Abstract
A number of different processes that impact on telomere length dynamics have been identified but factors that affect the turnover of repeats located proximally within the telomeric DNA are poorly defined. We have identified a particular repeat type (CTAGGG) that is associated with an extraordinarily high mutation rate (20% per gamete) in the male germline. The mutation rate is affected by the length and sequence homogeneity of the (CTAGGG)n array. This level of instability was not seen with other sequence-variant repeats, including the TCAGGG repeat type that has the same composition. Telomeres carrying a (CTAGGG)n array are also highly unstable in somatic cells with the mutation process resulting in small gains or losses of repeats that also occasionally result in the deletion of the whole (CTAGGG)n array. These sequences are prone to quadruplex formation in vitro but adopt a different topology from (TTAGGG)n (see accompanying article). Interestingly, short (CTAGGG)2 oligonucleotides induce a DNA damage response (γH2AX foci) as efficiently as (TTAGGG)2 oligos in normal fibroblast cells, suggesting they recruit POT1 from the telomere. Moreover, in vitro assays show that (CTAGGG)n repeats bind POT1 more efficiently than (TTAGGG)n or (TCAGGG)n. We estimate that 7% of human telomeres contain (CTAGGG)n repeats and when present, they create additional problems that probably arise during telomere replication.
INTRODUCTION
All vertebrate telomere repeat arrays are composed of tandem arrays of TTAGGG repeats and when active, the enzyme telomerase can add novel repeats onto the terminus of each telomere so maintaining its length. While the regulation of telomere length and control of telomerase activity is being studied extensively, the maintenance and turnover of repeats located more internally in the arrays are rarely considered. Telomeric DNA sequences are tandem repeat arrays that share some properties with other microsatellite or simple tandem repeat (STRs) sequences (e.g. di-, tri-, tetra- and penta-nucleotide repeat arrays) within the human genome. STRs are distributed throughout the human genome and some show length variation between unrelated individuals and consequently they have formed the basis of comprehensive genetic maps of the genome (1,2). The germline mutation dynamics of STRs (not associated with human disease) have been studied and, although the average mutation rate for such loci is ∼2 × 10−3 per meiosis, the mutation rates vary between loci (3–5). Factors that are known to affect mutation rates are the length and sequence homogeneity of the array (4). However, there is a complex relationship between the frequency and size of mutations resulting in gains or losses of repeats that varies between repeat types (6). The mutation mechanism underlying such microsatellites is generally accepted as being intra-allelic and may arise from slippage during replication (5).
A subset of trinucleotide repeats (TNRs) are associated with human neurological, neurodegenerative and neuromuscular disorders and as such they have been studied extensively. The mutation dynamics of these TNRs is particularly interesting because once the array is above a certain threshold length (that differs between loci) it becomes highly unstable in the male or female germline and usually with a bias towards large expansions (7,8). This expansion bias underlies the phenomenon known as anticipation within families. Interestingly, some of these disease-associated TNRs also show somatic instability in some but not all tissues, including mitotically inert tissues such as parts of the brain. It is therefore likely that the underlying mutation mechanism is dependent on replication and other factors such as the activity of different DNA repair pathways (e.g. mismatch repair) that have been implicated in the instability of some disease associated TNRs (8–11).
In humans, the majority of telomeres contain sequence-variant repeats interspersed with the consensus TTAGGG repeat at the proximal end of the array (12–15). It is assumed that these sequence-variant repeats arise randomly within the telomere repeat array but persist at the proximal end of the telomere repeat array because these regions are rarely influenced by telomerase in the germline. It is unknown how these sequence-variant repeats affect telomere function though many retain the G3 motif. However, it is thought that the presence of a large number of sequence variant repeats at the start of the telomere is likely to reduce the contribution these regions make to some aspects of telomere function. By using allele specific primers in the telomere-adjacent DNA to map the interspersion of TTAGGG and sequence variant repeats (by Telomere Variant Repeat mapping by PCR, TVR-PCR,) it is possible to compare telomeres (alleles) between individuals (13). This has shown that telomere repeat arrays are highly variable with a high mutation rate and that the underlying mutation mechanism is dominated by intra-allelic events in the normal germline. Furthermore, telomere repeat arrays are often mutated in colon carcinomas compared with the surrounding normal colon tissue and as this instability is even higher in colon cancers that are known to be defective in DNA mismatch repair (MMR), it suggests that MMR is required for telomere stability (16). Despite this, little is known about the mutation dynamics of human telomeres or the effect of sequence-variant telomere repeats. Here we have identified one particular repeat-type (CTAGGG) that, when present as a short contiguous array within the Xp/Yp telomere, causes an extraordinarily high level of localized telomere instability in vivo. We demonstrate that these repeats are prone to quadruplex formation in vitro and adopt a different folding topology from the classical TTAGGG repeats. We also show that (CTAGGG)2 and (TCAGGG)2 oligonucleotides induced a DNA damage response as efficiently as (TTAGGG)2 in normal fibroblast cells (17) and that (CTAGGG)n repeats bind to POT1 (18) more efficiently than (TTAGGG)n or (TCAGGG)n repeats in vitro. Finally, we discuss how the instability of (CTAGGG)n repeats may arise.
MATERIALS AND METHODS
DNA samples
Human DNAs derived from lymphoblastoid cell lines comprising the Centre d’Etude du Polymorphism Humain (CEPH, Paris, France) panel of DNAs from families with Northern or Western European ancestry were used for telomere mutation analysis. The Russian family DNAs, extracted from venous blood samples, were kindly donated by Prof. Y.E. Dubrova (19). DNA samples from normal colon and colon carcinomas had been collected previously from patients with sporadic colorectal cancer at the Leicester Royal Infirmary (16).
SNP analysis in telomere adjacent DNA
Assays to determine the genotypes at SNPs in the telomere-adjacent DNA at Xp/Yp and 12q were conducted as described previously (13,15). Telomere analysis was conducted on heterozygous or homozygous DNA samples from the Xp/Yp −30 base, using the allele specific primers TS-30A or TS-30T (13) and from the 12q-197 base, using the allele-specific primers 12q-197A or 12q-197G (15).
Identification of the CTAGGG repeats
To identify the sequence of the repeats associated with germline instability in the CEPH family 1362, TVR-PCR was conducted at the Xp/Yp telomere. Radioactively labelled TVR-PCR fragments were excised from dried polyacrylamide gels using a scalpel. The polyacrylamide gel slices were rehydrated and DNA eluted in 30 μl elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate tetrahydrate, 1 mM EDTA (pH 8.0), 0.1% SDS) at 37°C for 5 h. The eluate was centrifuged, transferred to a fresh tube and ethanol precipitated. The precipitated DNA was dissolved in 10 μl TE (10 mM Tris, 1 mM EDTA) and 2 μl taken for reamplification in 40 μl PCR reaction using a primer nested in the telomere-adjacent DNA and the TAG primer. The reamplified product was further purified by electroelution from an agarose gel or by excising the DNA band from the agarose gel and using QIAquickTM gel extraction kits (Qiagen). Once an amplicon of a single length was obtained, it was sequenced from the TAG primer using the ABI Big DyeTM Terminator Cycle Sequencing Ready Reaction Kit. The unknown repeats in 1362 family members were identified as CTAGGG repeats and primers (TAG-TelCTA and TAG-TelCTA2, below) designed to amplify them specifically.
Telomere variant repeat mapping by PCR (TVR-PCR)
The distribution of TTAGGG and sequence variant repeats at the start of Xp/Yp or 12q telomeres was determined using allele specific primers (TS-30A/T or 12q-197A/G) in conjunction with one of the following TVR-PCR primers: TAG-TelW, TAG-TelX, TAG-TelY and TAG-TelJ that detect the TTAGGG, TGAGGG, TCAGGG or TTGGGG repeats types, respectively (13,14). In addition the primer TAG-TelCTA2 5′-TCATGCGTCCATGGTCCGGACCCTTACCCTTRCCCTARCCCTAG -3′ (and occasionally primer TAG-TelCTA 5′-TCATGCGTCCATGGTCCGGACCCTTACCCTTACCCTNACCCTAG-3′ that preferentially detects CTAGGG repeats located next to TTAGGG repeats) was used in TVR-PCR to detect CTAGGG repeat types within the telomere repeat array. Resolution of the TVR-PCR amplified products on acrylamide gels was described previously with detection of the 32P labelled products using a phosphorimager (Typhoon 9400, G.E. Healthcare).
Detection and analysis of the polymorphic 16p/16q telomere
To determine the presence or absence of the polymorphic telomere on chromosome 16, genomic DNA was amplified with the primers Nitu14eA and TelC using 96°C for 20 s, 68°C for 30 s and 68°C for 5 min for 20 cycles. The amplified products were resolved on 1.2% agarose gels, blotted onto a nylon membrane and hybridized in phosphate-SDS solution to a radioactively labelled probe from the telomere-adjacent DNA sequence. TVR-PCR at the polymorphic 16p/16q telomere was carried out using the telomere-adjacent primer Nitu14eD in conjunction with the TVR-PCR primers described above (14).
Single-molecule STELA
To determine the somatic mutation frequency in DNA extracted from normal colon and colon carcinomas samples the single telomere length analysis (STELA) method was used to amplify full length telomeres from very small aliquots of DNA such that each reaction contained only a single STELA product derived from one telomere molecule (single molecule-STELA or sm-STELA) at either the Xp/Yp or 12q telomere. Subsequently the interspersion of TTAGGG and sequence-variant repeats was determined for approximately 100 different sm-STELA products by TVR-PCR and the somatic mutation frequency determined (20).
Circular dichroism measurements
Circular dichroism (CD) spectra were recorded on a JASCO-810 spectropolarimeter using a 1 cm path length quartz cuvette in a reaction volume of 580 µl, as previously described (21). Oligonucleotides were prepared as a 4 µM solution in 10 mM lithium cacodylate pH 7.2, 100 mM NaCl or KCl buffer and annealed by heating to 90°C for 2 min, followed by slow cooling to 20°C.
UV melting assays for G-quadruplexes
The following oligonucleotides were used for UV melting analysis: CTA-22mer: AGGGCTAGGGCTAGGGCTAGGG, TCA-22mer: AGGGTCAGGGTCAGGGTCAGGG and TTA-22mer: AGGGTTAGGGTTAGGGTTAGGG. Oligonucleotides were synthesized by Eurogentec (Seraing, Belgium) at the 40 or 200 nmol scale and used without further purification. Concentrations were estimated using extinction coefficients provided by the manufacturer. Melting assays were performed on a Uvikon 840 spectrophotometer in a 10 mM lithium cacodylate pH 7.2 buffer (supplemented with either 0.1 M KCl or NaCl, hereafter referred to as potassium and sodium conditions, respectively), as previously described (22). Quadruplex formation may be evidenced by an inverted transition at 295 nm (23,24). Melting experiments were typically performed at a concentration of 4 μM per strand. All transitions were reversible, as shown by superimposable heating and cooling profiles at a fixed rate of 0.2°C/min.
Exposure of MRC5 cells to single-stranded oligonucleotides
The MRC5 human lung fibroblast cell line was obtained from ATCC and was grown in MEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine and non-essential amino acids. MRC5 cells at PD 25 were seeded at 105 cells/ml (200 µl per well) in 8-well culture slide chambers (BD Biosciences) and treated for 48 h with 40 µM of the indicated 12-mer oligonucleotides: TTAGGG (5′-TTAGGGTTAGGG-3′), CTAGGG (5′-CTAGGGCTAGGG-3′), TCAGGG (5′-TCAGGGTCAGGG-3′), CCCTAA (5′-CCCTAACCCTAA-3′), CCCTAG (5′-CCCTAGCCCTAG-3′), CCCTGA (5′-CCCTGACCCTGA-3′) and random (5′-NNNNNNNNNNNN-3′).
Detection of γH2AX by immunofluorescence
After oligonucleotide treatment, the MRC5 cells were washed with 1X PBS (pH 7.2), fixed with 4% paraformaldehyde, and permeabilized with 20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose and 0.5% v/v Triton X-100 for 15 min at room temperature. Cells were then washed twice with PBS (pH 7.2). Cells were blocked with 10% goat serum in 1× PBS for 1 h and incubated with anti-phosphorylated γ-H2AX antibody (mouse, Upstate Biotechnology) at 1/2000 in blocking buffer for 1 h at 37°C. After washing with 1× PBS, cells were incubated for 30 min with Alexa fluor 568 goat anti-mouse IgG (Molecular probes) at 1/3000 in blocking buffer, then washed with 1× PBS. The nuclear DNA was stained with 0.1 µg/mL DAPI in PBS (pH 7.2) for 4 min. Cells were mounted in Shandon Immu-Mount medium (Thermo Scientific). Samples were observed with a DMR Leica microscope and images were captured with a Cool Snap HQ camera (Roper Scientific) controlled by Metamorph software (Roper Scientific). For the quantification of the DNA damage signal, >200 nuclei were analysed and results represent the mean ± SD of four independent experiments, except as indicated.
TRF2 and POT1 binding assays
Purified recombinant hPOT1 and TRF2 prepared from baculovirus expression were a generous gift from Dr D. Gomez (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France). An electrophoretic mobility shift assay using hPOT1 was performed on the consensus TTA-22mer telomeric sequence (5′-AGGGTTAGGGTTAGGGTTAGGG-3′), on the CTA-22mer variant-repeat sequence (5′-AGGGCTAGGGCTAGGGCTAGGG-3′) or on the TCA-22mer variant-repeat sequence (5′-AGGGTCAGGGTCAGGGTCAGGG-3′). Oligonucleotides were labelled at the 5′ end with [γ-32P]-ATP using T4 Polynucleotide Kinase (New England BioLabs®). The mobility shift assay was performed in 10 µl of the following solution: 50 mM HEPES, pH 7.9, 100 mM NaCl, 0.1 mM EDTA, 4% w/v sucrose, 2% v/v glycerol, 0.1 mg/ml BSA, 0.02% w/v bromophenol blue, 20 nM labelled oligonucleotides and different concentrations of hPOT1 (6, 20, 40 and 60 nM). The reaction mixture was incubated at room temperature for 30 min. Each individual mixture was separated immediately by electrophoresis on 1% agarose gels in 0.5× Tris–Borate–EDTA buffer. The gels were run at 80 V for 45 min, dried on Whatman DE81 paper and visualized by a phosporimager (Typhoon 9210, Amersham). Analysis of the data was carried out by ImageQuant software (Amersham) and results were expressed as the fraction of DNA bound to hPOT1. Values corresponded to the mean value of three independent experiments ± SD.
For TRF2 assays, the G-strand consensus telomeric (5′-ACATGTTAGGGTTAGGGTTAGGGTTAG-3′) or CTAGGG variant-repeat (5′-ACATGCTAGGGCTAGGGCTAGGGCTAG-3′) or TCAGGG variant-repeat (5′-ACATGTCAGGGTCAGGGTCAGGGTCAG-3′) and their respective complemetary C-strands were used to form a double-stranded oligonucleotide. G-strands were labelled at the 5′ end with [γ-32P]-ATP using T4 Polynucleotide kinase (New England BioLabs®). Labelled G-strand and unlabelled C-strand, 20 nM of each were assembled in 50 mM HEPES, pH 7.9, 100 mM NaCl, 0.1 mM EDTA, 4% w/v sucrose, 2% v/v glycerol, 0.1 mg/ml BSA, 0.02% w/v bromophenol blue, heated at 50°C for 10 min and cooled at room temperature for 15 min. Different concentrations of TRF2 (30, 60, 100 and 200 nM) were added to the double-stranded substrate and further incubated for 30 min at room temperature. Gel electrophoresis and analysis was performed as for hPOT1 and corresponded to the mean ± SEM of two independent experiments.
RESULTS
Analysis of the Xp/Yp telomere repeat arrays in the CEPH family panel identified two families that showed clustering of germline mutations within the proximal portion of the telomeres containing an interspersion of TTAGGG and sequence-variant repeats. Sequence analysis of the progenitor telomeres revealed that they both contain (CTAGGG)n repeat sequences. The effect of individual sequence-variant telomere repeats on human telomere repeat arrays has not been characterized in detail and therefore we have sought to determine whether the presence of CTAGGG repeats within the array affects telomere stability in vivo.
Prevalence of CTAGGG repeats within human telomeres and distribution within alleles
In order to detect CTAGGG repeats within individual telomere repeat arrays, two PCR primers were designed to identify all CTAGGG repeats and to identify CTAGGG repeats adjacent to TTAGGG repeat types (TAG-TelCTA2 and TAG-TelCTA, respectively). The Xp/Yp telomeres were screened for the presence of CTAGGG repeats in a population of northern and western European descent (CEPH parental DNAs) and in a population from Russia (19) (Figure 1). Among the 1068 telomeres screened 6.6% contained five or more CTAGGG repeats (17/160 or 10.6% in CEPH panel; 53/908 or 6% in Russian population).
Figure 1.
Comparison of the Xp/Yp telomeres containing CTAGGG repeats. The distribution patterns of TTAGGG (T), TGAGGG (G), TCAGGG (C), TTGGGG (J) CTAGGG (O) and null (N) repeats are shown from the first repeat in the array (on the left). These telomere maps were obtained from individuals in the CEPH family panel (identifying numbers in black) and from a Russian population (identifying numbers in red). The telomere maps associated with haplotype A, B or C across the SNPs in the telomere adjacent DNA have been grouped separately and further subdivided by similarity between telomere maps of individual alleles. Individuals in which the telomere maps that were determined from diploid codes (because the individual is homozygous at SNPs in telomere adjacent sequence) are shown in italics. The individuals that share telomere maps that are identical by descent (because the individuals are related) are shown in bold. The telomere maps in individuals 13292.01 and 13293.01 contained CTAGGG repeats further into the array but it was not possible to generate a detailed map beyond the 110th repeat because of poor size resolution. Some of the CTAGGG containing telomeres were transmitted to offspring within families. The sex of the transmitting parent is shown (M, male; F, female) with the number of offspring that inherited the CTAGGG containing allele. The telomeres that mutated upon germline transmission are identified with an asterisk.
We have shown previously that there is a high density of SNPs in strong linkage disequilibrium adjacent to the Xp/Yp telomere. Analysis of four SNPs (located 30, 176, 427 and 540-bp from the start of the Xp/Yp telomere repeat array) can be used to determine the haplotype status across the SNPs thus allowing identification of individuals that are heterozygous for the two common haplotypes (A and B) in the telomere adjacent DNA (13,15). Haplotype analysis was conducted in individuals that contain CTAGGG-repeats within their Xp/Yp telomeres. Then the interspersion pattern of TTAGGG with sequence-variant telomere repeats (including CTAGGG) was determined for single telomere alleles by TVR-PCR. In this method an allele specific primer, at the SNP 30 bp from the start of the Xp/Yp telomere repeat array, was used in conjunction with a primer that anneals to the TTAGGG or to one of the sequence variant repeats. The amplicons from the TVR-PCRs were size separated in a polyacrylamide gel so allowing the order of the repeats in the ladder to be determined and converted to a telomere code (Figure 2). In individuals that were homozygous for haplotypes in the telomere adjacent DNA, telomere codes were deduced from diploid telomere interspersion patterns and where possible using offspring to verify them.
Figure 2.
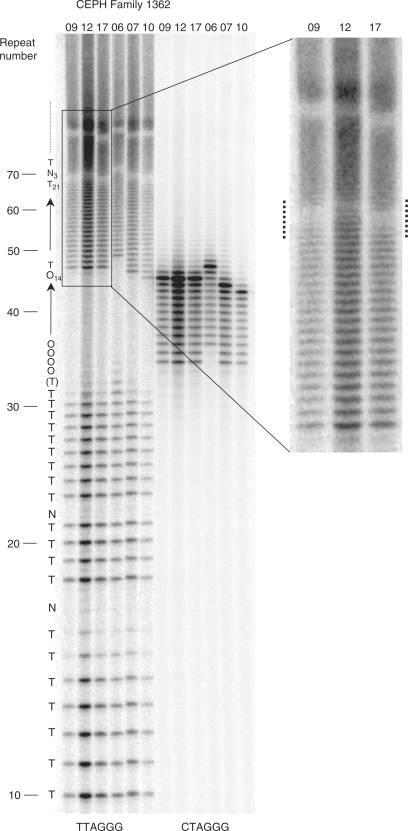
Identification of germline telomere mutations in CEPH family 1362. TVR-PCR was used to compare telomere maps between the father 1362.01 and the six children that inherited his Xp/Yp CTAGGG containing telomere. Haplotype A associated allele specific telomere maps are shown for 1362.09 (daughter), 1362.12 (daughter), 1362.17 (son), 1362.06 (daughter), 1362.07 (daughter) and 1362.10 (daughter) who are all heterozygous for different haplotypes in the telomere adjacent sequence. The daughter 1362.09 inherited a non-mutant copy of the (CTAGGG)n containing allele from her father but the other five children all show mutations in the CTAGGG or adjacent repeats. The insert on the right shows and enlarged image of part of the TVR-PCR gel to highlight the region of the telomeres that show mutations in 1362.12 and 17 (flanked by dotted lines, also see Figure 3a). The letters on the left side of the image show the telomere code derived from the TVR-PCR of individual 1362.09.
In the majority of telomeres, the CTAGGG repeats were confined to the first 1 kb of telomere sequence but a few alleles e.g. haplotype A associated alleles in individuals 13292.01 (Figure 1) and 1377.02 (not shown) contained CTAGGG repeats further into the array but the distribution could not be mapped accurately because of limitations in acrylamide gel resolution. The CTAGGG containing Xp/Yp telomeres that were mapped, were grouped according to the haplotype in the telomere-adjacent sequence and then subdivided by visual inspection of similarities between the alleles (Figure 1). This revealed groups of related alleles that have evolved along haploid lineages as found previously (13,15). The number and distribution of CTAGGG repeats along the telomeres differed between groups of alleles. For example. the haplotype A associated telomeres represented in 1420.01 contain a single block of 9 or 10 CTAGGG repeats adjacent but distal to a block of TCAGGG repeats and most of these alleles contain additional sequence-variant repeats further into the array. Most of the alleles in the haplotype A-associated group represented by 3183 also contain a single block of CTAGGG repeats that is more variable in length (8–19 repeats) and it may be interrupted by a few N-type repeats (null repeats of unknown sequence that do not amplify with any of the primers used). Distal to the CTAGGG block there appears to be little sequence variation from the consensus TTAGGG repeat. The haplotype B associated telomeres containing CTAGGG repeats were subdivided depending on the presence of proximally located TCAGGG repeats.
To determine whether CTAGGG repeats are found in other human telomeres we screened a subset of telomeres that are located on chromosome 16 (14). Among the 32 telomeres screened across individuals from a variety of ethnic backgrounds, six telomeres (18.8%) that contained a small number (<5) of CTAGGG repeats were identified (data not shown).
Germline mutation analysis of CTAGGG containing Xp/Yp telomeres
Previously, using TVR-PCR analysis of telomeres that contain a variety of sequence variant repeats (but not CTAGGG repeats), we showed that the germline mutation rate in families is 0.006/kb/gamete [(16); unpublished data]. However analysis of (CTAGGG)n containing telomeres within families in this study revealed that some mutated at an extraordinarily high rate upon germline transmission. For example, the haplotype A-associated allele in CEPH father 1362.01 (Figure 1), that contains (CTAGGG)14 repeats in a homogenous array, was transmitted to six children and different mutations were identified in five of them [μ = 0.83/sperm (5/6); Figures 2 and 3]. A comparable germline mutation rate was identified upon transmission of a similar telomere present in CEPH father 1408.01 [μ = 0.6/sperm (3/5)] and pedigree analysis revealed three more germline mutations (Figure 3). The germline mutations identified appear to be intra-allelic gains and/or losses or occasional conversions of 1 or 2 repeats from the CTAGGG block or adjacent repeats. Nevertheless some (CTAGGG)n containing telomeres were transmitted to multiple children without mutation (e.g. haplotype A-associated allele in 23.01 and haplotype B associated telomere 1347.02a; Figure 1).
Figure 3.
Characterization of germline and somatic mutations in telomeres that contain (CTAGGG)n repeats. (a) Germline mutations were identified in families. The interspersion of TTAGGG and sequence-variant repeats along the array is shown for the parent (bold type) and children who inherited a mutated copy of the allele. The mutations are seen as small gains or losses of repeats within the (CTAGGG)n block or in adjacent repeats. In family 1362 each of the five children inherited a different mutation from their father. The telomere present in the male individual 1331.12 mutated upon transmission to his son 1331.01, but no further mutations were identified upon subsequent transmission from 1331.01 into five children. (b) Somatic mutations were identified in normal colon or colon carcinoma DNA samples. The progenitor telomere is shown in bold and the mutant molecules are shown below. As inthe interspersion patterns of TTAGGG (T), TGAGGG (G), TCAGGG (C), TTGGGG (J) CTAGGG (O Figure 1 ) and null (N) repeats are shown from the first repeat in the array (on the left). The mutations are summarized on the right e.g. +2 T indicates that the mutant contained an additional two TTAGGG repeats.
From analysis of the population and germline transmission data (Figures 1 and 3) of (CTAGGG)n containing telomeres, we have identified some factors that seem to influence the mutability of such telomeres in the germ-line. To date all the telomere mutations identified occurred upon transmission from the father indicating that (CTAGGG)n containing telomeres are more likely to mutate in the male than the female germline. Moreover, the number and sequence homogeneity of the (CTAGGG)n block within the telomere seem to influence stability. Such that longer and more homogenous (CTAGGG)n arrays are more unstable, as seen for other STRs (5,8). It is also possible that the repeats surrounding the (CTAGGG)n block influence its stability, however, it is difficult to disentangle the possible effect of surrounding repeats from other factors. In summary our data indicate that if a telomere contains a homogenous (CTAGGG) array of at least 11 repeats flanked by TTAGGG repeats then it is likely to be highly unstable in the male germline as a result of intra-allelic events that give rise to small gains or losses of repeats.
Instability of (CTAGGG)n repeats in somatic tissues
To determine whether the presence of CTAGGG repeats within a telomere causes instability in somatic tissues, DNA from 34 normal colon samples (16) were screened by TVR-PCR to identify individuals with Xp/Yp telomeres that contain CTAGGG repeats. Three donors that were heterozygous for SNPs adjacent to the Xp/Yp telomere, were selected for further analysis. To identify rare mutant copies of the telomere in DNA from the tissue sample, we used allele specific STELA to amplify full length telomeres from aliquots of DNA that contain a single molecule of the telomere of interest [SM-STELA (20)]. Individual SM-STELA products were then subjected to TVR-PCR to identify mutant telomeres.
Two telomeres that did not contain CTAGGG repeat blocks (12q haplotype A in sample 23 and Xp/Yp haplotype A in sample 2) did not show any mutations but the CTAGGG containing Xp/Yp telomeres in normal tissue from individuals 20 and 23 had mutation frequencies of 2 and 5.6%, respectively (Table 1). The combined somatic mutation frequency for the two CTAGGG containing telomeres is significantly different from the telomeres without (Fisher's exact test, two-tail P = 0.009). The mutation frequencies of the two (CTAGGG)n containing telomeres is not significantly different (Fisher's exact test, two-tail P = 0.12) but may reflect the different telomere maps as sample 23 contains a longer homogenous array of CTAGGG repeats. Furthermore, the Xp/Yp (CTAGGG)n containing telomere in individual 23 showed a similar mutation frequency in DNA from a colon carcinoma from the patient (Table 1). This tumour has been shown to contain a mutation in the A10 mononucleotide tract of TGFβRII gene (nucleotides 709 to 718), which is associated with microsatellite instability in 90% of colorectal cancers (16,25).
Table 1.
(CTAGGG)n associated telomere mutations in somatic tissues
| Sample number | Tissuea | Sex/age of donor (years) | Telomere analysed | CTAGGG repeats | Number of mutant molecules | Number of molecules analysed | Mutation frequencyc |
|---|---|---|---|---|---|---|---|
| 2 | N | M/66 | Xp/Yp | None | 0 | 92 | 0 |
| 23 | N | F/88 | 12q | None | 0 | 86 | 0 |
| 23 | N | F/88 | Xp/Yp | (CTAGGG)13 | 6 | 108 | 5.6% |
| 23 | T | F/88 | Xp/Yp | (CTAGGG)13 | 3 | 98 | 3.1% |
| 20 | N | F/81 | Xp/Yp | (CTAGGG)7b | 2 | 102 | 2% |
aN from normal colon; T from colon carcinoma.
bThe Xp/Yp telomere in this sample contains multiple CTAGGG repeats, the largest consecutive block comprising seven repeats.
cThe changes to the number and order of repeats in a single mutant molecule (Figure 3b) must have arisen at a single point in time i.e. from a single mutation event. Therefore the mutation frequency is the number of mutant molecules/the number of molecules analysed.
In summary, most of the somatic telomere mutations appear to comprise intra-allelic changes localized to the CTAGGG or adjacent repeats as seen in the germline (Figure 3a). However, mutant 5 in the normal tissues from donor 23 (Figure 3b) contains multiple changes that must have arisen simultaneously and include a deletion of at least 23 repeats that results in complete loss of the (CTAGGG)13 array and mutant 1 in donor 20 appears to comprise a large deletion. These two mutants could have arisen by a variety of mechanisms.
CTAGGG repeats can form G-quadruplex structures
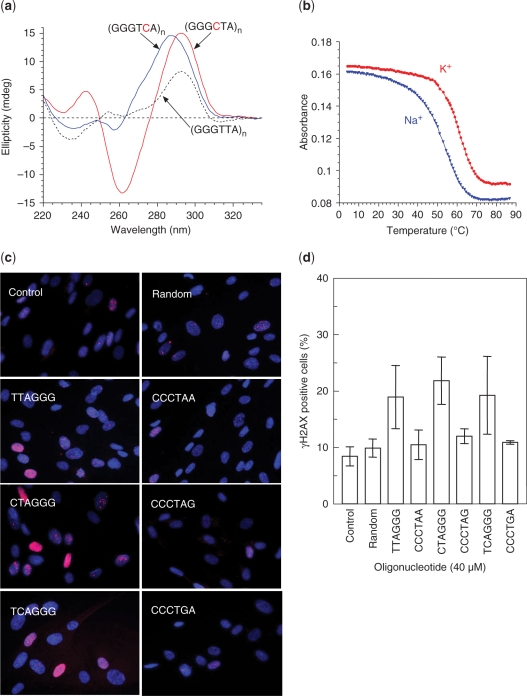
In order to explore why the (CTAGGG)n repeats are so unstable compared to other repeats within the telomere, we have assessed whether the CTAGGG repeats can form G-quadruplex structures in vitro. CD spectra using preformed G-quadruplexes on 22mer oligonucleotides of the CTAGGG, TCAGGG and TTAGGG repeats (‘Materials and Methods’ section) indicated that all three sequences form G-quadruplexes in 100 mM KCl (Figure 4a). Interestingly, the three CD spectra are quite distinct, and suggest that the CTA-22mer adopts a different G-quadruplex conformation than the TTA-22mer or the TCA-22mer (Figure 4a) (26,27). These three oligonucleotides form quadruplexes with similar stabilities with melting temperatures (Tm) in the same range (CTA-22 mer: Tm 62°C in K+ and 54°C in Na+: Figure 4b; TTA-22 mer: Tm 64°C in K+ and 56°C in Na+: TCA-22 mer: Tm 61°C in K+ and 51°C in Na+: not shown). Furthermore, the accompanying article shows that the (CTAGGG) repeats adopt a novel antiparallel G-quadruplex structure (doi:10.1093/nar/gkp630; 28).
Figure 4.
Properties of (CTAGGG)n repeats. (a) Circular dichroism spectra of the human single-stranded telomeric repeat [A(GGGTTA)3GGG] (TTA-22mer, dotted line) and telomere variant repeat sequences TCA-22mer (blue) and CTA-22mer (red) in a potassium buffer. (b) UV melting profiles (absorbance recorded at 295 nm) of CTA-22mer in potassium (red circles) or sodium (blue triangles) buffers. (c) Representative images of the DNA damage foci in MRC5 cells treated with 12-mer single-stranded oligonucleotides as indicated. γH2AX foci are shown in red, DAPI staining of DNA is shown in blue. (d) Quantification of the DNA damage foci in MRC5 cells after 48h treatment with G- or C-rich oligonucleotides corresponding to wild type or variant telomere repeats, as indicated. A 12-mer random oligonucleotide was used as a negative control. The data are mean ± SD of four independent experiments, except for the CCCTAG, CCCTGA and the random oligos that correspond to the mean ± standard error of two experiments.
Induction of DNA damage foci by single-stranded (CTAGGG)2 and (TCAGGG)2 oligonucleotides
It has been shown that single-stranded (TTAGGG)2 oligonucleotides induce the formation of DNA damage foci in MRC5 normal fibroblast cells (17) and so we have investigated whether the degenerate telomere repeats can induce the same response. Figure 4c and d show, that the C-rich oligonucleotides do not induce the formation of γH2AX foci above the background level in the untreated MRC5 (control) cells or above that seen for a random oligonucleotide. However, the G-rich (CTAGGG)2 and (TCAGGG)2 oligos induced γH2AX foci formation with a similar efficiency to the consensus (TTAGGG)2 oligo and significantly above the background level in the control cells (P-values = 0.0128, 0.0024 and 0.0198 for TTAGGG, CTAGGG and TCAGGG, respectively).
CTAGGG repeats bind to TRF2 and POT1
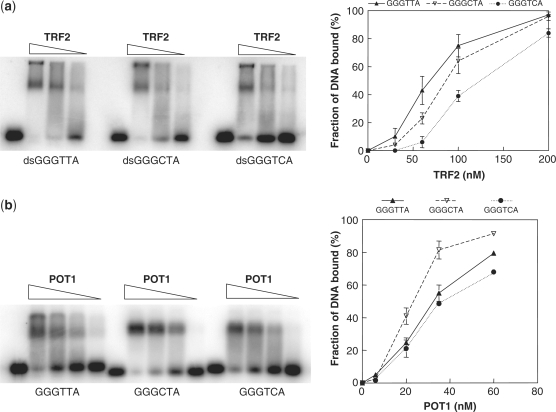
It has been proposed that the formation of DNA damage foci following exposure to (TTAGGG)2 oligonucleotides occurs because the oligos titrate factors away from telomeres. The resulting uncapped telomeres then signal DNA damage (17). Therefore, we next sought to determine whether telomere DNA binding proteins can also bind to CTAGGG and TCAGGG repeats. The in vitro double-strand DNA binding assays showed that CTAGGG and TCAGGG can bind to TRF2, though not as efficiently as the consensus TTAGGG repeat (Figure 5a). Similar results were obtained with TRF1 (Supplementary Figure 1). Unexpectedly, the in vitro single-stranded binding assay showed that POT1 binds to CTAGGG repeats (CTA-22mer) more efficiently than to the consensus TTAGGG (TTA-22mer) or TCAGGG(TCA-22mer) repeats (Figure 5b). In contrast and as a control, a C to A mutation in the minimal binding site for POT1 (5′-TAGGGATAG-3′) impairs POT1 binding [Supplementary Figure 2; (29)]. Thus, these results combined with the induction of γH2AX foci (above) strongly indicate that (CTAGGG)n repeats bind to POT1.
Figure 5.
Interaction of telomere binding proteins with sequence variant telomere repeats. (a) Bandshift assay of double-stranded GGGTTA, GGGCTA and GGGTCA repeats in the presence of purified TRF2. The image on the left shows a representative gel experiment and the graph on the right shows quantification from two independent experiments. (b) Bandshift assay of single-stranded GGGTTA, GGGCTA and GGGTCA repeats in the presence of purified POT1. The image on the left shows a representative gel experiment and the graph on the right the quantification of three independent experiments.
DISCUSSION
The data presented has shown that the presence of a short array of CTAGGG repeats within the Xp/Yp telomere repeat array can cause an extraordinarily high level of instability in the male germline with a mutation rate of 20% per gamete (10/49 across all CTAGGG containing alleles transmitted via the male germline). In contrast, no mutations were observed when CTAGGG carrying telomeres were transmitted via the female germline (0/52), thus the male and female germline mutation rates are significantly different (Fisher's exact test, two-tailed P = 0.0004). The male germline mutation rate varies widely between the (CTAGGG)n carrying alleles with the highest observed in this study being 80% per gamete (in 1362.01). It seems clear that the length and homogeneity of the CTAGGG repeat array within the telomere influence its mutability, such that longer, homogenous arrays are more unstable, as seen for other STR arrays (5,8). Furthermore in the male germline, instability results predominantly in small gains or losses of a few repeats within or adjacent to (CTAGGG)n arrays.
The fact that instability was only observed when the telomere carrying the CTAGGG array was transmitted via the male germline suggests that the underlying mechanism causing the instability is replication dependent. Consistent with this hypothesis, we found that CTAGGG carrying telomeres showed elevated instability when compared to other telomeres lacking CTAGGG repeats, in DNA samples derived from normal colon samples and in a colon carcinoma derived from one of the same individuals, even though the tumour showed microsatellite instability, suggestive of defective DNA MMR (16). Interestingly, although the majority of somatic mutations were small gains or losses of repeats as seen in the germline, two mutant molecules (mutant 5 in normal colon 23 and mutant 1 in normal colon 20, Figure 3b) showed larger changes that resulted in loss of all or the majority of CTAGGG repeats present in the progenitor telomere. Furthermore, the loss of the CTAGGG repeats in mutant 5 is accompanied by other changes.
Replication of telomeres is problematic for several reasons. Primarily telomere replication leads to incomplete lagging-strand synthesis and this, accompanied by resectioning of the C-rich strand, causes telomere erosion (30). In addition, the initiation of telomere replication is thought to occur only at an origin adjacent to the telomere repeat array. The replication fork then migrates in one direction through the telomere towards the terminus [reviewed in (31,32)]. As there is only one replication fork travelling through the telomeric DNA, it must be restarted if it pauses or stalls during replication (33) or the telomere will not be replicated fully. It has been proposed that as the replication fork approaches the t-loop, it may pause or even stall until the t-loop is released (32). Furthermore, there is increasing evidence that, as the telomeric DNA is unwound, the G-rich strand of the replicating telomere is likely to form unusual structures that can impede replication and these include G-quadruplex structures. We have shown that CTAGGG repeats can form G-quadruplex structures in vitro that are different from those formed by the consensus telomere sequence. TTAGGG repeats have been shown to present an important structural polymorphism for quadruplex folding in vitro, and several different folds that greatly differ by their loop arrangements and anti/syn guanine orientations in the quartets have already been structurally characterized [for a recent review see (34)]. Our results suggest a rather different topology for CTAGGG repeats and structural analysis by NMR confirms an unusual folding compared to TTAGGG repeats (PDB:2KM3; see doi:10.1093/nar/gkp630; 28). It is possible that this structural conformation represents an additional obstacle for normal telomere lagging-strand synthesis during replication.
The investigation of TRF1 and TRF2 binding to TTAGGG and sequence variant repeats in vitro suggests that CTAGGG and TCAGGG repeats show only a mild decrease in binding efficiency, in contrast to other degenerate repeats (TTGGGG or TTAGGC) that abolish TRF1 or TRF2 binding (35,36). In contrast, the in vitro POT1 binding assay showed that CTAGGG repeats (CTA-22mer) had a strikingly higher binding affinity than the TTAGGG or TCAGGG repeats. Previous studies have shown that hPOT1 has a minimal binding sequence (29,37). Moreover, it was shown that some bases within the TTAGGGTTAG binding motif are less essential for hPOT1 binding in vitro than others. For example, T to A substitutions in position 1 or 7 of the decamer substrate reduced but did not prevent binding (37). Our study supports and extends these observations as we show that there is a greater tolerance for substitutions at these positions and that some may in fact enhance binding. Although our interesting result needs to be confirmed by other methods, it is partially corroborated by our observation that short single-stranded oligos of the CTAGGG repeats induce the strongest DNA damage response of the three G-rich oligos (Figure 4d). Moreover, it has already been shown that DNA damage response induced by TTAGGG oligos appears to arise through the recruitment of telomere capping proteins (such as POT1) away from telomeres as they enter S-phase (17).
If present within a telomere, the CTAGGG repeats occur near the start of the repeat array. Therefore, they are only likely to be available for POT1 binding if the telomeric single-stranded overhang inserts into this region during t-loop formation or following strand separation during DNA replication. The reduced binding efficiency of sequence variant telomere repeats to TRF1 and TRF2 may increase the chance of forming G-quadruplex structures as the duplex DNA is unwound. If this occurs over CTAGGG repeats, the different topology adopted by these particular repeats may cause additional problems. POT1, like RPA has been shown to unfold telomeric G-quadruplexes (38,39). Therefore, it would be interesting to know whether the increased binding of POT1 to (CTAGGG)n repeats leads to quadruplex removal. In that case, POT1 binding to these repeats in t-loop formation or during replication would represent a mechanism to prevent fork stalling. When a replication fork is blocked, the DNA continues to unwind and bind to RPA until the replication checkpoint is activated and the replication helicase is stopped (40). There may be competition between RPA and POT1 for binding to the single-stranded telomeric DNA during replication, if this results in reduced quadruplex removal, it would lead to fork stalling and a possible slippage during replication. Whatever the mechanism, restarting a replication fork that has stalled at a (CTAGGG)n array will likely involve DNA repair that can cause localized mutation or even removal of the (CTAGGG)n. Finally, the possibility that the C-strand of the (CTAGGG)n repeats also forms an unusual structure is not excluded and has to be considered in regard to other single-stranded proteins involved in telomere replication.
In summary, when considered together the in vivo mutation data, the structural analysis, the DNA damage response and protein binding assays all indicate that homogenous arrays of CTAGGG repeats have unusual properties. We propose that such CTAGGG arrays are more likely to form abnormal structures than other telomere variant repeats and that these structures either impede t-loop release or telomere replication causing the replication fork to stall. The subsequent processing may involve a variety of repair processes that result in small gains/losses of repeats from the (CTAGGG)n array or more intricate repair leading to deletion of the CTAGGG repeats but facilitating restarting of the telomere replication fork. We have shown that a subset of Xp/Yp telomeres and telomeres on chromosome 16 contain CTAGGG repeats. By extrapolation we predict that 7% of all telomeres will contain (CTAGGG)n repeats that add another layer of complexity to the telomere length dynamics in the individual.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council (UK) (grant G0500336 to N.J.R.); Cancer Research-UK (grant C17992/A8641 to N.J.R.); the Ligue Nationale contre le Cancer (to J.F.R.); the E.U. FP6 ‘MolCancer Med’ grant (LSHC-CT-2004-502943 to J.L.M.) and CONACYT (Mexico) funded A.M.B. Funding to open access charge: University of Leicester CR-UK grant C17992/A8641.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Jennie Jeyapalan and Andrew Jones for their contributions toward this work. D. Gomez (Toulouse, France) and M.J. Giraud-Panis (Lyon, France) for generous gift of POT1, TRF1 and TRF2, A. Guédin and J.L. Lacroix (Muséum Paris) for helpful discussions.
Footnotes
Present addresses: Mark Hills, Terry Fox Laboratory, BC Cancer Agency, 675 West 10th Avenue, Vancouver, B.C., Canada V5Z 1L3. Hilda A. Pickett, Cancer Research Group, Children's Medical Research Institute, 214 Hawkesbury Road, Westmead NSW 2145, Australia.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 2.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl Acad. Sci. USA. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X, Peng M, Fang Z. The direction of microsatellite mutations is dependent upon allele length. Nat. Genet. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
- 5.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 6.Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- 7.Pearson CE. Slipping while sleeping? Trinucleotide repeat expansions in germ cells. Trends Mol. Med. 2003;9:490–495. doi: 10.1016/j.molmed.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 9.Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least 3 types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird DM, Jeffreys AJ, Royle NJ. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 1995;14:5433–5443. doi: 10.1002/j.1460-2075.1995.tb00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman J, Baird DM, Royle NJ. The plasticity of human telomeres demonstrated by a hypervariable telomere repeat array that is located on some copies of 16p and 16q. Hum. Mol. Genet. 1999;8:1637–1646. doi: 10.1093/hmg/8.9.1637. [DOI] [PubMed] [Google Scholar]
- 15.Baird DM, Coleman J, Rosser ZH, Royle NJ. High levels of sequence polymorphism and linkage disequilibrium at the telomere of 12q: implications for telomere biology and human evolution. Am. J. Hum. Genet. 2000;66:235–250. doi: 10.1086/302721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickett HA, Baird DM, Hoff-Olsen P, Meling GI, Rognum TO, Shaw J, West KP, Royle NJ. Telomere instability detected in sporadic colon cancers, some showing mutations in a mismatch repair gene. Oncogene. 2004;23:3434–3443. doi: 10.1038/sj.onc.1207477. [DOI] [PubMed] [Google Scholar]
- 17.Tsolou A, Passos JF, Nelson G, Arai Y, Zglinicki T. ssDNA fragments induce cell senescence by telomere uncapping. Exp. Gerontol. 2008;43:892–899. doi: 10.1016/j.exger.2008.08.043. [DOI] [PubMed] [Google Scholar]
- 18.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 19.Dubrova YE, Nesterov VN, Krouchinsky NG, Ostapenko VA, Vergnaud G, Giraudeau F, Buard J, Jeffreys AJ. Further evidence for elevated human minisatellite mutation rate in Belarus eight years after the Chernobyl accident. Mutat. Res. Fund. Mol. Mech. Mutagen. 1997;381:267–278. doi: 10.1016/s0027-5107(97)00212-1. [DOI] [PubMed] [Google Scholar]
- 20.Jeyapalan JN, Mendez-Bermudez A, Zaffaroni N, Dubrova YE, Royle NJ. Evidence for alternative lengthening of telomeres in liposarcomas in the absence of ALT-associated PML bodies. Int. J. Cancer. 2008;122:2414–2421. doi: 10.1002/ijc.23412. [DOI] [PubMed] [Google Scholar]
- 21.Guedin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 23.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 24.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 26.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 28.Lim KW, Alberti P, Guédin A, Lacroix L, Riou JF, Royle NJ, Mergny JL, Phan AT. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G•C•G•C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 30.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 31.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 32.Gilson E, Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 33.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 34.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broccoli D, Chong L, Oelmann S, Fernald AA, Marziliano N, vanSteensel B, Kipling D, LeBeau MM, de Lange T. Comparison of the human and mouse genes encoding the telomeric protein, TRF1: chromosomal localization, expression and conserved protein domains. Human Mol. Genet. 1997;6:69–76. doi: 10.1093/hmg/6.1.69. [DOI] [PubMed] [Google Scholar]
- 36.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 37.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 38.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nitani N, Yadani C, Yabuuchi H, Masukata H, Nakagawa T. Mcm4 C-terminal domain of MCM helicase prevents excessive formation of single-stranded DNA at stalled replication forks. Proc. Natl Acad. Sci. USA. 2008;105:12973–12978. doi: 10.1073/pnas.0805307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.