Abstract
The location of MLL translocation breakpoints within therapy-related acute myeloid leukemia linked to drugs targeting Topoisomerase II and infant acute leukemia (IAL) are biased towards the intron 11 - exon 12 region of MLL, though lacking a comprehensive explanation. To address this, blood samples were taken from breast cancer and lymphoma patients receiving Topoisomerase II inhibitor therapy. Inverse PCR analysis was used to interrogate the exon 12 region of MLL for rearrangements. Eleven of 19 observed translocations showed breakpoint junctions restricted to a single 5 bp location within exon 12. A similarly restricted distribution (11/20 breakpoint junctions) was observed in TK6 cells exposed to either estrogen (linked to IAL) or anti-CD95 antibody. The translocation hotspot was at the 5' edge of a 10 bp tract matched with a perfect palindrome, 101 bp distant. A high stringency Topoisomerase II consensus sequence binding site was noted at the geometric midpoint between palindromes. Using ligation-mediated PCR to screen TK6 cells exposed to anti-CD95 antibody showed 14/37 (38%) of DNA breaks were found adjacent to the 5' palindrome and 10/37 (27%) at the 3' partner. We propose a model whereby Topoisomerase II facilitates the organization of nuclease sensitive secondary structures, stabilized by palindrome association, which are prone to rearrangement.
Introduction
Translocations of the MLL gene are found in adult leukemia, especially in therapy-related acute myeloid leukemia (tAML) occurring in patients who received topoisomerase II inhibitors such as etoposide for a solid primary tumor or lymphoma (Super et al., 1993; Broeker et al., 1996; Martinelli et al., 1998). Also, 75–80% of patients with Infant Acute Leukemia (IAL) carry an aberration of the MLL gene (Cimino et al., 1997; Alexander et al., 2001). In support of a role for non-genotoxic agents affecting MLL biology, a large epidemiological study from Brazil found an association with the maternal use of oral contraceptives, either before or during the first trimester of pregnancy and the incidence of IAL (Pombo-de-Oliveira et al., 2006). Notably, the association was stronger for cases with MLL aberrations, and the authors raise the question whether estrogens could play a role in the etiology of IAL. Taken together these data indicate a likely multi-factorial process is involved in the generation of MLL translocations.
In the case of both t-AML and IAL MLL breakpoints, a bias in distribution is observed towards the 3' end of an 8.3 kbp tract of MLL, including exons 8 to 14, termed the breakpoint cluster region (BCR). This is the location of the majority of MLL translocation breakpoints, suggesting a common pathway or process is involved in their production (Broeker et al., 1996; Cimino et al., 1997). Substantial data are available that have associated exposure to topoisomerase II inhibitors to breaks occurring within the MLL gene suggesting a cause and effect relationship between the breaks and translocations (Stanulla et al., 1997a,b; Betti et al., 2001; Betti et al., 2003). Examination of translocations linked to prior Topoisomerase II inhibitor therapy show that, other than in rare cases, translocations do not occur at Topoisomerase II consensus binding sites (Lovett et al., 2001; Whitmarsh et al., 2003). Such an association might be expected if errors in the cleavage and/or strand passing activity of Topoisomerase II were directly involved in translocation generation (Whitmarsh et al., 2003; Libura et al., 2005; Felix et al., 2006). Others have also shown a discrete apoptosis related cleavage site within the 3' end of the BCR located close to the intron 11/exon 12 border that are also not uniquely linked to an increase in translocation breakpoints at that location (Betti et al., 2001; Betti et al., 2003). Also, in the latter case, it is not clear if this feature is mechanistically linked to translocation induction, which would also require subsequent suppression of apoptotic execution. In both cases, exonuclease processing of the breakpoint might move the site of eventual translocation some distance from the initial site of DNA damage.
In order to investigate the mechanism(s) involved in MLL translocation induction in more detail, factors affecting the bias towards translocation breakpoints within the 3' end of the breakpoint cluster region were studied using a range of conditions all linked to MLL driven disease. Thus, blood samples from both lymphoma and breast cancer patients exposed to Topoisomerase II inhibitor therapy as part of their treatment were examined for indications of MLL aberrations using an inverse PCR (iPCR) technique, examining a ∼700 bp region that included exon 12. In addition, both effects of estrogen hormone exposure, relevant to the incidence of MLL translocations found through epidemiologic studies and exposure to the apoptosis inducer anti-CD95 antibody were examined in terms of the MLL aberrations produced in vitro (Betti et al., 2001; Betti et al., 2003; Pombo-de-Oliveira et al., 2006).
For both in vitro and in vivo studies a substantial fraction (50% or greater) of all MLL translocations found showed a discrete distribution of breakpoints within a 5 bp tract of DNA within exon 12 of MLL. Further examination of this location showed that the translocation breakpoints were distributed adjacent to a 10 bp tract of palindromic DNA defining a potential 101 bp stem loop structure. A high stringency Topoisomerase II binding site was found at the geometric apex of the potential stem-loop. These data implicate DNA secondary structure in the formation of a sub-group of MLL translocations, some of which may progress to leukemogenic transformation.
Materials and Methods
Patient Samples and DNA Extraction
After informed consent had been obtained, 10 ml of blood were drawn from either volunteers, or patients undergoing treatment for lymphoma or breast cancer that included exposure to topoisomerase II inhibitors. The blood was placed into heparinized tubes either before the first treatment and subsequently 1, 3, 6 and 12 months later. In taking samples, care was taken to initiate DNA extraction within one hour of sample collection. This was done as others have shown that overnight storage of such material leads to apoptosis generating MLL rearrangements in vitro (Basecke et al., 2006). Blood was mixed with HBSS at a ratio of 1:1 and laid over the same volume of Ficoll and centrifuged at 2800g for 10 min (no brake). The white blood cells were removed from the buffy coat, washed with HBSS, and centrifuged at 2800 g for 10 min. Some cells were then aliquoted for freezing in FBS with 10% DMSO, the remainder immediately processed for DNA extraction. The DNA extraction was performed according to manufacturer’s protocol using wizard genomic DNA purification kit from Promega (Madison, WI).
Cell Line
The human lymphoblast cell line TK6 was obtained from American Type Culture Collection (Manassas, VA) and routinely maintained in RPMI 1640 (ATCC) supplemented with 10% fetal bovine serum.
Cell Treatment
TK6 cells were incubated at 37ºC for 20 minutes with zVAD-fmk (Promega, Madison, WI) at a final concentration of 20 µM prior to treatment with anti-CD95 antibody (eBioscience, San Diego, CA). Various concentrations of anti-CD95 antibody (0.25 µg/ml, 0.5 µg/ml, and 1.0 µg/ml) were used in the treatment and a four hour pulse delivered and removed by washing. Cells were harvested, from 4 h up to 29 days and processed for iPCR analysis. Some populations of cells were exposed to 0.01–1.0 µM estradiol (E2) or 0.01–1.0 µM 4-OH-E2. DNA samples were collected 1–3 days later.
Inverse PCR Analysis of Rearrangements
5 µg of DNA per group were incubated at 37 °C for 3–4 h with the restriction enzyme Sau3A1 (Promega,) and purified by Quickspin kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Digested DNA was eluted in 50 µl of ddH2O. Eluates were self-ligated with T4 DNA Ligase (Promega) at 4°C overnight, and the reaction terminated by incubating at 70 °C for 15 min. In addition, an aliquot of 20 µl of each ligated sample was further digested with PvuII (Promega) and the reaction was inactivated at 70 °C for 15 min. This step re-linearizes the native MLL fragment and therefore removes it as a template for the inverse PCR reaction that follows. A first round of semi-nested PCR was performed with 100 ng of ligated DNA or PvuII-redigested DNA as template. For iPCR, the following cycles were used: Denaturing for 10 min at 95 °C, 30 cycles of 94 °C (1 min), 58 °C (1 min) and 72 °C (1 min 30 sec), final extension at 72 °C for 10 min. The second round of iPCR was performed, using 1 µl of the first round PCR as a template, under the same conditions except with an annealing temperature of 60 °C and round 2 forward primer. Sequences of primers used for iPCR were as follows:
Round 1 forward primer: 5'-CACTCTTAGGTCACTTAGCATGTT-3'
Round 2 forward primer: 5'-TACTCTGAATCTCCCGCAGTGTCC-3'
Reverse primer: 5'-ACAGTTGTAAGGTCTGGTTTGTCC-3'.
All reactions were performed with GoTaq polymerase (Promega) and run using a 1.2 % agarose gel with ethidium bromide at 0.5 µg/ ml.
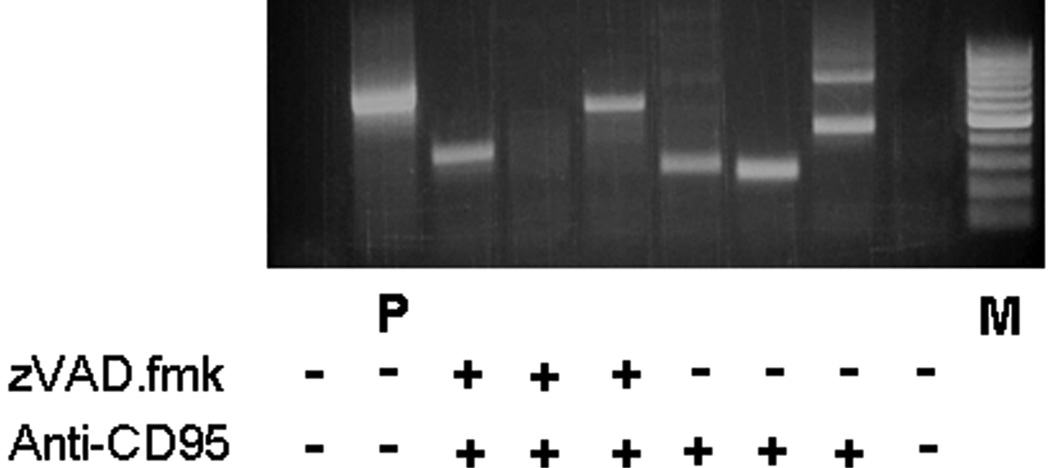
Aberrant bands were gel-extracted (Quickspin gel extraction kit, Qiagen, Valencia, CA) and cloned into TOPO-TA vector pCR2.1 (Invitrogen, Carlsbad, CA) according to the protocols provided by the manufacturers. Plasmids extracted from positive clones were sent for sequencing to Seqwright (Fisher Scientific, Pittsburgh, PA). Sequences were analyzed by comparison to the BLAST database of the NCBI. To reduce the possibility of artifacts being mistaken for rearrangements, primarily as a result of in vitro ligation of genomic DNA, all rearrangements had templates with confirmed re-ligation at the appropriate Sau3AI cleavage site. In addition, both secondary primer sites had to be present within the sequence analyzed. These precautions substantially reduce the possibilities of errors in interpretation (Marschalek 2008). It was found that the presence of a detectable PCR product did not always reflect the presence of an actual rearrangement after the analysis of sequenced material was complete (Fig. 1).
Figure 1.
IPCR Analysis: in vitro data. DNA gel of iPCR products – Sample iPCR product 16 days after exposure to both anti-CD95 antibody (0.5 µg/ml) and +/− 20 µM zVAD.fmk at the start of the experiment. All DNA extracts (except P) were digested with PvuII restriction enzyme that fragments native MLL and thus eliminates amplification of unmodified MLL sequence. P = MLL containing plasmid control. The presence of a PCR product, for example in the zVAD.fmk lanes, did not necessarily reflect the presence of rearrangements after sequencing and analysis was complete.
Ligation-Mediated PCR to Detect MLL Cleavage
In this technique, a blunt end linker is ligated to all available double-strand DNA breaks and then semi-nested PCR is carried out using linker and sequence specific primers adjacent to a pre-identified break-site. For these experiments, MLL cleavage was introduced in TK6 cells with anti-CD95 antibody. After a minimum of 4 hours treatment, genomic DNA was collected and ligated with linker.
Linker
Linker 11 (5'- GAATTCAGATC −3')
Linker 25 (5'- GCGGTGACCCGGGAGATCTGAATTC −3')
The detail on making of the linker and its ligation to genomic DNA has been described before (Betti et al., 2005). For the first round of PCR the reaction mixture uses linker ligated DNA as the template (1 µl from the ligation reaction) with Linker 25 primer plus 12.2 F primer. Prior to LM-PCR the reaction mixture was heated to 72 oC for 3 min to dissociate the 11-mer oligomer, leaving a 5', 25-mer overhang. Then Taq DNA Polymerase (2.5 Units) was added and LM-PCR executed with the following settings: 1× (72 oC for 5 min), 1× (95 oC for 4 min), 30× (95 oC for 30 s, 66 oC for 30 s, 72 oC for 45 s), 1× (72oC for 10 min) and left at 4 oC. For the second round, 1 µl from the first round was used for the template with Linker 25 and 12.3F primers and cycled for 1 × (95 oC for 4 min), 30 × (95 oC for 30 s, 66 oC for 30 s, 72 oC for 45 s), 1 × (72 oC for 5 min) and left at 4 oC. The PCR product generated was ∼290 bp in size allowing its extraction from the appropriate gel location with a standard kit (Qiagen, Valencia, CA). Subsequently, the extracted material was cloned into pCR2.1 Topo vector (Invitrogen, Carlsbad, CA) and sequenced using a vector specific primer. The site of cleavage in the MLL-BCR was defined by the location of linker in the gene sequence with numbering following the convention of Gu et al, primer locations noted below use this numbering system (Gu et al.,1992).
MLL Semi-nested Primers
12.2 F (5'- ATGCCCAAGTCCCTAGACAAAATGGTG −3') [7458 – 7484]
12.3 F (5'- GTCTGTTCACATAGAGTACAGAGGCAACTA −3') [7029 – 7058]
Results
Effect of Treatment on MLL Rearrangements
In vivo
For the patient based data, samples were taken both prior to therapy and at 1, 3, 6 and 12 months thereafter (Table 1&Table 2). Of five normal volunteers, each analyzed twice (one sample lost), none showed any aberrations of the MLL gene (0/9). At this time, 9 lymphoma patients and 13 breast cancer patients are available for study, of these, 5/22 contained translocations prior to therapy reducing the possibilities the rearrangements detected were secondary to therapy-linked DNA damage. For the 13 breast cancer patients 2/13 exhibited a translocation before treatment and 4/13 contained identifiable MLL translocations of any type, some with more than one translocation, giving 6 MLL aberrations total. For the individuals with lymphoma, 3/9 presented samples with translocations before treatment and 6/9 overall were positive for MLL rearrangements comprising 13 rearrangements in total. In the lymphoma treated patients 4/19 translocations detected were >3 months. after treatment was complete. The translocations detected here are clearly more common than the incidence of secondary leukemia in such patients and they thus likely represent alterations with restricted proliferative potential.
TABLE 1.
Treatment regimen following diagnosis of lymphoma
| Patient # | Gender | Age at first draw (years) | Regimens |
|---|---|---|---|
| 1001 | M | 56 | B |
| 1002 | M | 60 | B1 |
| 1003 | F | 56 | A2 |
| 1004 | F | 72 | A1 |
| 1005 | M | 61 | A3 |
| 1006 | M | 49 | A1 |
| 1007 | F | 66 | A4 |
| 1008 | F | 38 | C |
| 1009 | M | 58 | A5 |
| 1010 | M | 71 | A1 |
A, CHOP; A1, CHOP-R (rituxan) × 6; A2, CHOP × 7; R × 8; A3, CHOP × 6; Bexxar (radioimmunotherapy) × 1; A4, CHOP-R × 4; CVP-R × 1; GaRD (× 5); A5, CHOP-R × 4; Etoposide; Stem Cell Transplant; B, HyperCVAD; B1, HyperCVAD; Rituximab; C, ABVD × 10. Regimen Key: CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone); HyperCVAD (cyclophosphamide, cytarabine, doxorubicin, etoposide, methotrexate, vincristine). CVP-R (cyclophosphamide, vincristine, prednisone, rituxan); GaRD (gallium, rituxan, dexamethasone). ABVD (adriamycin, bleomycin, decarbazine, vinblastine). Drugs targeting Topoisomerase II shown in bold.
TABLE 2.
Treatment regimen for women with initial diagnosis of breast cancer
| Age at first draw (years) | Patient # | Regimens |
|---|---|---|
| 58 | 2001 | A |
| 47 | 2002 | B |
| 46 | 2003 | B |
| 60 | 2004 | A1 |
| 49 | 2005 | A1 |
| 56 | 2006 | C1 |
| 64 | 2007 | A1 |
| 49 | 2008 | C2 |
| 41 | 2009 | C |
| 46 | 2010 | C3 |
| 62 | 2011 | E |
| 47 | 2012 | B |
| 47 | 2013 | D |
A, Dox and Oral Cyc × 15; Pac × 12; A1, Dox & Oral Cyc × 15; Pac × 6; B, Dox & Cyc (IV) × 4; Pac × 4; C, Dox & Oral Cyc × 15; Pac × 12; Tras × 20; C1, Dox & Cyc × 6; Pac × 12; Tras ; C2, Dox & Cyc × 4; Pac × 4; Tras ; C3, Dox & Cyc × 4; Pac; Tras ; D, Dox & Cyc q 2 × 6; Pac × 12 ; E, Oral Cyc; Dox & Fluorouracil w/Pac. Regimen Key: Drugs: Dox= Doxorubicin; Cyc = Cyclophosphamide; Pac = Paclitaxel; Tras = Trasuzumab;
In vitro
In order to investigate factors contributing to MLL aberration induction, TK6 cells were exposed to anti-CD95 antibody with or without the broad spectrum caspase inhibitor zVAD.fmk, or left untreated in continuous culture (controls). IPCR analysis was performed as described generating discrete PCR products (Fig. 1). Unlike previous shorter duration experiments, detectable MLL translocations were found in all groups over the month duration of the experiment (Table 3). Interestingly, the majority (7/8) of translocations occurred 1–2 weeks after challenge with anti-CD95 antibody and only a single translocation was noted in the group treated with the anti-apoptotic compound zVAD.fmk. This provides some support for previous suggestions that apoptotic nucleases may be activated, and cause rearrangements, in cells capable of surviving (Betti et al., 2001, 2003, 2005). To explore alternative, non-apoptotic exposure conditions, TK6 cells were also briefly exposed to either 0.01–1µM of estradiol (E2) or 4-OH-E2 that generated no detectable apoptotic response as seen by Annexin V staining and flow cytometry analysis (data not shown). In this case, over a three day period, 7 translocations were seen in the E2 group and 2 translocations in the 4-OH-E2 group. In the case of estrogen these data indicate that MLL aberrations may be induced by agents that lack direct DNA damaging capacity
TABLE 3.
Distribution of MLL rearrangements in TK6 cells treated in-vitro
| Analysis Time | Control | E2 | 4-OH-E2 | Anti-CD95 ab | Anti-CD95 ab + zVAD.fmk |
|---|---|---|---|---|---|
| 24 h | TTTT | ||||
| 48 h | |||||
| 72 h | TTT | TT | |||
| 4 h | T | ||||
| 6 d | TT | TTTTT | |||
| 16 d | TT | T |
Data gathered from TK6 cells after exposure to either 0.01–1 µM E2 (estrogen), 4-OHE2 or anti-CD95 antibody (0.5 µg/ml) +/− 20µM of zVAD.fmk. T = Translocation. Drug treatments were applied at the start of culture only.
Distribution of Breakpoints within MLL Rearrangements
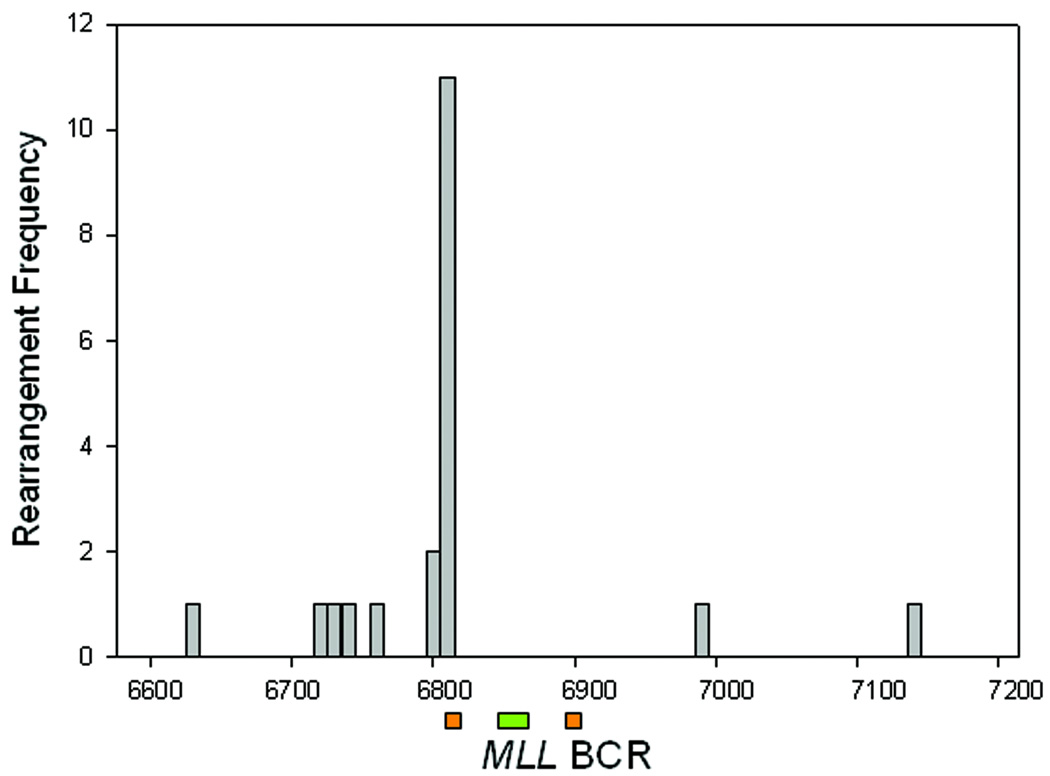
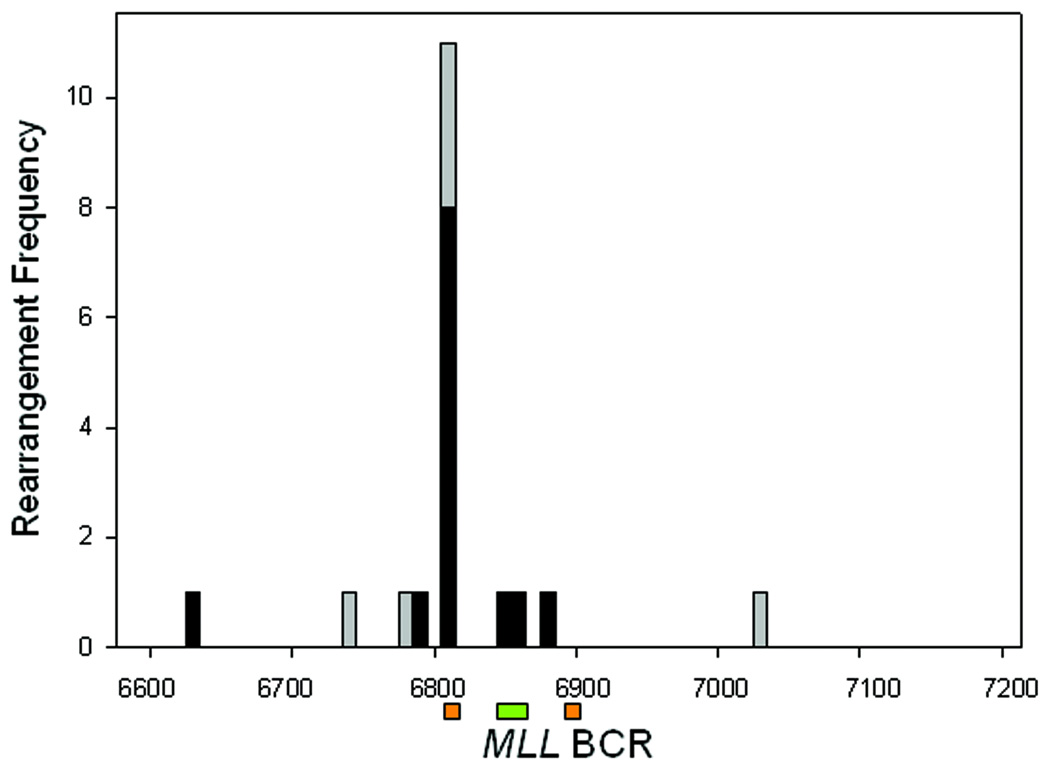
The majority of translocations detected both in vitro (16/20) and in vivo (16/19) showed microhomology at the site of fusion (Table 4 & Table 5). This indicates that the NHEJ repair pathway was likely involved in the formation of such rearrangements, as has been previously suggested. Examination of the translocations detected within patient blood samples, and detected in vitro, showed that the breakpoints were distributed in a non-random fashion. While breakpoint junctions were observed throughout the region analyzed, a specific clustering of events was observed at one major location, within exon 12 (Fig. 2). Altogether, of the 19 MLL rearrangements detected in both breast cancer and lymphoma groups, 11/19 contained a translocation located at or between MLL sequences 6804–6807. Examination of the in vitro generated rearrangements showed 11/20 breakpoints were localized at 6803-6, closely matching the distribution found within the patient samples. As the distribution of aberrations in patients tracked closely with that observed in vitro, it is possible that a common mechanism may be involved for their production.
TABLE 4.
Rearrangements detected in peripheral blood of lymphoma patients undergoing treatment
| Patient | Draw | Site | Chromosome | Partner location (bp) | MLLmicrohomology partner |
|---|---|---|---|---|---|
| 1001 | 1st | 6805 | 1 | 78875267 | TGTTGTGAGCCTCTGTTTAACCCTTAA |
| 1st | 6872 | 18 | 17998961 | ATGGTTGATACCCAGCTCC | |
| 6 month | 6805 | 10 | 69640793 | CTGTTGTGAGCCACATAATAAGGTGTCAAA | |
| 1002 | 6 month | 6805 | 8 | 119562654 | TCTGTTGTGAGCCACTGCGCCCAGCCATT |
| 6 month | 6805 | 18 | 70836192 | TGTTGTGAGCCGTGGCACCTTAGGGC | |
| 6 month | 6804 | 4 | 133117976 | TGTTGTGAGCTTCCCTGTCCCACACAGGT | |
| 1003 | 3 month | 6846 | 9 | 97627491 | TCTGGAGGACCCCACATGGAGGAGTGG |
| 6 month | 6804 | 18 | 4240167 | ATTGCCAAGTCTGTTGTGAGCAGAAGAGAAA | |
| 6 month | 6625 | X | 41763674 | CTGTACTTACAGCAGGTGAGGTGGGG | |
| 1005 | 1st | 6804 | 12 | 11664603 | CTGTTGTGAGCCAAGACTGCGCCA |
| 1st | 6853 | 5 | 70816135 | GAGGACCTCATCTGAAGGAATGATAGA | |
| 1007 | 1 month | 6804 | 4 | 168956757 | TGTTGTGAGCTACAATGCTTTGGGGACCCAGA |
| 1009 | 1st | 6781 | 6 | 71299271 | AGTTTGGCTCGAACCCAGGAGG |
Draw refers to the time of blood collection after treatment was initiated. Most individuals exhibited more than one rearrangement per sample. Site is breakpoint location within MLL BCR (see Fig. 2) Translocation breakpoint highlights region of microhomology (bold). Partner location (breakpoint site) identified by BLAST using build 36.3 and refers to individual chromosome numbering in base pairs.
TABLE 5.
Rearrangements detected in peripheral blood of breast cancer patients undergoing treatment
| Patient | Draw | Site | Chromosome | Partner location (bp | MLL microhomology partner |
|---|---|---|---|---|---|
| 2002 | 1st | 6807 | 14 | 67141929 | GTCTGTTGTGAGCCCTTGAGCCCAGGAATTTGAAGTTATAG |
| 2002 | 1 month | 6771 | 1 | 70342304 | TATCTTCCCATGTTCTTAAATGTGTTCAGTTTCTTATA |
| 2002 | 1 month | 5735 | 9 | 139711966 | GGTTGATTATGTTTTTCTACAAGAATAAAAACTACTTTG |
| 2003 | 1 month | 6805 | 19 | 59007420 | TGTATTGCCAAGTCTGTTGTGAGCCACTGCACTTAGCCC |
| 2005 | 1st | 7023 | 11 | 117863937 | TGGGAAAAAATCTACTAAAAATACAAAAAATTAGCCAGGTGTGG |
| 2010 | 1 month | 6805 | 22 | 23673419 | TGCCAAGTCTGTTGTGAGCCACCGCGCCTGGCCTGGT |
Draw refers to the time of blood collection after treatment was initiated. Most individuals exhibited more than one rearrangement per sample. Site is breakpoint location within MLL BCR (see Figure 2) Translocation breakpoint highlights region of microhomology (bold). Partner location (breakpoint site) identified by BLAST using build 36.3 and refers to individual chromosome numbering in base pairs.
Figure 2.
Breakpoint regions within MLL BCR. (A) Site of MLL breakpoints from TK6 cells treated in vitro after either pro-apoptotic anti-CD95 stimuli or exposure to E2 or 4-OH-E2. (B) Blood samples analyzed from patients with primary diagnosis of lymphoma showing number of events and approximate locations of breakpoints for lymphoma (open bar) or breast cancer patients (closed bar). Notation below represents the 8.3kbp MLL BCR after Gu et al. (Gu et al., 1992). Green bar - Topoisomerase II binding site, Orange bar- each half of a 10 bp palindromic DNA sequence.
Site of MLL Cleavage
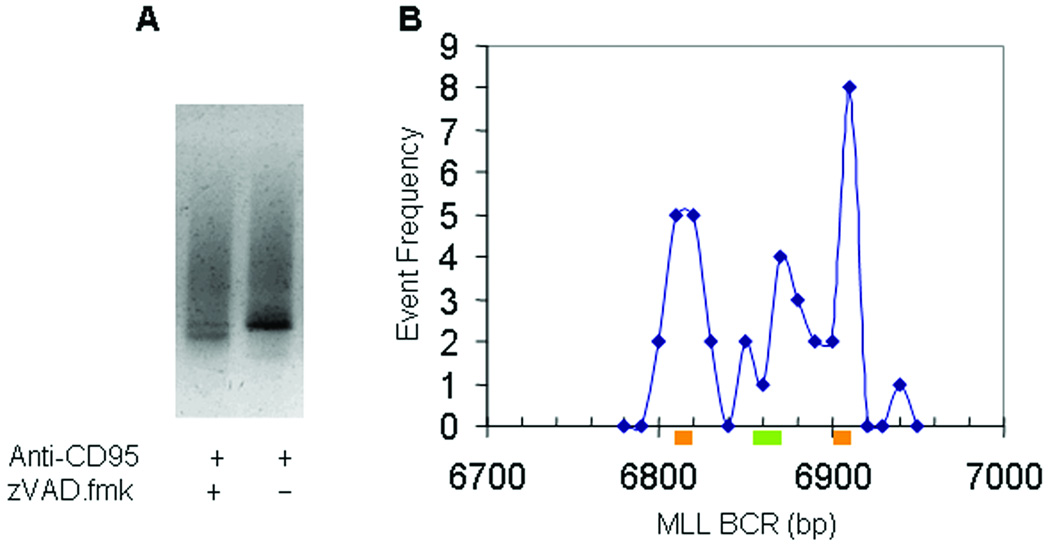
It has been reported by a number of groups that MLL is a specific target for DNA cleavage, with the most sensitive location being adjacent to the intron 11/exon 12 border (Stanulla et al., 1997a,b; Betti et al., 2003). This target is cleaved as a consequence of exposure to a variety of agents that have in common the ability to induce apoptosis. The actual cleavage itself may therefore potentially arise from nucleases related to the apoptotic program. To determine if the focus of MLL rearrangements seen in Fig. 2 coincided with DNA cleavage events, a ligation mediated PCR reaction was performed using an adapter oligonucleotide linker ligated to all accessible DNA breaks. Using MLL specific primers, PCR products were obtained indicating fragmentation of MLL close to the intron 11/exon 12 border, as shown before (Betti et al., 2005) (Fig. 3A). Such data is normally presented by use of Southern blotting techniques which provides relatively imprecise indications of break site location. To better locate the precise sites of cleavage, the hot spot as identified by Southern was used as a guide to excise LM-PCR product from a fresh DNA gel followed by Topo-TA cloning of the result (Fig. 3B). This allowed the precise identification of cleavage sites. As shown in Fig. 3B, the cleavage sites occupied two principle locations, the most 5' of which overlapped with the predominant site of translocations detected by iPCR (Fig. 2).
Figure 3.
Cleavage of MLL in TK6 cells detected by LM-PCR. (A) DNA gel after LM-PCR to detect MLL specific fragmentation after exposure to 0.5µg/ml anti-CD95 antibody. Evidence for increased fragmentation ∼300bp is observed that, after Southern blotting with a MLL specific probe (not shown), confirms MLL specificity. DNA from this region extracted from gel, cloned and sequenced to determine precise location of cleavage. (B) Results of cloning and sequencing DNA cleavage sites within the region demonstrated in Fig. 2 and plotted using the same MLL notation. Data plotted in 10bp bins. Shown to scale are sites of Palindromic DNA (Orange) and Topoisomerase II consensus binding site (Green).
Role of DNA Secondary Structure
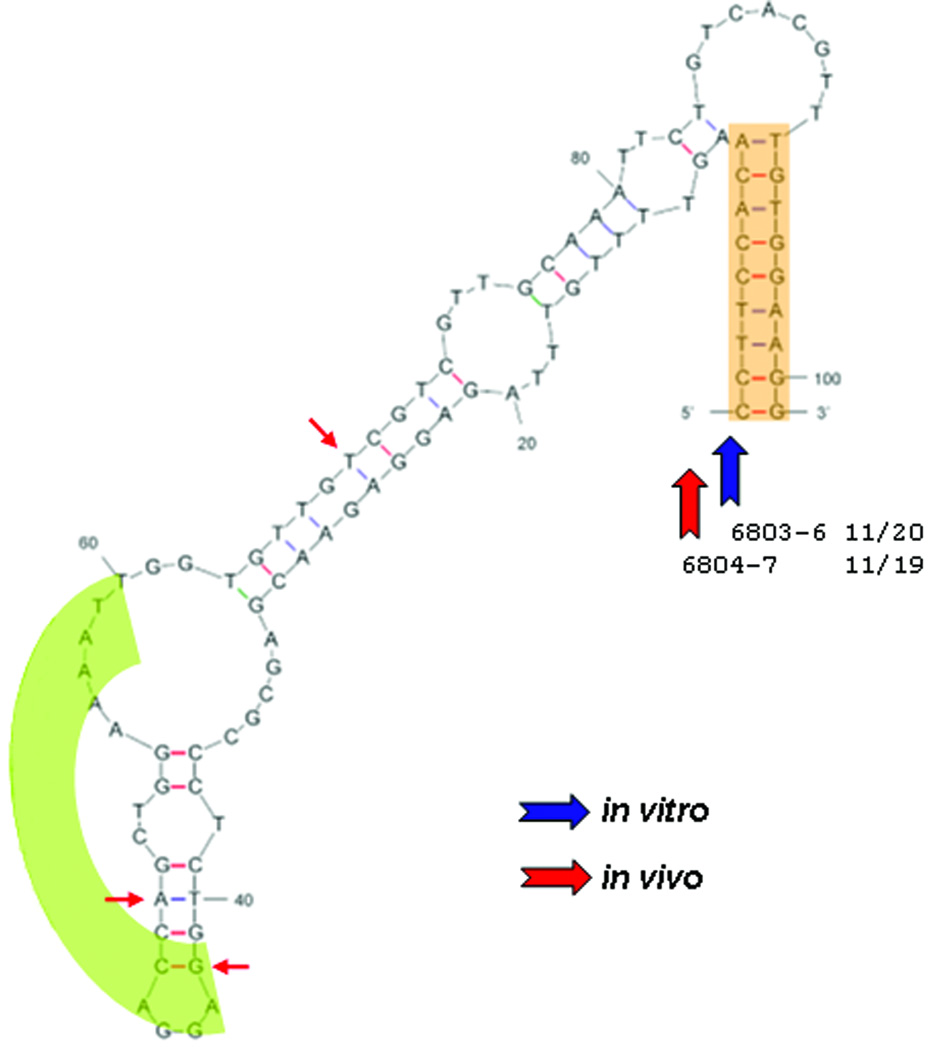
As shown in Fig. 2, MLL aberrations, generated both in vitro and in vivo, showed a restricted distribution within exon 12. Further analysis of the region surrounding the individual rearrangements showed the highest frequency of breakpoints co-localized with a 10 bp tract of DNA that matched a palindromic partner, approximately 101 bp distant and 3' to the site of rearrangement. Analysis using the mfold program of Zuker showed a structure similar to a stem loop structure that exhibits weak stability (ΔG = −11.87 kcal) (Zuker, 2003). This is in contrast to the breaks detected by LM-PCR where cleavage was observed to peak at both the 3' and 5' palindrome locations. Further analysis of the included region between palindromes showed a high affinity site of topoisomerase II binding at the geometric mid-point of the palindromic DNA, previously identified by others (Broeker et al., 1996). This feature, discussed later, has the potential to exert bending stress on the DNA molecule that is not accounted for in the DNA stability calculation above. These locations are presented in the derived orientation in Fig.4.
Figure 4.
MLL rearrangements mapped onto proposed DNA secondary structure. Shown is the result of a putative formation (ΔG of −11.87 kCal) of the breakpoint region highlighting the palindrome sequence (orange) and potential binding site for Topoisomerase II (green) using the mfold routine. In vitro data blue arrows – in vivo data in red. The majority of breakpoints for translocation for both sets of data are at the base of the palindrome as shown. Additional in vitro breakpoints are either more 5' (7/20) or 3' (2/20) to the structure shown. For the patient data, three are located in the secondary structure, 4/19 are 5' and 1/19 are 3' of the proposed secondary structure. In both data sets the predominant site of MLL breakpoint fusion is at the 5′ palindrome base.
Discussion
MLL rearrangements implicated in leukemia’s are found throughout the 8.3 kbp of the MLL BCR, covering exons 8–14. Others have noted that therapy related breakpoints, primarily those involving Topoisomerase II inhibitors, and those linked to IAL are preferentially found at the 3' end of the BCR. Key to an understanding of therapy related AML is the nature of the link between topoisomerase II inhibitors and development of the disease (Lovett et al., 2001; Whitmarsh et al., 2003; Libura et al., 2005). Despite the strong clinical association it has been difficult to unambiguously assign a direct cause (Topoisomerase II inhibitor linked DNA damage) to an effect (a MLL translocation at the break location). As discussed by a number of authors, both Topoisomerase II mediated damage and apoptotic nuclease attack can be detected experimentally in cells at locations that do not coincide with the MLL breakpoints identified in clinical disease (Mirault et al., 2006; Scharf et al., 2007). In addition, neither do the sites of topoisomerase II induced cleavage after etoposide exposure always correlate with the local canonical site of topoisomerase II binding (Mirault et al., 2006). As the role of topoisomerase II inhibitors in tAML is unambiguous, it remains to be determined how exposure to such drugs is translated into MLL translocations linked to the disease.
The study reported here has focused on a specific region of MLL at the intron 11and exon 12 border that is both a DNAse hypersensitivity target and falls within a region showing increased frequency of MLL fusions within tAML and IAL (Broeker et al., 1996; Stanulla et al., 1997a,b; Betti et al., 2001; Betti et al., 2003). First, in the in vivo study, blood samples were examined from patients treated with Topoisomerase II inhibitors as part of their therapy for either breast cancer or lymphoma (Table 1&Table 2). Second, cells in culture were challenged with either estrogen or its 4-OH derivative or anti-CD95 antibody with or without addition of the broad range caspase inhibitor, zVAD.fmk. The former follows an earlier epidemiological report linking intake of contraceptive formulations with infant acute leukemia (Pombo-de-Oliveira et al., 2006). Though the treatments and target cells for the in vitro and in vivo studies were quite different, a number of findings in common were obtained. Firstly, except for two translocations in the control group from the in vitro study, MLL translocations were restricted to treatment groups only. The presence of translocations in control samples in vitro may simply reflect the fact that all cell samples have the potential to access apoptosis, a process known to induce MLL rearrangements (Betti et al., 2001; Betti et al., 2003). Of note, in the in vitro study, MLL translocations were found predominately in the anti-CD95 antibody treatment group (8 events), 6–16 days after treatment and were lacking in the anti-CD95 antibody + zVAD.fmk treatment group (1 event) (Table 3). This protracted delay in MLL translocation detection, suppressed by the pan caspase inhibitor, zVAD.fmk, provides some support for the contention that transient apoptotic triggers may be overcome and cells survive with apoptotic nuclease mediated rearrangements (Betti et al., 2001; Betti et al., 2003). Also, the locations of the breakpoints within identifiable translocations were tightly clustered at 6803–6807 bp in both the in vitro (11/20) and in vivo (11/19) data sets. This hot spot for MLL rearrangements is the same location identified by other groups using similar techniques in the analysis of break aberrations induced using in vitro data only (Mirault et al., 2006; Scharf et al., 2007).
In order to track the induction of MLL aberrations from the point at which the MLL gene is cleaved, ligation mediated PCR was executed on a population of TK6 cells that had been exposed to the apoptotic trigger, anti-CD95 antibody for a 4h period. In this “shotgun” approach, all LM-PCR products identified were used so as to reduce selection bias. After isolation and sequencing, the site of each break was plotted with reference to the MLL gene. With the semi-nested primers located in intron 12, the assay can detect the presence of DNA breaks within exon 12 extending through the adjacent intron border(s). As can be seen from Fig. 3, the distribution of breaks followed a bi-modal distribution with the most 5' peak mapping to the same location as the MLL rearrangements. No similar congruence between MLL rearrangements and apoptotic cleavage sites were observed for the 3' group of cut sites – no rearrangements were noted here. In order to analyze the event distributions in more detail, both cut sites and rearrangements were mapped with reference to DNA secondary structure and topoisomerase II consensus binding sites. This was achieved using the “mfold” structural analysis program (Zuker 2003). As seen in Fig. 4, the sites of DNA cleavage map to the base of a large potential stem loop structure with a consensus binding site for Topoisomerase II at the geometric mid-point between palindromes. In this conformation, the DNA cleavage sites are aligned at the base of the stem, formed by a 10bp palindrome, with the MLL translocations located at the 5' but not 3' side (Spitzner & Muller, 1988; Broeker et al., 1996). The lack of any MLL translocation junctions at the 3' side was not likely due to any technical difficulty in their measurement at this location as rearrangements were observed even further 3' (distal to the primers used) to this site in both the in vitro and patient data sets (Fig. 3).
The presence of a topoisomerase II binding site within this region of both targeted DNA MLL cleavage and rearrangement raises the question of the role of the enzyme in tAML. As noted before, a direct role for topoisomerase II in mediating DNA breaks linked to tAML is lacking. However it is possible that dietary consumption of topoisomerase II inhibitors may contribute to IAL linked to MLL rearrangements, though the data are controversial (Specter et al., 2005, Alexander et al., 2001). Thus it is possible that the role of topoisomerase II is indirect, rather than direct (Betti et al., 2005). Such an indirect effect may be related to the physical association of Topoisomerase II binding to DNA that interferes with normal DNA processing. It has been demonstrated that on binding duplex DNA, Topoisomerase II introduces a bend at the binding site that would, in this case, potentially bring the palindromic sequences of the stem loop closer together (Vologodskii et al., 2001; Dong & Berger, 2007). Such an effect may promote the transient formation of a stem-loop structure as described here, which otherwise would be marginally stable (ΔG = −11.87 kcal), that could occur with or without drug-induced stabilization of the enzyme. However drug stabilized topoisomerase II is likely to exert a bending stress over a longer period of time and thus promote longer palindromic association. Therefore the act of topoisomerase II binding may introduce a transient stem-loop organization that, once formed, is permissive for DNA nuclease attack (Elborough & West, 1990; Germe & Hyrien, 2005; Cote & Lewis, 2008). Viewed in this way, the role of topoisomerase II and its inhibitors within tAML is part of a process whereby the DNA structure facilitates rearrangements, rather than directly playing a part in their creation.
An additional possibility to explain such a biased distribution of breakpoints is the involvement of either a transcription or DNA duplication event that collides with the transient stem loop structure and contributes to a rearrangement. Scharf et al. have described an internal gene promoter site within the intron 11 adjacent (∼75 bp) to the exon 12 region discussed here (Scharf et al., 2007). The site was shown by the authors to be active indicating that this region may be a site for transcriptional initiation. Activation of the transcriptional complex in this region with the subsequent DNA unwinding by helicases would generate a protein complex which collides with a partially or fully formed stem loop structure in exon 12. Subsequent attempts at resolution of the stalled transcriptional complex may lead to nicking of the stem-base and the potential for translocation or further nuclease processing. Scharf et al. also detected rearrangements at the location shown here to be a Topoisomerase II binding site, though we found few such rearrangements in this study, their direct formation by a failed Topoisomerase II cleavage/ligation step is clearly possible (Scharf et al., 2007).
A finding from these data was possible support for aberrations that persist through time, either in vitro or in vivo, as has been found in other studies (Libura et al., 2005). In the in vitro experiment using anti-CD95 antibody, aberrations were seen 6–16 days after treatment was complete. In addition, the patient data from the lymphoma study group, translocations were found in 2/9 patients, 3 months after cessation of treatment. It is not implied that the aberrations detected are of themselves leukemogenic, but that they may represent a class of rearrangements that share a common process of formation. The process of selection of individual aberrant cells for clonal expansion clearly requires additional, rare, events before leukemia develops. An additional issue concerning the rearrangements detected in exon 12, also brought out by other studies in this field, is that the locations discussed here lie outside the mainstream sites of MLL rearrangements (Mirault et al., 2006; Scharf et al., 2007). Specifically, the majority of MLL aberrations linked to IAL or tAML are found distributed within the most 3' one third of the MLL BCR, not within exon 12 itself (Broeker et al., 1996; Cimino et al., 1997). Once formed, however, such DNA breaks may be susceptible to exonucleolytic digestion secondary to attempts at repair. In these cases repair associated enzymes such as MRE-11 and Artemis may provide such 5'>3' exonuclease function that initiate DNA digestion (Germe & Hyrien, 2005; Yannone et al., 2008). The exonuclease processing by such systems may remove multiple base pairs of DNA prior to rearrangement or repair. Also, the hot spot for rearrangements described here is limited in extent to the length of DNA between the Sau3AI restriction sites used to generate the iPCR template – approximately 750 bp in this case. Other sites of palindomic DNA adjacent to toposomerase II binding sites may trigger similar rearrangements at other sites within the MLL BCR. A screen for such sites shows many potential stem-loop structures throughout the MLL BCR that is itself the location for multiple Topoisomerase II binding sites (Broeker et al., 1996). We suggest a model for a class of MLL rearrangements whereby Topoisomerase II facilitates formation of a secondary structure that is permissive for site-specific DNA strand breaks that are efficiently processed into translocations.
Acknowledgments
Supported by: The Work was supported by a NIH Grant CA10504
REFERENCES
- Alexander FE, Patheal SL, Biondi A, Brandalise S, Cabrera ME, Chan LC, Chen Z, Cimino G, Cordoba JC, Gu LJ, Hussein H, Ishii E, Kamel AM, Labra S, Magalhães IQ, Mizutani S, Petridou E, de Oliveira MP, Yuen P, Wiemels JL, Greaves MF. Transplacental chemical exposure and risk of infant leukemia with MLL gene fusion. Cancer Research. 2001;61:2542–2546. [PubMed] [Google Scholar]
- Basecke J, Karim K, Podleschny M, Becker A, Glass B, Trumper L, Griesinger F. MLL rearrangements emerge during spontaneous apoptosis of clinical blood samples. Leukemia. 2006;20:1193–1194. doi: 10.1038/sj.leu.2404211. [DOI] [PubMed] [Google Scholar]
- Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic triggers initiatetranslocations within the MLL gene involving the nonhomologous end joining repair system. Cancer Research. 2001;61:4550–4555. [PubMed] [Google Scholar]
- Betti CJ, Villalobos MJ, Diaz MO, Vaughan AT. Apoptotic stimuli initiate MLL-AF9 translocations that are transcribed in cells capable of division. Cancer Research. 2003;63:1377–1381. [PubMed] [Google Scholar]
- Betti CJ, Villalobos MJ, Jiang Q, Cline E, Diaz MO, Loredo G, et al. Cleavage of the MLL gene by activators of apoptosis is independent of topoisomerase II activity. Leukemia. 2005;19:2289–2295. doi: 10.1038/sj.leu.2403966. [DOI] [PubMed] [Google Scholar]
- Broeker PL, Super HG, Thirman MJ, Pomykala H, Yonebayashi Y, Tanabe S, Zeleznik-Le N, Rowley JD. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996;87:912–1922. [PubMed] [Google Scholar]
- Cimino G, Rapanotti MC, Biondi A, Elia L, Lo Coco F, Price C, Rossi V, Rivolta A, Canaani E, Croce CM, Mandelli F, Greaves M. Infant acute leukemias show the same biased distribution of ALL1 gene breaks as topoisomerase II related secondary acute leukemias. Cancer Research. 1997;57:2879–2883. [PubMed] [Google Scholar]
- Coté AG, Lewis SM. Mus81-dependentdouble-strand DNA breaks at in vivogenerated cruciform structures in S. cerevisiae. Molecular Cell. 2008;31:800–812. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- Elborough KM, West SC. Resolution of synthetic Holliday junctions in DNA by an endonuclease activity from calf thymus. EMBO Journal. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology ofchromosomal translocations. DNA Repair (Amst) 2006;8:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Reports. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce CM, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- Lovett BD, Lo Nigro L, Rappaport EF, Blair IA, Osheroff N, Zheng N, Megonigal MD, Williams WR, Nowell PC, Felix CA. Near precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9802–9807. doi: 10.1073/pnas.171309898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R. Etoposide-treatment and MLL rearrangements. European Journal of Haematology. 2008;81:481–482. doi: 10.1111/j.1600-0609.2008.01139.x. [DOI] [PubMed] [Google Scholar]
- Martinelli G, Testoni N, Zinzani PL, Biondi A, Cimino G, Tura S. Therapy-related acute leukemia associated with involvement of 11q23 after high grade non-Hodgkin lymphoma. Haematologica. 1998;83:283–284. [PubMed] [Google Scholar]
- Mirault ME, Boucher P, Tremblay A. Nucleotide-resolution mapping oftopoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot. American Journal Human Genetics. 2006;79:779–791. doi: 10.1086/507791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo-de-Oliveira MS, Koifman S Brazilian Collaborative Study Group of Infant Acute Leukemia. Infant acute leukemia and maternal exposures during pregnancy. Cancer Epidemiology Biomarkers & Prevention. 2006;15:2336–2341. doi: 10.1158/1055-9965.EPI-06-0031. [DOI] [PubMed] [Google Scholar]
- Scharf S, Zech J, Bursen A, Schraets D, Oliver PL, Kliem S, Pfitzner E, Gillert E, Dingermann T, Marschalek R. Transcription linked to recombination: a gene-internal promoter coincides with the recombination hot spot II of the human MLL gene. Oncogene. 2007;26:1361–1371. doi: 10.1038/sj.onc.1209948. [DOI] [PubMed] [Google Scholar]
- Spitzner JR, Muller MT. A consensus sequence for cleavage byvertebrate DNA topoisomerase II. Nucleic Acids Research. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanulla M, Wang J, Chervinsky DS, Thandla S, Aplan PD. DNA cleavage within the MLL breakpoint cluster region is a specific event which occurs as part of higher-order chromatin fragmentation during the initial stages of apoptosis. Molecular Cell Biology. 1997a;17:4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanulla M, Wang J, Chervinsky DS, Aplan PD. Topoisomerase II inhibitors induce DNA double-strand breaks at a specific site within the AML1 locus. Leukemia. 1997b;11:490–496. doi: 10.1038/sj.leu.2400632. [DOI] [PubMed] [Google Scholar]
- Super HJ, McCabe NR, Thirman MJ, Larson RA, Le Beau MM. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA topoisomerase II. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh RJ, Saginario C, Zhuo Y, Hilgenfeld E, Rappaport EF, Megonigal MD, Carroll M, Liu M, Osheroff N, Cheung NK, Slater DJ, Ried T, Knutsen T, Blair IA, Felix CA. Reciprocal DNA topoisomerase II cleavage events at 5'-TATTA-3' sequences in MLL and AF-9 create homologous single-stranded overhangs that anneal to form der (11) and der (9) genomic breakpoint junctions in treatment-related AML without further processing. Oncogene. 2003;22:8448–8459. doi: 10.1038/sj.onc.1207052. [DOI] [PubMed] [Google Scholar]
- Yannone SM, Khan IS, Zhou RZ, Zhou T, Valerie K, Povirk LF. Coordinate 5' and 3' endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Research. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]