Abstract
We live in a macroscopic three-dimensional world, but our best description of the structure of matter is at the atomic and molecular scale. Understanding the relationship between the two scales requires that we bridge from the molecular world to the macroscopic world. Connecting these two domains with atomic precision is a central goal of the natural sciences, but it requires high spatial control of the 3D structure of matter.1 The simplest practical route to producing precisely designed 3D macroscopic objects is to form a crystalline arrangement by self-assembly, because such a periodic array has only conceptually simple requirements: [1] A motif whose 3D structure is robust, [2] dominant affinity interactions between parts of the motif when it self-associates, and [3] a predictable structures for these affinity interactions. Fulfilling all these criteria to produce a 3D periodic system is not easy, but it should readily be achieved by well-structured branched DNA motifs tailed by sticky ends.2 Complementary sticky ends associate with each other preferentially and assume the well-known B-DNA structure when they do so;3 the helically repeating nature of DNA facilitates the construction of a periodic array. It is key that the directions of propagation associated with the sticky ends not share the same plane, but extend to form a 3D arrangement of matter. Here, we report the crystal structure at 4 Å resolution of a designed, self-assembled, 3D crystal based on the DNA tensegrity triangle.4 The data demonstrate clearly that it is possible to design and self-assemble a well-ordered macromolecular 3D crystalline lattice with precise control.
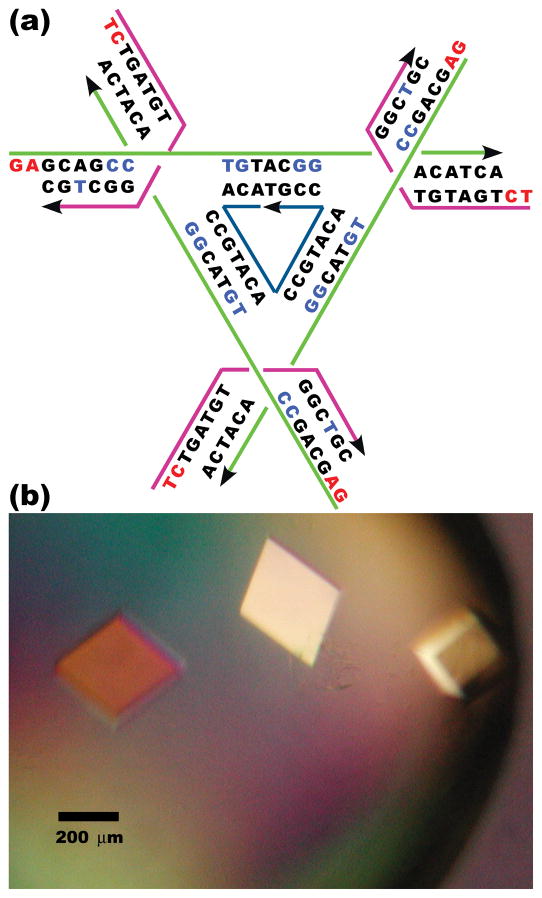
The tensegrity triangle is a rigid DNA motif with three-fold rotational symmetry, consisting of three helices that are directed along linearly independent vectors, i.e., their helix axis directions do not all share the same plane. The helices are connected pair-wise by three four-arm branched junctions so as to produce the stiff alternating over-and-under motif shown schematically in Figure 1a. Thus, there are three helical domains, each containing two double helical turns (21 nucleotide pairs, including sticky ends). There are seven strands in the molecule, three that partake in a crossover near the corners (magenta in Figure 1a), three that extend for the length of each helix (green in Figure 1a), and a final nicked strand at the center (blue in Figure 1a), completing the crossovers and the double helices between the crossovers; the green and magenta strands indicate an over-and-under motif. By tailing the three helices in short single-stranded cohesive segments (‘sticky ends’) the helices can be directed to connect with helices belonging to six other molecules in six different directions, thereby yielding a 3D periodic lattice, i.e., a crystal; the complementary GA and TC sequences of the sticky ends used here are indicated in red letters in Figure 1a. In this case, we have worked with a three-fold symmetric system, because that design has produced the best crystals of this motif (~4 Å resolution). Thus, each of the magenta and green strands present three times contains the same sequence; the central 21-mer strand has a triply repeating sequence, and its nicked site is threefold rotationally averaged, occurring with 1/3 occupancy in each edge; in this design, the six sticky ends form three identical complementary pairs. Triangles lacking this symmetry produced crystals that diffracted to somewhat lower resolution. Indexing the crystal in a rhombohedral lattice yields a unit cell edge of 69.22 Å and an angle between the edges of 101.44°. Pictures of the rhombohedral crystals are shown in Figure 1b. Unlike self-assembled 2D crystals, typically a few microns in extent,5 or 3D DNA-nanoparticle crystals,6,7 these self-assembled 3D crystals are macroscopic objects, exceeding 250 microns in dimension.
Figure 1. Schematic Design, Sequence, and Crystal Pictures.
(a) Schematic of the Tensegrity Triangle. The three unique strands are shown in magenta (strands restricted to a single junction), green (strands that extend over each edge of the tensegrity triangle) and dark blue (one unique nicked strand at the center passing through all three junctions). Arrowheads indicate the 3′ ends of strands. Nucleotides with A-form-like characteristics are written in bright blue. Cohesive ends are shown in red letters. (b) An Optical Image of Crystals of the Tensegrity Triangle. The rhombohedral shape of the crystals and the scale are visible.
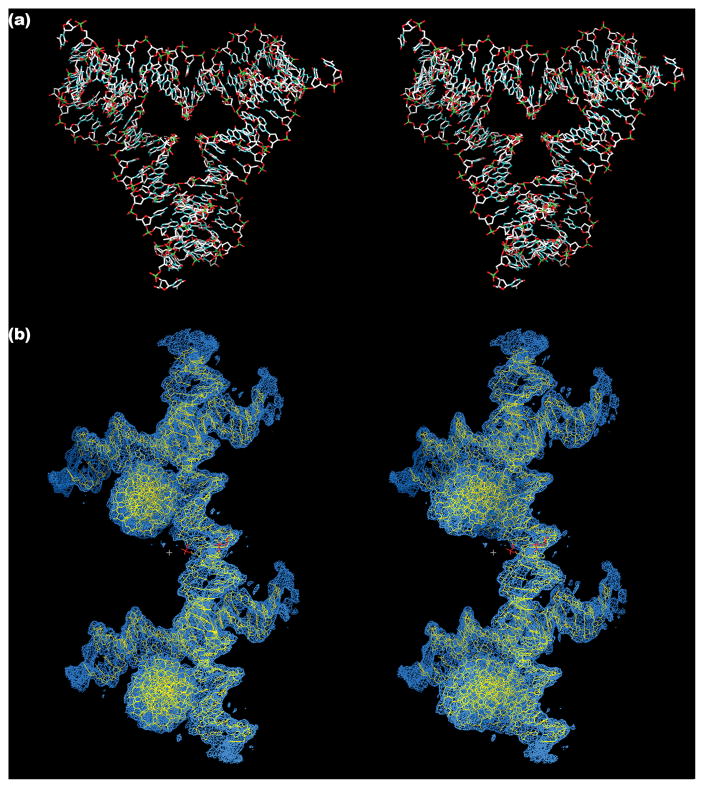
Although the crystallographic asymmetric unit of the unit cell is one third of the triangle, it is most useful for comprehending the structure to think about the whole triangular structure. The triangle is shown in a stereoscopic view in Figure 2a, where its over-and-under motif is readily visible: For example, the base pairs of the horizontal double helix are in front at the left, but are seen at the rear on the right. Figure 2b shows a view of two of the triangles in electron density, with emphasis on their connection by sticky ended cohesion. Molecular details are not very reliable at this resolution, but it is clear from the electron density that the DNA is largely in the B-form, with C2′-endo nucleosides. However, seven of the independent nucleotides have characteristics closer to A-form nucleotides rather than B-form, estimated by comparisons with Ho et al.’s A-B series of crystal structures;8 these nucleotides are indicated by blue lettering in Figure 1a. The view of the structure perpendicular to a helix axis seen Figure 2b, shows unmistakable double helical features, such as the major and minor grooves. Holliday junctions, similar to the four-arm junctions that comprise the corners of the triangle, have been examined previously by crystallography9 and by AFM.10,11 The angle between the axes in those cases is about 40–60°, somewhat smaller than the 78° angle enforced by the structure of this tensegrity triangle. A Holliday crossover between two helices is visible at the bottom of Figure 2a.
Figure 2. Views of the Tensegrity Triangle.
(a) Stereoscopic View of the Triangle Down its 3-Fold Axis. It is in the same orientation as the schematic in Figure 1a. The helix on the top edge starts above the mean plane of the molecule at the left and proceeds to the rear as it moves to the right. (b) Stereoscopic View of Two Triangles in Electron Density. This image is perpendicular to an edge of the rhombohedron, showing the connection of two triangles by sticky ends. Sticky ends are magenta for emphasis. Some density features belong to neighboring molecules not depicted.
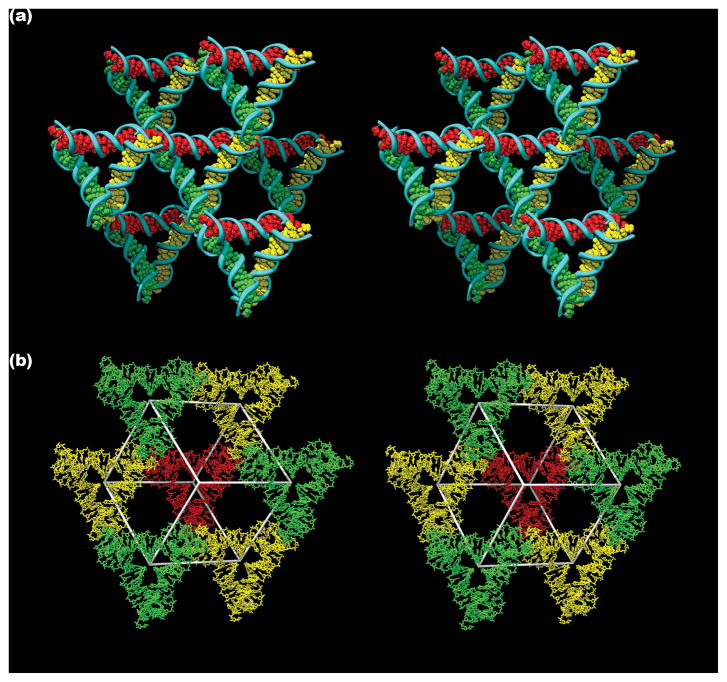
This crystal structure demonstrates the viability of designing periodic nucleic acid structures in three dimensions. The three directions that define the lattice are evident from the red, green and yellow color coding in Figure 3a, which shows the surroundings of a given tensegrity triangle. The open nature of this stick-like lattice is shown in Figure 3b, which illustrates the rhombohedron that is flanked by eight of the triangles. The red triangle is at the rear, bonded by sticky ends to the three yellow triangles that flank it lying in a plane closer to the viewer. The yellow triangles are bonded to the green triangles lying in a plane yet closer to the viewer. An eighth triangle lying even closer to the viewer and directly above the red triangle has been excluded for clarity. The volume of this rhombohedral cavity is ~103 nm3, and its open cross section has an area of ~23 nm2.
Figure 3. Lattice Formed by Tensegrity Triangles.
(a) Surroundings of a Triangle. This stereoscopic image distinguishes three independent directions by base pair color. The central triangle is flanked by six other triangles. (b) Rhombohedral Cavity Formed by Tensegrity Triangles. This stereoscopic image shows seven of the eight triangles that comprise the rhombohedron’s corners. The cavity outline is drawn white. The rear red triangle connects through one edge each to the three yellow triangles in a plane closer to the viewer. The yellow triangles are connected through two edges each to two different green triangles that are even nearer the viewer.
The arrangement of the molecules in this crystal is not fortuitous: It is clearly the result of sticky-ended cohesion as a consequence of our design; indeed, these are the only direct intermolecular contacts in the crystal. As a further demonstration of the ability to program crystalline DNA arrangements in 3D using sticky-ended cohesion, we have constructed eight other rhombohedral lattices from related tensegrity triangles. The crystals are summarized in Table 1, and their structures will be described elsewhere; Table 1 also indicates that these crystals contain cavities that can exceed 1000 nm3 (one zeptoliter). The unit cells, lengths and angles are approximately what would be predicted12 from the design. The resolution of the crystals decreases with increasing edge length; this is possibly a consequence of constructing stick-like lattices that lack the contacts that stabilize the more common ball-like lattices found in most biomacromolecular crystals. The presence of three-fold rotational symmetry also seems to improve resolution; this is possibly because only three unique strands, rather than seven, need to be purified and mixed with appropriate stoichiometry. Thus, the system described here is a robust basis for crystalline design. A previous system that diffracts well has been reported;13,14 however, it does not contain rationally designed Watson-Crick pairing in all directions of cohesion. Designed inorganic15 3D periodic systems and coordination networks16 have been reported, but unlike DNA with sticky ends,5,17,18 those systems cannot be used conveniently to design an arbitrary number of components in the asymmetric unit.
Table 1. Crystalline Tensegrity Triangle Lattices.
The cross-sectional area and cavity size are derived from the lattice parameters. Cross-sections and cavity sizes are estimated by subtracting two radii of the double helix (~10 Å) from the unit cell dimensions. The Space Group indicates whether deliberate three-fold rotational averaging has been performed; it has for those in R3, not for those in P1. Edge lengths and inter-junction distances (within triangles) are given in nucleotide pairs. Crystal 1 is the work reported here. The structures of crystals 3 and 7 have been determined by molecular replacement; others are in progress.
| Crystal No. | Edge Length | Space Group | Inter-junction Pairs | Rhombohedral Cell Dimensions | Resolution (Å) | Cross Section (nm2) | Cavity Size (nm3) |
|---|---|---|---|---|---|---|---|
| 1 | 21 | R3 | 7 | a = 68.3 Å, α = 102.4° | 4.0 | 23 | 103 |
| 2 | 21 | P1 | 7 | a = 68.0 Å, α̣= 102.6° □.0 23 | 101 | ||
| 3 | 31 | R3 | 17 | a = 102.0 Å, α = 112.7° | 6.1 | 62 | 366 |
| 4 | 31 | P1 | 17 | a = 100.9 Å, α = 111.6° | 6.3 | 61 | 373 |
| 5 | 32 | R3 | 18 | a = 103.6 Å, α = 113.6° | 6.5 | 64 | 367 |
| 6 | 32 | P1 | 18 | a = 103.3 Å, α = 112.2° | 6.5 | 64 | 395 |
| 7 | 42 | R3 | 17 | a = 134.9 Å, α = 110.9° | 11.0 | 123 | 1104 |
| 8 | 42 | P1 | 17 | a = 133.7 Å, α = 111.3° | 14.0 | 120 | 1048 |
| 9 | 42 | R3 | 28 | a = 134.9 Å, α = 117.3° | 10.0 | 117 | 643 |
The applications that have been suggested previously for designed 3D nucleic acid crystalline systems include the scaffolding of biological systems for crystallographic structure determination,19 as well as the organization of nanoelectronics.20 Both of these applications will likely be most usefully realized with scaffolding that is not 3-fold rotationally averaged. Guests that can be accommodated three at a time in the unit cell would be acceptable for structure determination, but they would need to be small enough to fit; threefold rotational averaging of a single guest would not be optimal for determining structures. Thus, the crystallographic application will likely require somewhat higher resolution and perhaps the larger unit cells noted above, so as to accommodate larger guest molecules. The nanoelectronics application probably requires only the larger unit cells, so as to accommodate large components. Nevertheless, following this beginning, the other steps apparently needed for these applications are likely to prove incremental and feasible. It seems reasonable that many of the applications of DNA arrays that have been prototyped in two dimensions can be extended to three dimensions. For example, it should be possible to incorporate nanomechanical devices17 into a 3D lattice, perhaps capable of being programmed to capture specific molecules at specific times.14,21 The ~250 micron-sized crystals used in this study contain an estimated 4.5 × 1013 unit cells; metallic nanoparticles have already been used to form patterns on 2D DNA arrays,18 so extending that capability to 3D could provide a very dense ordered arrangement of nanoparticles. In addition to periodic assembly, aperiodic algorithmic assembly22 in three dimensions is also conceivable, but crystallography will not be used readily to establish that the assembly has occurred flawlessly.
Methods Summary
Synthesis, purification and crystallization
DNA sequences were designed using program SEQUIN.23 DNA strands, including iodinated derivatives were synthesized by standard phosphoramidite techniques on an Applied Biosystems 394 DNA synthesizer. Strands were doubly purified by reverse-phase HPLC using a C-18 column (Waters). Crystals were grown from 80 μL sitting drops in a thermally-controlled incubator containing 0.25 μg/μL DNA, 30 mM sodium cacodylate, 50 mM magnesium acetate, 50 mM ammonium sulfate, 5 mM magnesium chloride, 25 mM Tris (pH 8.5), equilibrated against a 1.5 mL reservoir of 1.7 M ammonium sulfate. Rhombohedral-shaped crystals with dimensions as large as 250×250×250 μm were obtained by slow annealing, in which the temperature was decreased from 60 °C to room temperature (~20 °C) with a cooling rate of 0.2 °C per hour over a period of 7 days, during which the volume of the drop diminished by about 90%. Crystals were obtained at the end of the cooling step, and appeared full-sized within a day. Both native and iodinated derivative crystals were produced under the same conditions.
Data Collection
Crystals were transferred to a cryosolvent of 30% glycerol, 100 mM ammonium sulfate, 10 mM MgCl2, and 50 mM Tris and were frozen by immersion into liquid nitrogen. X-ray data diffraction data were collected from crystals of iodinated derivatives (12 iodine atoms per triangle -- on the fourth and thirteenth nucleotide of each green strand and on the sixth and eleventh nucleotide of each red strand) at 1.7 Å on beamlines X6A and X25 at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, New York, USA). A complete sphere of native x-ray data were collected at the APS beamline 19ID.24
Structure Determination
Complete crystallographic details and associated references are available in the supplementary information in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
This research has been supported by grants to NCS: from the National Institute of General Medical Sciences, the National Science Foundation, the Army Research Office, the Office of Naval Research and a grant from the W.M. Keck Foundation. It has also been supported by NSF grant CCF-0622093 and NIH grant 1R21EB007472 to CM. We thank Dr. William Sherman for assistance in establishing the likely structural features of tensegrity triangles. We would like to thank Drs. R. Sweet, M. Allaire, H. Robinson, A. Saxena and A. Heroux at the BNL-NSLS at beamlines X6A and X25 of the National Synchrotron Light Source, respectively. BNL-NSLS is supported principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. The use of the 19ID beamline at the Structural Biology Center/ Advanced Photon Source supported by the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Author contributions: Jianping Zhang grew crystals, collected data, analyzed data and wrote the paper; Jens Birktoft collected data, analyzed data, and wrote the paper; Yi Chen grew crystals, collected data and analyzed data; Tong Wang grew crystals, collected data, analyzed data and wrote the paper; Ruojie Sha grew crystals, analyzed data and wrote the paper; Pamela Constantinou grew crystals and analyzed data; Stephen Ginell collected data and analyzed data; Chengde Mao devised the motif, analyzed data and wrote the paper; Nadrian C. Seeman initiated the project, analyzed data and wrote the paper.
Atomic coordinates and experimental structure factors have been deposited within the Protein Data Bank and are accessible under the code 3GBI.
References
- 1.Whitesides GM, Mathias JP, Seto CT. Molecular self-assembly and nanochemistry: A chemical strategy for the synthesis of nanostructures. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC. Nucleic Acid Junctions and Crystal Formation. J Biomol Struct & Dyns. 1985;3:11–34. doi: 10.1080/07391102.1985.10508395. [DOI] [PubMed] [Google Scholar]
- 3.Qiu H, Dewan JC, Seeman NC. A DNA Decamer with a Sticky End: The Crystal Structure of d-CGACGATCGT. J Mol Biol. 1997;267:881–898. doi: 10.1006/jmbi.1997.0918. [DOI] [PubMed] [Google Scholar]
- 4.Liu D, Wang W, Deng Z, Walulu R, Mao C. Tensegrity: Construction of rigid DNA triangles with flexible four-arm junctions. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 5.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 6.Nykypanchuk DMM, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 8.Vargason JM, Henderson K, Ho PS. A crystallographic map of the transition from B-DNA to A-DNA. Proc Nat Acad Sci (USA) 2001;98:7265–7270. doi: 10.1073/pnas.121176898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichmann BF, Vargason JM, Mooers BHM, Ho PS. The Holliday junction in an inverted repeat DNA sequence: Sequence effects on the structure of four-way junctions. Proc Nat Acad Sci (USA) 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao C, Sun W, Seeman NC. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy . J Am Chem Soc. 1999;121:5437–5443. [Google Scholar]
- 11.Sha R, Liu F, Seeman NC. Atomic force measurement of the inter-domain angle in symmetric Holliday junctions. Biochem. 2002;41:5950–5955. doi: 10.1021/bi020001z. [DOI] [PubMed] [Google Scholar]
- 12.Birac JJ, Sherman WB, Kopatsch J, Constantinou PE, Seeman NC. GIDEON, a program for design in structural DNA nanotechnology. J Mol Graphics & Modeling. 2006;25:470–480. doi: 10.1016/j.jmgm.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paukstelis PJ, Nowakowski J, Birktoft JJ, Seeman NC. The crystal structure of a continuous three-dimensional DNA lattice. Chem & Biol. 2004;11:1119–1126. doi: 10.1016/j.chembiol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Paukstelis PJ. Three dimensional DNA crystals as molecular sieves. J Am Chem Soc. 2006;128:6794–6795. doi: 10.1021/ja061322r. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa H, Kim HJ, Ockwig NW, O’Keeffe M, Yaghi OM. Control of vertex geometry, structure dimensionality, functionality, and pore metrics in the reticular synthesis of crystalline metal-organic frameworks and polyhedra. J Am Chem Soc. 2008;130:11650–11651. doi: 10.1021/ja803783c. [DOI] [PubMed] [Google Scholar]
- 16.Kawano M, Kawamichi T, Haneda T, Kajima T, Fujita M. The modular synthesis of functional porous coordination networks. J Am Chem Soc. 2007;129:15418–15419. doi: 10.1021/ja0752540. [DOI] [PubMed] [Google Scholar]
- 17.Ding B, Seeman NC. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. 2D nanoparticle arrays show the organizational power of robust DNA motifs. NanoLett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 20.Robinson BH, Seeman NC. The design of a biochip: A self-assembling molecular-scale memory device. Protein Eng. 1987;1:295–300. doi: 10.1093/protein/1.4.295. [DOI] [PubMed] [Google Scholar]
- 21.Gu H, Chao J, Xiao SJ, Seeman NC. Dynamic patterns programmed by DNA tiles captured on a DNA origami substrate. Nature Nanotech. 2009;4:245–249. doi: 10.1038/nnano.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothemund PWK, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLOS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seeman NC. De Novo Design of sequences for nucleic acid structure engineering. J Biomol Str & Dyns. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 24.Rosenbaum G, et al. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Rad. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.