: 10.1371/journal.pone.0005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T, Cornelis GR. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, et al. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog. (3rd) 2006;2:e76. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu T, Minamino T, Macnab RM, Kutsukake K. Peptidoglycan-hydrolysing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu T, Inagaki Y, Kutsukake K. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet Syst. 2006;81:381–389. doi: 10.1266/ggs.81.381. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Chaudhuri RR, Henderson IR. Genomic analysis of secretion systems. Curr Opin Microbiol. 2003;6:519–527. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Penn CW, Chaudhuri RR. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 2005;13:143–149. doi: 10.1016/j.tim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, et al. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael BH, Monteville MR, Klena JD, Joens LA, Konkel ME. Interactions of Campylobacter jejuni with non-professional phagocytic cells. In: Ketley JM, Konkel ME, editors. Campylobacter: Molecular and Cellular Biology. Norfolk NR: Horizon Bioscience; 2005. pp. 397–413. [Google Scholar]
- Rivera-Amill V, Konkel ME. Secretion of Campylobacter jejuni Cia proteins is contact dependent. Adv Exp Med Biol. 1999;473:225–229. doi: 10.1007/978-1-4615-4143-1_23. [DOI] [PubMed] [Google Scholar]
- Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- Schmiel DH, Wagar E, Karamanou L, Weeks D, Miller VL. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiel DH, Young GM, Miller VL. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J Bacteriol. 2000;182:2314–2320. doi: 10.1128/jb.182.8.2314-2320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YC, Jin S, Louie H, Ng D, Lau R, Zhang Y, et al. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol Microbiol. 2004;53:541–553. doi: 10.1111/j.1365-2958.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, Lindeman J. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut. 1985;26:945–951. doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM., Jr Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol. 2002;44:479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- Taylor DE, Hiratsuka K, Ray H, Manavathu EK. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J Bacteriol. 1987;169:2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Hultgren SJ. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol. 2000;12:420–430. doi: 10.1016/s0955-0674(00)00111-3. [DOI] [PubMed] [Google Scholar]
- Warren SM, Young GM. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J Bacteriol. 2005;187:6075–6083. doi: 10.1128/JB.187.17.6075-6083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrell T, Ciampa N, Boelaert F, Helwigh B, Korsgaard H, Chriel M, et al. Zoonotic infections in Europe in 2007: a summary of the EFSA-ECDC annual report. Euro Surveill. 2009;14:1–3. [PubMed] [Google Scholar]
- Wosten MM, Boeve M, Koot MG, van Nuenen AC, van der Zeijst BA. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM, Wagenaar JA, van Putten JP. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Young BM, Young GM. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol. 2002;184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


