Summary
Man-made molecular “computers” that operate inside live cells will enable unprecedented level of control over cellular physiology. A promising approach to building these computers uses RNA molecules and RNA-based regulation. RNA naturally lends itself to create “digital” molecular networks that embody standardized (normal) forms of logic functions. The network’s inputs, that may or may not be inverted by single-input NOT logic gates, feed into multi-input AND gates whose outputs are in turn integrated in a multi-input OR gate. Below I review recent steps that have been taken toward implementing these networks with allosteric riboswitches and ribozymes in bacteria and yeast, and RNAi in mammalian cells. I also propose how to co-opt recently-discovered additional RNA regulation mechanisms into future construction efforts.
Introduction
Biological pathways and networks “compute” developmental decisions and environmental responses using molecules as information cues. Accordingly, our ability to rationally control and manipulate biological systems will take a quantum leap once we succeed in creating and embedding in biological hosts computational biomolecular networks that process molecular information in a desired fashion. For example, in medicine “diagnostic biocomputers” could lead to the next generation of therapy whereby the amount, location and instance of drug release will be determined in real time based on a diagnostic computation using disease-related molecular cues in each cell and organ of the body [1].
Unlike a silicon computer that can be reprogrammed with a few hits on a keypad, a biomolecular computer is really a set of design tools which, when provided with a description of a computational task, generates a blueprint of a molecular network that can implement this task. One important challenge is to make sure that these tools are flexible enough to enable a sufficient variety of tasks. In theoretical computer science this problem was solved by the invention of universal models of computation, such as the Turing machine. While it seems unlikely that similarly universal approaches could be realized with biomolecular building blocks anytime soon, biocomputer architectures could, and should, aspire to at least some degree of generality in the computational sense.
A blueprint of a biomolecular computational network subsequently needs to be translated into actual system operating in a living cell. In the current state of the art, network’s conceptual design takes only a fraction of time spent on its experimental construction, and the major challenge is the extensive fine-tuning of network’s components and interactions at the implementation stage. Among several alternatives, RNA has recently emerged as an attractive material for biocomputer construction [2-5], likely to support a wide range of computational tasks on one hand, and behaving in a relatively predictable way in engineered systems on the other. This review discusses the latest results and points toward future directions in the field of engineered RNA-based computational networks that work in live cells.
Molecular logic
An example of a computational task is “if concentration of protein A is above A0, and concentration of protein B is above B0, and concentration of metabolite C in less than C0, then activate a biological pathway X (e.g. apoptosis)”. While different approaches to biocomputation will lead to different network blueprints, they will all have a stereotypical structure: sensors that read out individual signals (i.e. concentrations of proteins A, B and metabolite C); computational core that determines a value of a certain function that manipulates these concentrations, such as: if (A > A0 and B > B0 and C < C0) then output signal Y; and the actuation component: if Y, activate pathway X.
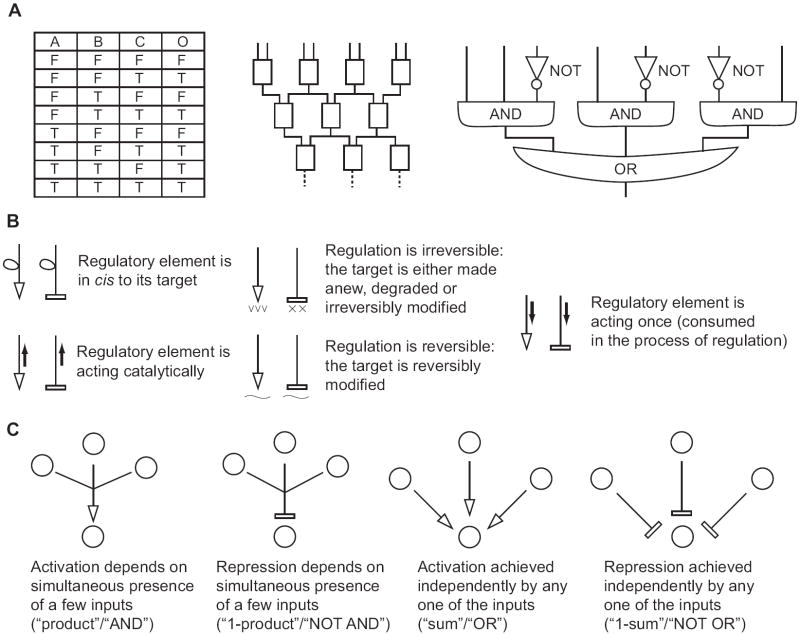
The above example is a special case of what is known as Boolean, or logic, functions. These functions are of singular importance for computer science and engineering, as all modern digital computers operate by calculating and acting upon the truth values of various logic functions. Moreover, logic functions faithfully describe many important biological processes, for example during development of a multicellular organism, and as shown above they can be used to guide rationally-designed intervention. A formal definition of a Boolean function on N inputs (N = 3 in the above example) is a mapping that assigns either a True or False value to each of 2N specific combinations of input values, where each input can also be either True or False (Fig. 1A, left panel). In a biological “computational” network one needs to convincingly define what does it mean for a molecular input or output to be True or False. The most common way to relay information within and between cellular modules is to use concentrations of molecules, or related notions of their absolute numbers or activity. Therefore it is natural to correlate a molecule’s concentration to its interpretation as True or False in the context of a Boolean computational network. Of course, intracellular concentrations of individual molecular species can take any non-negative value from zero to millimolar. Plausible physiological concentrations of certain biomolecules can be more restricted – for example, a certain transcription factor can be either completely turned off, or fully induced. Were such a factor used as an input for a computational network, its Boolean interpretation would be straightforward, with the off state corresponding to False, and fully induced state corresponding to True. However, if a physiological concentration of a given molecule spans a range of values, one could ask if there is a way to define upper and lower thresholds so that increasing the input concentration above the upper threshold, or decreasing it below the low one, will make no effect on the network’s output. (On a side note, this discussion only considers concentrations of inputs and output in the steady state, when neither of them changes over time. Dynamic behavior that may arise when one or more of the inputs change suddenly cannot be directly predicted from the Boolean description and is related to kinetic parameters of interactions between network’s components). In this case, any concentration above the upper threshold would be interpreted as True, and below the low threshold as False. The intermediate values will necessarily eliminate the digital, Boolean features of the computation and turn it into a gradual, analog response. Those values might either be ignored if they are physiologically unimportant or explicitly dealt with, in which case one will need to consider the behavior of the computational network in this analog regime and understand the consequences of this behavior. Neither alternative has been explicitly addressed by the field but it will become increasingly important in the future.
Figure 1. Basic definitions and schematics of logic networks.
A, A given logic function as represented by input/output mapping (left panel, F=False, T=True) can be implemented by cascading universal gates such as NAND (middle panel) or by constructing a circuit in a normal form (right panel). B, Basic regulatory links that create complex molecular logic networks. Generic interactions of activation and inhibition are subdivided into classes based on the fine details of the interactions. These details, while not altering the high-level logic behavior of a network, can significantly influence its quantitative properties [17] and must be carefully considered in the design process. C, Designation of different ways in which concurrent regulation of a biological target by multiple effectors gives rise to different basic logic relations and their “analog” counterparts in an unsaturated regime.
To test the Boolean molecular network in the steady state we assign all possible 2N True/False combinations to the inputs, and measure the network’s output. The output is an ultimate readout of the engineering system we aim at constructing, so a convincing definition of the output’s truth value should be based on the absolute concentration of the output molecule. In other words, all computations that are supposed to produce a True logic output should generate sufficiently uniform high levels of the output molecule, and those with False output should lead to distinctly lower uniform output values. The definition of “high” and “low” here can be somewhat arbitrary; the ability of the downstream natural processes that are controlled by the computational network to distinguish between them and generate distinct physiological responses will ultimately define the success of the implementation.
A good biocomputer toolkit should ideally be able to lay out a computational core for any pre-defined logic function, or at least a wide range of useful functions. It should also tell us how to build sensors that feed various molecular inputs into these functions. While the core should be able to compute the truth value of a Boolean expression such as (A > A0 and B < B0 and C > C0 and D > D0) between four molecular signals, sensors will substitute specific inputs for abstract placeholders A, B, C, D, such as the protein NFkB for A, the metabolite cAMP for B, and so on. There are two popular ways to compute a logic function in a molecular context. The first recurrently employs certain two-input logic functions called universal gates such as NOT AND, or NAND, that outputs False when both inputs are True and True otherwise [6]. However, in practice this leads to deep, narrow cascades of the physical elements implementing the NAND gates whose outputs feed into the next layer (Fig. 1A, middle panel). The second uses disjunctive and conjunctive normal logic forms: certain combinations of multi-input AND and OR gates, and single-input NOT gates. Disjunctive Normal Form (DNF) consists of the OR function integrating multiple AND gates, and the Conjunctive Normal Form (CNF) consists of the AND function integrating multiple OR gates. Normal forms translate to wide circuits of at most three layers (Fig. 1A, right panel, Fig. 2A).
Figure 2. Finite-depth layered logic circuits.
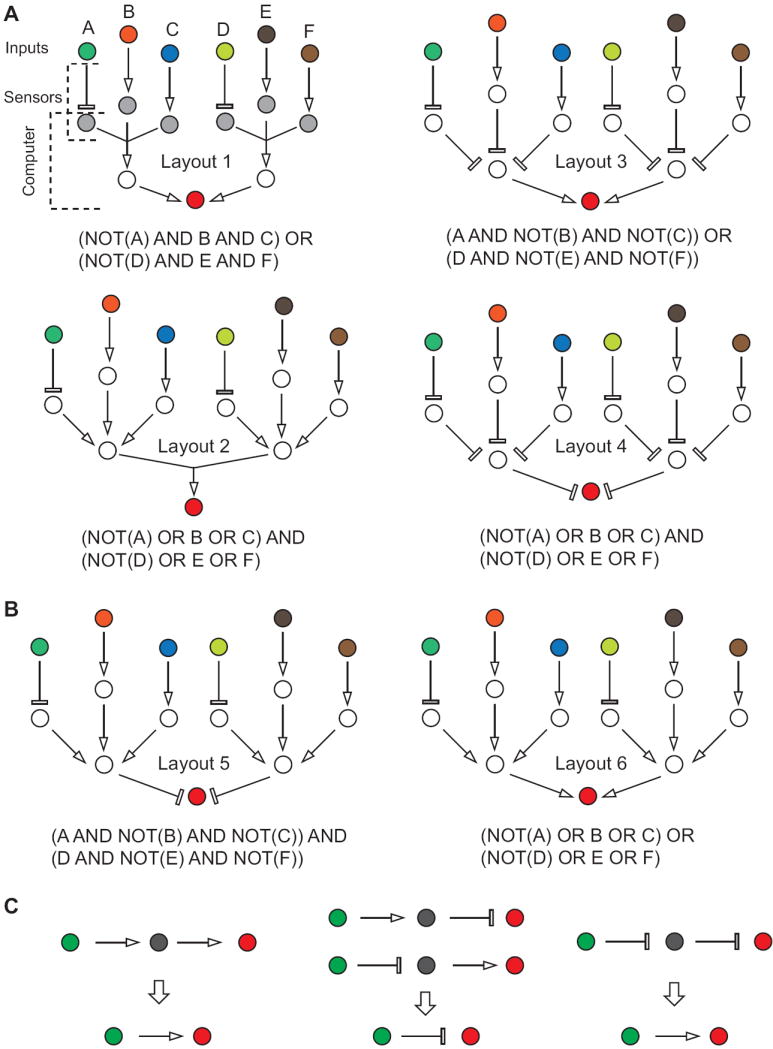
A, Circuits that implement normal logic forms (Layouts 1-4). Colored circles represent external molecular inputs. They are linked by individual activating or inhibitory links to the immediate inputs of the molecular computations (gray color in top left diagram). Sub-networks that convert external inputs into the inputs for a computational core (collapsed into single activating or inhibitory links in the scheme) are called “sensors”. A purpose of sensors is to convert various external inputs into a uniform molecular format to promote scalability and modularity. The computational inputs are converted into the network’s output (red color) via a computational core implementing a particular mathematical relation, here a logic function. Layouts 1 and 2 use not-trivial converging regulatory links to implement disjunctive (DNF) and conjunctive (CNF) normal forms, respectively. Layouts 3 and 4 only use links that act independently of each other and hence are easier to design, but by virtue of their respective actions implement similar logic functions. B, Examples of layered circuits that, while being superficially similar to Layouts 1-4, do not implement normal forms but only one kind of logic operation. C, Collapsing sequential regulatory links for the purpose of circuit analysis. In a regulatory chain with no convergence two sequential links can be replaced by a single interaction according to the schematics in the figure.
In biology, the construction of logic gates usually involves connecting molecular components via activating and inhibitory regulatory links (Figs. 1B, 1C) with independent components being the inputs, and the targets of regulation representing the result of intermediate logic processing or the global network output. (Mutual activation or inhibition, or long-range feedbacks will complicate the description and are not a part of this review. In general their introduction could significantly alter the functions computed by a network in the steady state, or eliminate the steady state altogether and lead to oscillations). If one decides to use universal logic gates for network construction, it is critical that the output of a gate be of the same molecular class as its input. Only then the gates can be cascaded for an unlimited number of times; all this, assuming that additional mechanisms prevent signal dissipation as information travels along the cascades [5]. On the other hand, in normal-form circuits there is no need for indefinite cascading of layers; and therefore one could design layer-specific ways to relay information so that the output of upstream gates can serve as an input for the downstream gates. It so happens that in most RNA-based approaches the inputs to the molecular gates are radically different from their outputs. For example, the inputs are often small RNA molecules or motifs, while the output is mRNA. This poses a problem for cascading approaches but leaves the door open to circuits in normal forms. Moreover, RNA regulation mechanisms naturally integrate multiple activating or inhibitory inputs as required by AND and OR gates required for normal-form circuits (Fig. 2). Therefore in what follows I will examine how to create circuits that fully or partially implement normal forms of logic functions using RNA. (Additional RNA-based regulation mechanisms, including those that may not be readily scaled, or those that can be used in synthetic biology project not directly related to biological computation, are discussed in a number of excellent reviews [7,8]). I will classify the different approaches according to the type of a host organism and consider bacteria, yeast and mammalian cells.
RNA-based computational networks in bacteria
RNA computation in bacteria is in its infancy. Most of the relevant mechanisms have only recently been discovered, muss less tinkered with from engineering perspective. Although there has truly been an explosion of discovery in the field [9], a few “winners” emerge that have the potential to support complex molecular programming. Two of them, intrinsic terminators controlled by riboswitches and sRNAs, are analyzed below.
Intrinsic terminators are irreversible cis-inhibitory stem-loop RNA structures with poly-U tract found in the 5’- or 3’-UTR of an mRNA [10]. Properly folded terminators act by causing dissociation of the RNA polymerase/mRNA complex from the transcribed gene. The folding of intrinsic terminators can be altered – induced or destroyed – by “riboswitches”, regulatory sequences that are themselves remodeled in the presence of molecular ligands [11]. Using our terminology, a gene/s with terminators constitute a computational core, while the ligand-binding motifs constitute the sensors (in this arrangement the sensors are physically connected to the core but this makes no difference from a conceptual standpoint). In a few cases in nature adjacent terminators are controlled by different riboswitches that respond to two different ligands [12]. It is conceivable that the same mRNA can accommodate more than two terminators, resulting in multi-input regulatory programs. In particular, the network will implement parts or a whole of a DNF circuit based on Layout 3 (Figs. 2A, 3A). I note that engineering de novo riboswitches that remodel an intrinsic terminator stem-loop is challenging, because the switches require a combination of binding activity with a particular secondary structure remodeling at a relatively remote location. Indeed, to the best of my knowledge engineered riboswitches of this type have not been reported yet.
Figure 3. Modes of RNA computation in bacteria based on riboswitches and sRNAs.
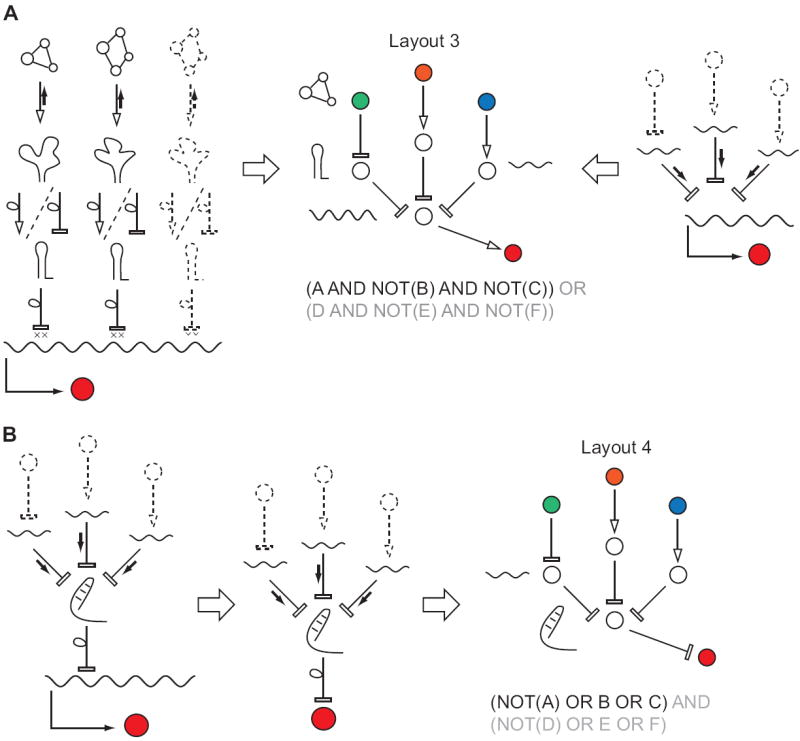
A, A DNF-like computations (central panel) that involve multiple intrinsic terminators controlled by aptamers (left panel) or multiple sRNAs targeting the same transcript (right panel). Depicted diagrams only implement multi-input AND gates, but an OR operation could be included by adding a parallel mRNA expressing the same gene but targeted by a different set of inputs. B, A multi-input OR gate, a portion of a CNF circuit implemented by sRNAs that target a transcription silencing element. Extending this architecture to a full-fledged CNF requires that a number of cis silencing elements control the same mRNA. Such arrangement has not yet been reported but is conceivable.
Another mode of RNA regulation in bacteria is based on small RNA (sRNA) regulation [9]. These are relatively short non-coding transcripts that regulate other genes by sequence-dependent binding to certain mRNA regions. The binding can repress translation by interference with the ribosome binding site (RBS), or activate it by altering conformation of a different cis negative regulatory structure. While reminiscent of RNA interference (RNAi, see below) in higher eukaryotes, inhibitory sRNAs cannot be designed against arbitrary region in the mRNA. Instead the target is limited to the close vicinity of RBS and perhaps five N-terminal codons [13]. Besides, most sRNAs act irreversibly in a stoichiometric fashion. If multiple sRNA inhibit the same element on an mRNA and are themselves controlled by external signals (which is the case in nature), the network will confer with Layout 3 and as above implement parts of, or an entire DNF circuit [14,15]. An implementation where sRNAs target a cis-inhibitory element is less amenable to scaling and it will constitute a multi-input OR gate of a CNF circuit (Fig. 3B). First steps in engineering networks using sRNA-inspired ideas have already taken place, for example in a reported switch by Isaacs et al. [16] or by fusing an sRNA target to a reporter gene [17]. However, scaling these approaches to perform complex computations has not been shown yet.
RNA computations in yeast
Yeast is an important model organism and they are widely used in biotechnology applications, from alcoholic beverage industry to biofuels and fine chemicals biosynthesis. Reprogramming yeast via RNA-based computational networks could therefore have immediate consequences in these and other areas. However, as opposed to prokaryotes and metazoans (see below), yeast do not seem to have simple, clean-cut RNA-based regulatory mechanisms. For example, a pathway analogous to RNAi is used in fission yeast to remodel chromatin rather than knockdown individual genes in a modular fashion [18], while regulation by sRNA and riboswitches is all but non-existent (or waits to be discovered). One case of a riboswitch-based regulation has been reported by Breaker lab, and it involved induction of an alternative splicing pathway in the presence of a ligand, creating a translationally inactive transcript [19]. It is feasible that a number of similar switches could be inserted in the same gene and result in complex multi-input logic programs.
Despite the relative dearth of endogenous RNA regulation, yeast has proven as a promising platform for incorporation of exogenous RNA regulatory elements. Switches based on a combination of an antisense RNA and an aptamer moieties have been successfully shown to repress or induce a reporter gene in the presence of a ligand [20]. If scaled, these switches could enable DNF-like circuits similar to those in Figures 3A and 3B. A different approach explored allosterically-controlled self-cleaving hammerhead ribozymes fused in the 3’-UTR of a reporter gene [21, 22]. These cis-elements were design to be either active or inactive in their baseline state, with ligand binding respectively inactivating or activating the cleavage and hence modulating the reporter gene expression. Here the logic function was encoded by appropriate placement of the ribozymes moieties in the gene’s 3’-UTR, the placement of the aptamer moieties relative to the ribozymes, and the baseline ribozyme activity. In their core the networks are functionally similar to the multi-input riboswitches in prokaryotes and their logic functions can be appropriately described by Disjunctive Normal Forms (Fig. 4A).
Figure 4. RNA computations in eukaryotes.
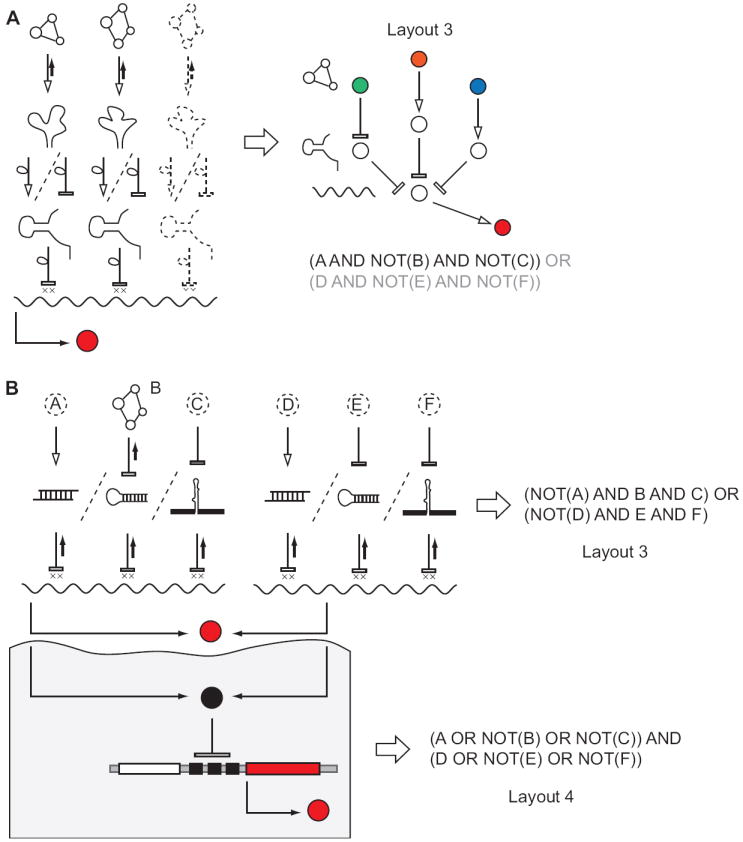
A, Yeast-based networks. Arrays of allosteric riboswitches in a gene’s 3’-UTR enables an AND clause of a DNF circuit, while combining two reporter constructs with an identical output can add the OR gate and extend the network to a full DNF. B, RNAi-based mammalian DNF and CNF circuits. A regulatory link between a small molecule metabolite B and an shRNA has been shown in a number of reports, while other classes of inputs (dotted lines) yet need to be demonstrated. In the DNF circuit the small RNAs target the output directly, while in the CNF layout they target a transcriptional repressor that controls the output via engineered promoter.
RNA computations in mammalian systems
Mammalian systems arguably offer the most raw materials for constructing complex computation networks in the form of numerous regulatory mechanisms. Regulation on the RNA level that involves RNA effectors includes alternative splicing, RNA interference (RNAi) and antisense RNA, to name a few. Exogenous regulatory elements such as riboswitches have also been shown to function in these cells. In particular, a report showed a ligand-regulated riboswitch that regulated gene expressed when incorporated in this gene’s intron [23]. While in that work the ribozyme itself, its placement in the mRNA transcript, and the ligand effector were designed with the help of extensive screening, it is not infeasible that rational approaches could work in mammalian context to generate multiple ON/OFF regulatory elements and hence enable complex DNF-like logic. More recently, an endogenous ribozyme-like motif has been discovered in the 3’-UTR of a mammalian gene [24]. Interestingly, the motif is discontinuous and it forms through long-range secondary interactions. White the question of allosteric regulation of the motif is left open, it is plausible that this approach could be co-opted into the biocomputer engineer toolkit to provide a basis for scalable logic computations.
Full-length antisense RNA transcripts are being discovered at an increasing pace with the help of new high-throughput methods, and their role in gene regulation and specifically repression of gene expression is becoming evident [25]. While the relative importance of antisense RNA in the big picture of biological regulation is still being debated, effective antisense regulation is amenable to engineering as demonstrated by a recent report on a synthetic mammalian oscillator [26]. Multiple effective antisense RNAs could potentially be used as mammalian counterparts of bacterial sRNAs and support similar logic networks.
RNAi is perhaps the most widespread, or at least best-studied, RNA-based regulation mechanism in mammalian cells. In its most basic, RNAi is an inhibitory interaction between a short (19-27 nt) RNA strand and an mRNA that contains a subsequence of either full or partial complementarity to this strand in its coding region or 3’-UTR. Short RNA strand can be delivered exogenously as small interfering RNA (siRNA), expressed from exogenously-introduced DNA as short hairpin RNA (shRNA) or expressed from endogenous genes as micro RNA (miRNA). (Synthetic constructs can also be built to mimic the endogenous miRNA genes.) siRNA, shRNA and engineered miRNA can be made into very efficient repressors that act via mRNA degradation when they meet certain sequence requirements. Moreover, since the sequences that respond to small RNAs are short, they can be engineered in large numbers to control a single gene. This has led to the realization that RNAi can be used to assemble large-scale normal form logic circuits [27]. Using RNAi alone can afford DNF circuitry, while using an extra transcriptional layer enables CNF-like circuits (Fig. 4B). Experiments showed that indeed up to five siRNA inputs can be integrated in a DNF circuit and up to two inputs in CNF circuits [27]. The next challenge is transducing a variety of molecular signals into small RNA format, in other words building the sensory component of the networks. Encouraging first steps have been made in a number of research groups who showed that small molecule ligands can inhibit shRNA molecules via fused aptamers [28-30]. Expanding the repertoire of eligible inputs will greatly increase the applicability of RNAi networks.
Conclusions
RNA-based regulation with its effectors and targets consists of a large variety of different mechanism in different biological hosts, but many of these mechanisms possess common features that make them attractive building blocks for the implementation of complex molecular computations. The effectors are often small RNAs that act based on sequence complementarity, and they target small motifs in mRNA transcripts. Therefore de novo RNA regulators could be engineered from scratch using rational design or in vitro selection [31], and subsequently integrated into large networks. A particular approach to computation, namely normal-form logic circuits, seems best suited to guide the construction of these networks. Indeed, both natural and synthetic examples of such circuits have been shown in bacteria, yeast and mammalian cells. Future challenges include putting these initial results on solid engineering foundation, demonstrating complex logic computations with a large number of inputs, expanding the repertoire of input signals, and solving real-life problems. Maintaining the digital behavior of large circuits is another challenge. Overall, RNA networks offer an exciting direction towards programmable cells and cell populations, and promise to become a workhorse of large synthetic systems.
Acknowledgments
I thank I. Benenson and M. Leisner for critical reading of the manuscript. This work was supported by the Bauer Fellows Program and NIGMS grant GM068763 for National Centers for Systems Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shapiro E, Benenson Y. Bringing DNA computers to life. Scientific American. 2006;295:44–51. [PubMed] [Google Scholar]
- 2.Faulhammer D, Cukras AR, Lipton RJ, Landweber LF. Molecular computation: RNA solutions to chess problems. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1385–1389. doi: 10.1073/pnas.97.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nature Biotechnology. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 4.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 5.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 6.Weiss R, Homsy GE, Knight TF. Toward in vivo digital circuits. In: Landweber LF, Winfree E, editors. Evolution as Computation: DIMACS Workshop. Springer; 1999. pp. 275–295. DIMACS. [Google Scholar]
- 7.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nature Biotechnology. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 8.Davidson EA, Ellington AD. Synthetic RNA circuits. Nature Chemical Biology. 2007;3:23–28. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- **9.Waters LS, Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043.. This review very thoroughly describes the recently-discovered RNA-triggered regulatory mechanisms in bacteria. It can serve as an excellent starting point to an aspiring molecular engineer.
- 10.Henkin TM. Control of transcription termination in prokaryotes. Annual Review of Genetics. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 11.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Current Opinion in Structural Biology. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- *12.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716.. The authors describe a naturally-occurring gene control element that contains two riboswitches, each responding to a different ligand. The regulatory function is approximated by a NOR gate, where a presence of at least one ligand repressed gene expression.
- **13.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA Binding to 5 ’ mRNA Coding Region Inhibits Translational Initiation. Molecular Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027.. This paper delineates structural requirements behind sRNA regulation and comes to a conclusion that the binding should occur within the first five codons of the N-terminal.
- 14.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- **15.Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes & Development. 2007;21:221–233. doi: 10.1101/gad.1502407.. The authors report that a bacterial gene for quorum sensing in Vibrio harveyi is controlled by multiple sRNAs in a complex, additive manner. The system can inspire the construction of both logic circuits and analog switches.
- 16.Isaacs FJ, Dwyer DJ, Ding CM, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nature Biotechnology. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- *17.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. Plos Biology. 2007;5:1998–2010. doi: 10.1371/journal.pbio.0050229.. The paper provides a theoretical analysis and quantitative experimental measurements of sRNA action in a model reporter system. The authors highlight the qualitative differences between transcriptional and sRNA regulation due to an irreversible nature of sRNA action.
- 18.Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–U497. doi: 10.1038/nature05769.. The paper describes an endogenous riboswitch found in a filamentous fungus Neurospora crassa Upon biding of the ligand thiamine pyrophosphate (TPP), the intron-placed switch “tricks” the splicing machinery into using an incorrect splicing signal and producing a translationally inactive splicing variant.
- 20.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nature Biotechnology. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- *21.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104.. The authors report a modular approach to construction of allosterically-regulated self-cleaving ribozymes that control gene expression when placed in the 3’-UTR of a reporter gene in yeast. Ligand-binding aptamers are fused to ribozyme moieties through a transmission linker that allows the binding event to translate into active, or inactive, ribozyme.
- **22.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311.. The devices from reference [21] are combined by the same research team into networks that compute two-input logic functions using small molecule ligands as inputs.
- *23.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D’Amato RJ, Mulligan RC. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844.. Gene regulation by an engineered allosteric ribozyme is demonstrated in mammalian cells
- *24.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–U857. doi: 10.1038/nature07117.. The first example of an endogenous ribozyme controlling gene expression in mammalian (rodent) cells The ribozyme forms upon interaction of discontinuous RNA motifs and act constitutively by cleaving the transcript and downregulating the gene that harbors it.
- 25.Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: A critical look in both directions. Cellular and Molecular Life Sciences. 2009;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616.. The authors describe a mammalian oscillating circuit using antisense RNA regulation as a key network element.
- **27.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnology. 2007;25:795–801. doi: 10.1038/nbt1307.. The authors show that RNAi can enable complex logic computations in human cell culture. Computational component of the networks is experimentally demonstrated to work with up to five different siRNA inputs.
- *28.An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. Rna-a Publication of the Rna Society. 2006;12:710–716. doi: 10.1261/rna.2299306.. The authors show how an shRNA fused to an aptamer can be inhibited by a small molecule ligand.
- 29.Tuleuova N, An C-I, Ramanculov E, Revzin A, Yokobayashi Y. Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochemical and Biophysical Research Communications. 2008;376:169–173. doi: 10.1016/j.bbrc.2008.08.112. [DOI] [PubMed] [Google Scholar]
- *30.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Molecular Systems Biology. 2008;4 doi: 10.1038/msb.2008.62.. The authors describe a general approach toward modulating an shRNA via fused aptamer, and propose a modeling framework for rational design on these switches.
- 31.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]