Abstract
Previous studies have shown that treatment of mammalian cells with phospholipase A2 (PLA2) antagonists cause the normally interconnected Golgi ribbon to break up into large fragments of stacked Golgi cisternae (“mini-stacks”) that remain located in the juxtanuclear region. Using the reversible PLA2 antagonist, ONO-RS-082 (ONO) and live-cell, time-lapse microscopy to image the Golgi reassembly process, we found that Golgi mini-stacks underwent a burst of membrane tubule formation following washout of ONO: before washout only 4.3 ± 3.8 tubules/cell/10 min were formed, whereas after washout 29.9 ± 11.9 tubules/cell/10 min formed. These membranes tubules formed bridges between physically separate mini-stacks, thus mediating their coalescence into intact Golgi ribbons. Formation of inter-stack tubules and an intact Golgi ribbon was also facilitated by microtubules because treatment with nocodazole significantly inhibited both processes. This microtubule-dependent process was also dependent on dynein because the dynein inhibitor nordihydroguaiaretic acid (NDGA) inhibited reassembly. These studies show that a late stage of Golgi assembly occurs via membrane tubules, whose formation is dependent on PLA2 activity and microtubules. Considering these results together, we concluded that the maintenance and assembly of normal Golgi architecture is dependent on the PLA2-mediated, dynamic formation of inter-Golgi membrane tubules.
Keywords: Golgi complex, membrane tubules, phospholipase A2, PLA2, microtubules, dynein
INTRODUCTION
The Golgi complex in many mammalian cell types forms a large interconnected ribbon, usually situated in a juxtanuclear region. The mammalian Golgi complex also reversibly disassembles during mitosis [1; 2], which can be mimicked to some extent by a variety of compounds including brefeldin A (BFA) [3], nocodazole [4], and PLA2 antagonists [5; 6]. In all of these cases, reassembly involves a late step during which physically disconnected Golgi mini-stacks coalesce into an intact ribbon. Reassembly from mitotic breakdown appears to involve thin membrane tubules of uniform diameter (60-80 nm) and varying lengths that help to bridge physically separate mini-stacks [7]. Similarly, static fluorescence images suggest that during recovery from reversible PLA2 antagonists, such as ONO-RS-082 (ONO), membrane tubules sprout from Golgi mini-stacks and appear to be responsible for bringing mini-stacks together into an intact ribbon [6]. These results strongly suggested that maintenance and reassembly of an intact Golgi complex requires PLA2-mediated formation of membrane tubules[8]. Precisely how dynamic these membrane tubules are during reassembly is unclear.
The architecture of the mammalian Golgi complex is influenced by interactions with microtubules and their associated motor proteins. The juxanuclear positioning of the Golgi complex near the microtubule-organizing center (MTOC) is dependent on constant centripedal movement of Golgi membranes by the minus-end directed motor dynein [9; 10]. In addition, the formation of membrane tubules from the Golgi complex is facilitated by microtubules and associated motor proteins. For example, BFA-stimulated membrane tubules are greatly facilitated by microtubules and kinesin [11; 12; 13]. It is unknown if microtubules play a role in the late membrane tubule-dependent coalescence of Golgi mini-stacks into intact ribbons. The extent to which membrane tubules formed during reassembly of the Golgi complex utilize microtubules is unknown.
Here we show by live cell imaging that following washout of ONO, Golgi mini-stacks undergo a notable burst of membrane tubule formation, which is greatly facilitated by microtubules and dynein activity.
MATERIALS AND METHODS
HeLa cells stably expressing GFP fused to galactosyltransferase (GalT-GFP) were used for all experiments and were the kind gift of Brian Storrie. Cells were grown in minimal essential media (MEM) containing 10% Nu-Serum (BD Biosciences) in an atmosphere of 95% air, 5% CO2. ONO-RS-082 (ONO) and nordihydroguaiaretic acid (NDGA) were obtained from Biomol, Plymouth, PA). ONO and NDGA were prepared as 10 mM stock solutions in DMSO and stored at −20·C. They were freshly prepared in MEM to 10 μM before use. Nocodazole was used at 6 μg/ml in MEM. For all experiments, cells were washed 3 times in MEM without serum and incubated with freshly prepared 10 μM ONO-RS-082 in MEM for varying periods of time at 37°C. Cells were then allowed to recover as indicated and viewed immediately. For the nocodazole experiments, a frozen stock was prepared and freshly diluted to 6 μg/ml before each experiment. All imaging was done on a Perkin-Elmer Ultraview (Waltham, MA) or Intelligent Imaging Innovations (Denver, Colorado) spinning disk confocal microscope. Golgi membrane tubules were quantified by frame-by-frame analyses through 10 min movies. Quantifying the fragmentation of intact Golgi ribbons into separate mini-stacks and expression as a Fragmentation Index was performed as described [14].
RESULTS AND DISCUSSION
ONO-Induced Golgi Fragmentation and Recovery by Membrane Tubule Formation
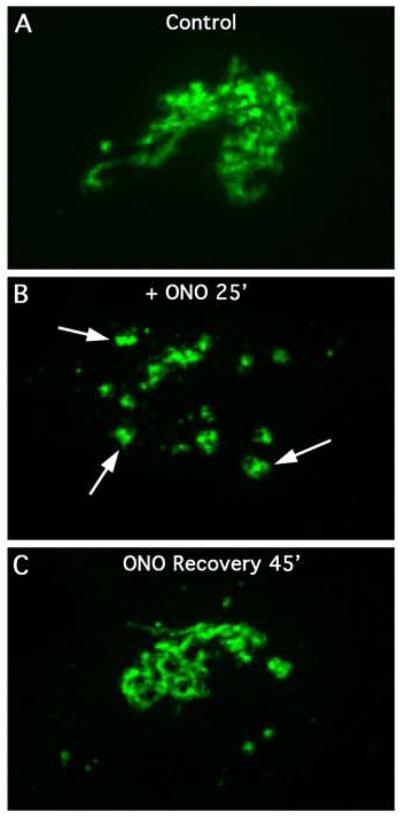
As previously shown [5; 6], treatment with the reversible PLA2 antagonist ONO for 45 min caused the intact Golgi ribbon to break up into fragments of disconnected mini-stacks (Fig. 1A, B). Time-lapse movies from live cell imaging reveal that the intact ribbon slowly comes apart into separated fragments; however, the mini-stacks mostly retain their juxtanuclear positions (Supplemental video 1). To image the recovery process, cells were treated with 10 μM ONO for 45 min, and then washed free of the drug (or kept in its presence) for various periods of time. Full recovery of an intact Golgi ribbon was seen by 45 min of washout (Fig. 1C). As observed by live cell imaging, at all time points during washout, multiple tubules were seen sprouting from Golgi mini-stacks, sometimes joining adjacent ones, and other times traveling unconnected from one stack to another (Fig. 2; Supplemental movie 2). These tubules were seen to extend up to 1-2 μm in length. In contrast, when cells were maintained in the presence of ONO, very few tubules were seen (Fig. 3A-C; Supplemental movie 3). From the time-lapse movies we determined the number of membrane tubules that formed from the Golgi mini-stacks/unit time and found that washout of ONO resulted in a substantial burst of tubule formation (Table I). Additional experiments have revealed the presence of more tubules (by binning the images and adjusting the exposure levels) than was initially found, leading us to conclude that the actual number of tubules formed during recovery is likely to be higher (data not shown).
Figure 1.
The reversible PLA2 antagonist, ONO, causes the Golgi complex to fragment into mini-stacks. HeLa cells stably transfected with GalT-GFP were used for these and all subsequent figures. (A) Control cells (B) Cells treated with ONO for 25 min. (C) Cell treated with ONO for 25 min and then washed free of the drug for 45 min. A time-lapse video of ONO-induced Golgi fragmentation in live cells treated is provided as Supplemental Movie 1.
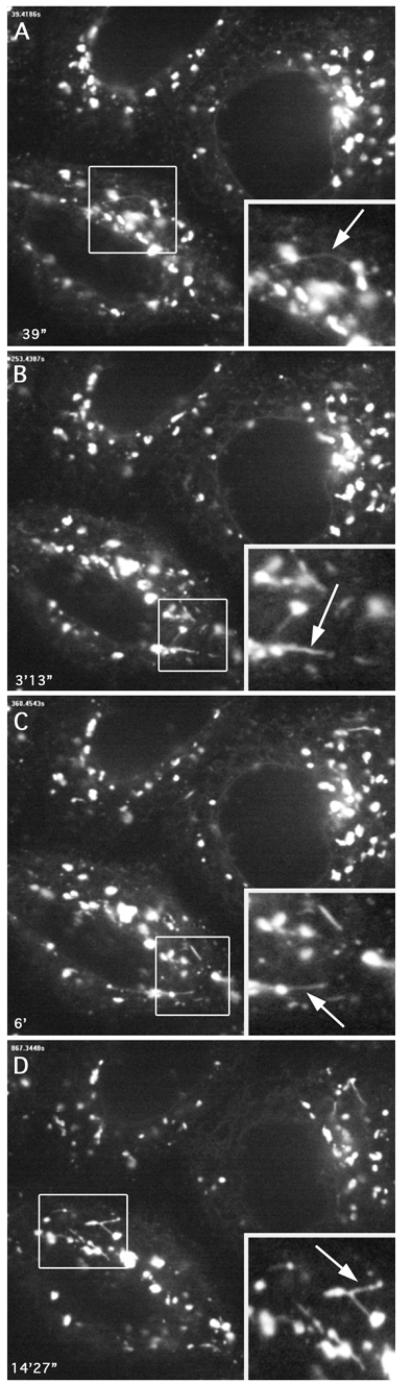
Figure 2.
Recovery from ONO induces a burst of Golgi membrane tubule formation. Still images taken from live cell movies of cells recovering from ONO treatment show a dramatic increase in membrane tubules forming from Golgi mini-stacks (see Supplemental Movie 2). HeLa cells stably transfected with GalT-GFP were treated with ONO for 45 min and then washed free of ONO. The cells were placed back in MEM + serum for 20 min and then imaged. Numerous tubules were observed (arrows). Insets show enlargement of boxed regions. Time intervals are shown in the lower left.
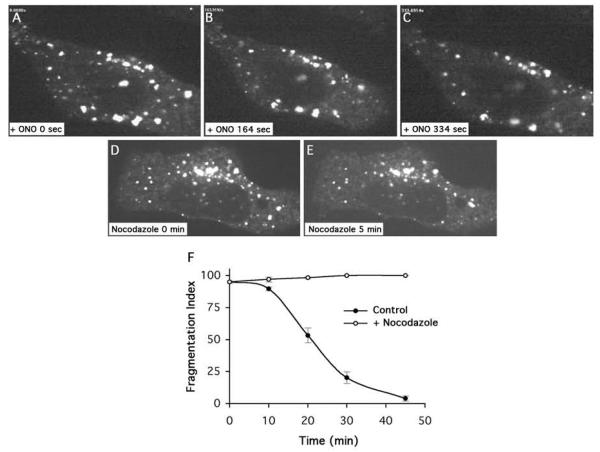
Figure 3.
Golgi membrane tubule formation during reassembly requires both PLA2 activity and microtubules. (A-C) Cells maintained in the continuous presence of ONO produce few tubules. Panels A-C were reproduced from Supplemental Movie 3. (D, E) Cells treated with ONO, but then allowed to recover in the presence of nocodazole. Panels were reproduced from Supplemental movie 4. In this experiment cells were treated with ONO for 45 min, nocodazole (6 μg/ml) was added during the last XX min of ONO incubation, and then cells were washed free of ONO but kept in the continuous presence of nocodazole. (D) Quantitation of Golgi recovery expressed as a Fragmentation Index (100% = 100% of cells with fragmented Golgi complexes; 0% = cells with normal Golgis.
Table 1.
Number of Golgi membrane tubules formed
| Condition/Treatment | Membrane tubules formed/cell/10′ |
|---|---|
| Continuous presence of ONO | 4.3 + 3.8 |
| Washout from ONO | 29.9 + 11.9 |
| Washout + Nocodazole | None detected |
These studies are consistent with previous work, which suggested that following washout of ONO, Golgi mini-stacks coalesce into an intact ribbon via membrane tubules [6]. However, our live cell imaging studies underscore the remarkable capacity of Golgi membranes to form tubules. The burst of membrane tubule formation following washout of ONO is consistent with previous ideas that the assembly and maintenance of an intact Golgi ribbon requires the dynamic formation of membrane tubules, which continuously probe the cytoplasm to “search and capture” for other Golgi membranes [6; 8]. Fusion of membrane tubules with their targets would facilitate the reassembly process. Currently, we do not know if the tubules are restricted to homotypic fusion with targets, i.e., medial Golgi tubules only with medial Golgi targets, or heterotypic fusion with other cisternae. Likewise, it is unclear if Golgi membrane fusion during recovery from mitosis is restricted to homotypic fusion, or if more relaxed interactions are permitted [15].
Microtubules are Required for Golgi Reassembly During Washout from ONO
In several cases the formation of Golgi membrane tubules is greatly facilitated by microtubules and microtubule motor proteins [4]. In addition, other studies have suggested that reassembly of disrupted Golgi ribbons is dependent on microtubules [16; 17]. Therefore, we asked if Golgi reassembly and membrane tubule formation following washout from ONO is affected by the microtubule depolymerizing drug nocodazole. Cells were first treated with ONO to fragment the Golgi, and then washed free of the drug in the presence or absence of nocodazole. In the presence of nocodazole, reassembly of an intact ribbon was significantly retarded (Fig. 3D, E), similar to just maintaining cells in ONO. Live cell imaging was done to determine if membrane tubules were similarly inhibited. Indeed, nocodazole treatment significantly reduced the number of tubules seen to form during washout from ONO (Table 1; Supplemental Movie 4).
The microtubule-facilitated movement of Golgi membrane tubules during reassembly strongly suggests the additional role of a microtubule motor. Given that interphase Golgi ribbons are positioned near the MTOC by the activity of cytoplasmic dynein [9; 10], we asked if dynein might be involved in membrane tubule-mediated reassembly.
NDGA, a Dynein Inhibitor, Slows Reassembly Following ONO Washout
Previous studies have shown that NDGA inhibits anterograde trafficking from the ER thus causing relocation of the Golgi complex and trans Golgi network (TGN) to the ER [18]. Originally, these affects were attributed to NDGA's activity as a lipoxygenase inhibitor. However, more recent studies have shown that NDGA has pleiotropic effects on cells because it causes abnormal, microtubule-dependent accumulation of dynein-dynactin complexes and Golgi membranes at the MTOC [19]. We hypothesize that NDGA could influence reassembly in opposite ways. Since it causes abnormal accumulation of dynein-dynactin complexes and Golgi membranes at the MTOC, NDGA could accelerate recovery during ONO washout. Conversely, NDGA could inhibit reassembly because dynein-dynactin complexes get stuck at the MTOC and are then unavailable for normal function. Therefore, we asked if Golgi reassembly during recovery from ONO treatment was sensitive to NDGA. Cells were treated with ONO to fragment the Golgi and then allowed to recover in the presence or absence of NDGA (Fig. 4). In solvent controls, the Golgi reassembled with normal kinetics (Fig. 4C). In contrast, in the presence of NDGA recovery was almost completely inhibited, as the Golgi remained in fragmented mini-stacks (Fig. 4B, D). This inhibition by NDGA was reversible following washout of the drug (data not shown).
Figure 4.
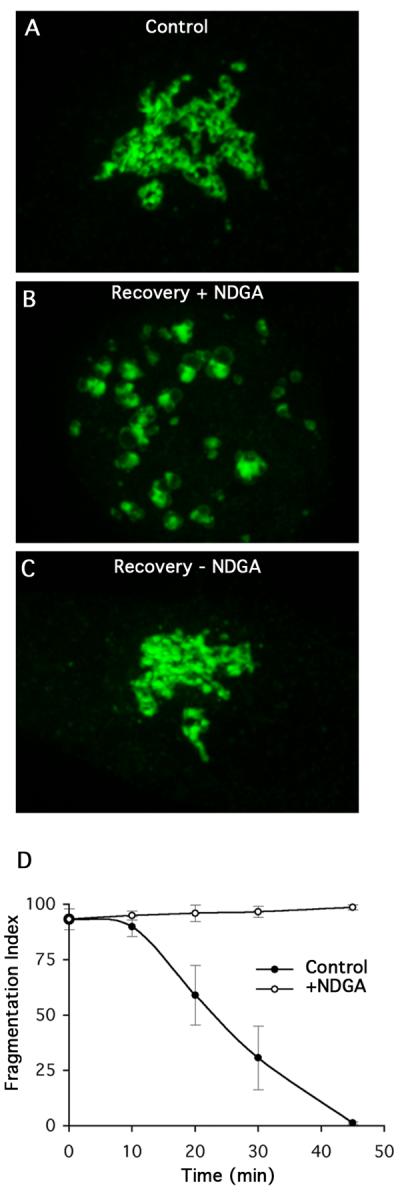
The dynein modulator NDGA inhibits reassembly of the Golgi complex during washout from ONO treatment. HeLa cells stably transfected with GalT-GFP were treated with ONO for 45 minutes and then washed free of ONO in the presence or absence of NDGA (10 μM). (A) Control cells before ONO treatment. (B) Cells were allowed to recover from ONO in the presence of NDGA for XX min. (C) Recovery from ONO in the absence of ONO for XX min. (D) Quantitation of Golgi recovery in the presence or absence of NDGA.
We conclude from these experiments that reassembly of physically separate mini-stacks into an intact Golgi ribbon requires PLA2 activity to induce membrane tubule formation and dynein-dependent microtubule transport to complete the process. These results are also consistent with the hypothesis that NDGA causes an accumulation of dynein-dynactin complexes that are rendered unavailable for reassembly.
A role for phospholipid remodeling enzymes in regulating the structure and function of the Golgi complex is becoming increasingly evident [20]. As we have shown here and elsewhere [8], cytoplasmic PLA2 enzymes appear to play an important role in Golgi structure and function by regulating membrane tubule formation. In addition, PLA2 enzymes have been shown to be important for endosome tubule formation and endocytic trafficking [21; 22] They could do so by generating positive-curve inducing lysophospholipids for membrane bending [23] and by providing for continual turnover of phospholipids to make phosphatidic acid and diacylglycerol, which recruit effector proteins to Golgi membranes [24]. In addition, other studies have shown that consumption of lysophospholipids by integral membrane lysophospholipid acyltransferases also contribute to control of Golgi tubule formation [25; 26]. More specifically, lysophosphatidic acid acyltransferase 3 (LPAAT 3), which converts lysophosphatidic acid to phosphatidic acid, negatively regulates the formation of Golgi membrane tubules [14]. Thus, a continual cycle of PLA2- and LPAAT-mediated remodeling of Golgi membranes appears to be important for normal structure and function [20].
Supplementary Material
HeLa cells stably expressing GalT-GFP were grown on coverslips, transferred to MEM + 10 μM ONO, and imaged immediately.
HeLa cells stably expressing GalT-GFP were treated with ONO for 45 min and then washed free of ONO. The cells were placed back in MEM + serum for 20 min and then imaged.
HeLa cells stably expressing GalT-GFP were treated with ONO for XX min and then imaged in the continuous presence of ONO for the times indicated.
HeLa cells stably expressing GalT-GFP were treated with ONO for 45 min, nocodazole (6 μg/ml) was added during the last XX min of ONO incubation, and then cells were washed free of ONO but kept in the continuous presence of nocodazole and imaged.
Acknowledgments
We thank Brian Storrie for providing the HeLa cells used in this study, and Marie E. Bechler for helpful comments on the manuscript. This work was supported by NIH grant DK51596.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Barr FA. Golgi inheritance: shaken but not stirred. J Cell Biol. 2004;164:955–8. doi: 10.1083/jcb.200402011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–93. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J. Cytoskeletal proteins and Golgi dynamics. Curr Opin Cell Biol. 1998;10:52–59. doi: 10.1016/s0955-0674(98)80086-0. [DOI] [PubMed] [Google Scholar]
- 5.de Figueiredo P, Drecktrah D, Katzenellenbogen JA, Strang M, Brown WJ. Evidence that phospholipase A2 activity is required for Golgi complex and trans Golgi network membrane tubulation. Proc Natl Acad Sci U S A. 1998;95:8642–8647. doi: 10.1073/pnas.95.15.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Figueiredo P, Polizotto RS, Drecktrah D, Brown WJ. Membrane tubule-mediated reassembly and maintenance of the Golgi complex is disrupted by phospholipase A2 antagonists. Mol Biol Cell. 1999;10:1763–1782. doi: 10.1091/mbc.10.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabouille C, Misteli T, Watson R, Warren G. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J Cell Biol. 1995;129:605–618. doi: 10.1083/jcb.129.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown WJ, Chambers K, Doody A. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic. 2003;4:214–221. doi: 10.1034/j.1600-0854.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- 9.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–84. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corthesytheulaz I, Pauloin A, Pfeffer SR. Cytoplasmic Dynein Participates in the Centrosomal Localization of the Golgi Complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri HP, Yuan LC, Klausner RD. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 12.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A's effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 13.Wood SA, Park JE, Brown WJ. Brefeldin A causes a microtubule-mediated fusion of the trans-Golgi network and early endosomes. Cell. 1991;67:591–600. doi: 10.1016/0092-8674(91)90533-5. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt JA, Brown WJ. Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function. J Cell Biol. 2009;186:211–8. doi: 10.1083/jcb.200904147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- 16.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drecktrah D, Brown WJ. Phospholipase A2 antagonists inhibit nocodazole-induced Golgi ministack formation: evidence of an ER intermediate and constitutive cycling. Mol Biol Cell. 1999;10:4021–4032. doi: 10.1091/mbc.10.12.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drecktrah D, de Figueiredo P, Mason RM, Brown WJ. Retrograde trafficking of both Golgi complex and TGN markers to the ER induced by nordihydroguaiaretic acid and cyclofenil diphenol. J Cell Sci. 1998;111:951–65. doi: 10.1242/jcs.111.7.951. [DOI] [PubMed] [Google Scholar]
- 19.Arasaki K, Uemura T, Tani K, Tagaya M. Correlation of Golgi localization of ZW10 and centrosomal accumulation of dynactin. Biochem Biophys Res Commun. 2007;359:811–6. doi: 10.1016/j.bbrc.2007.05.188. [DOI] [PubMed] [Google Scholar]
- 20.Bankaitis VA. The Cirque du Soleil of Golgi membrane dynamics. J Cell Biol. 2009;186:169–71. doi: 10.1083/jcb.200907008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Figueiredo P, Doody A, Polizotto RS, Drecktrah D, Wood S, Banta M, Strang M, Brown WJ. Inhibition of transferrin recycling and endosome tubulation by phospholipase A2 antagonists. J. Biol. Chem. 2001;276:47361–47370. doi: 10.1074/jbc.M108508200. [DOI] [PubMed] [Google Scholar]
- 22.Doody AM, Antosh AL, Brown WJ. Cytoplasmic Phospholipase A2 Antagonists Inhibit Multiple Endocytic Membrane Trafficking Pathways. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.08.067. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallop JL, McMahon HT. BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp. 2005:223–31. doi: 10.1042/bss0720223. [DOI] [PubMed] [Google Scholar]
- 24.Bard F, Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol. 2006;22:439–55. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- 25.Chambers K, Brown WJ. Characterization of a novel CI-976-sensitive lysophospholipid acyltransferase that is associated with the Golgi complex. Biochem Biophys Res Commun. 2004;313:681–686. doi: 10.1016/j.bbrc.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Drecktrah D, Chambers K, Racoosin EL, Cluett EB, Gucwa A, Jackson B, Brown WJ. Inhibition of a Golgi complex lysophospholipid acyltransferase induces membrane tubule formation and retrograde trafficking. Mol Biol Cell. 2003;14:3459–3469. doi: 10.1091/mbc.E02-11-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HeLa cells stably expressing GalT-GFP were grown on coverslips, transferred to MEM + 10 μM ONO, and imaged immediately.
HeLa cells stably expressing GalT-GFP were treated with ONO for 45 min and then washed free of ONO. The cells were placed back in MEM + serum for 20 min and then imaged.
HeLa cells stably expressing GalT-GFP were treated with ONO for XX min and then imaged in the continuous presence of ONO for the times indicated.
HeLa cells stably expressing GalT-GFP were treated with ONO for 45 min, nocodazole (6 μg/ml) was added during the last XX min of ONO incubation, and then cells were washed free of ONO but kept in the continuous presence of nocodazole and imaged.