Abstract
Background
We previously reported the existence of CXCR4-using HIV-1 in 6–14 week-old Ugandan infants. Whether these viruses were transmitted from the mother perinatally or evolved after transmission is not known. In the current study, we investigated the origin of the CXCR4-using viruses in these infants by comparing HIV-1 envelope clones from the infants to those from their mothers at or near the time of delivery.
Methods
Envelope clones were isolated from five Ugandan infant plasma samples that harbored CXCR4-using viruses, collected at the time of HIV diagnosis (four at birth, one at week 6), and from their mothers at delivery. Coreceptor usage and phylogenetic relatedness of HIV-1 populations in mother–infant pairs were analyzed in detail using the Trofile assay and sequence analysis of envelope clones, respectively.
Results
X4-tropic clones were identified in two mother–infant pairs and dual-tropic clones were found in three pairs, either alone or in combination with R5-tropic viruses. Dual-tropic clones varied in their ability to infect CXCR4-expressing cells. In each mother–infant pair, X4-tropic or dual-tropic clones shared similar phenotypic profiles and V3 sequence patterns; gp160 sequences of X4-tropic and dual-tropic clones from infants were phylogenetically indistinguishable from those of their mothers. The virus populations were phylogenetically homogenous in three infants and segregated according to coreceptor tropism in the remaining two infants.
Conclusions
This study demonstrates that X4-tropic and dual-tropic HIV-1 can be transmitted from mother to infant, before, during or shortly after delivery, and establishes vertical transmission as an important source of CXCR4-using viruses in infants.
Keywords: coreceptor tropism, CXCR4, HIV, mother-to-child, transmission, X4
Vertical transmission of HIV-1 from infected mothers to their children accounts for the vast majority of pediatric AIDS cases. Available reports indicate that most infants are infected with R5-tropic viruses [1–4]. Rare cases of CXCR4-using viruses in HIV-1-infected infants have been reported [3,5]; however, it is unclear whether the CXCR4-using viruses in these infants were transmitted directly from their mothers or rapidly evolved from transmitted R5-tropic viruses in the infants. To date, detailed genotypic and phenotypic analyses of CXCR4-using virus populations in mother–infant pairs have not been reported, which is needed to definitively establish the origin of CXCR4-using viruses in vertically infected infants and clearly define their phylogenetic relationship to the mother’s virus. Emergence and transmission of CXCR4-using viruses have been associated with rapid disease progression in adults and children. However, the origin, natural history and clinical relevance of the presence of CXCR4-using viruses in infants are less clear [6,7]. We recently identified CXCR4-using viruses in five (9%) of 57 HIV-infected Ugandan infants at 6–14 weeks of age and in their mothers [8]. This provided a unique opportunity to ask whether the CXCR4-using viruses in the infants were transmitted or evolved from transmitted R5-tropic viruses.
Plasma-derived envelope (env) clones were obtained from these five infants at HIV-1 diagnosis, four at birth (infants 185, 197, 632 and 827) and one at 6 weeks of age (infant223, HIV-1 negative at delivery), and from all mothers at delivery. A total of 140 env clones (12–16 clones from each mother or infant sample) were evaluated for coreceptor tropism using the Trofile assay (Monogram Biosciences, SouthSan Francisco, USA) [9]. Env clones were classified as R5-tropic, X4-tropic and dual-tropic, based on luciferase activity in U87/CD4/CCR5 and U87/CD4/CXCR4 cells, and whether the luciferase activity was specifically inhibited by a CCR5 or CXCR4 antagonist [9]. The term ‘CXCR4-using’ refers to X4-tropic and dual-tropic viruses. The results of this analysis provided accurate qualitative and quantitative assessments of the coreceptor tropism of the env clones that highlights the phenotypic similarities of paired mother–infant virus populations, and illustrates the variation in CCR5-mediated and CXCR4-mediated infectivity of dual-tropic viruses (Table 1).
Table 1.
Characterization of coreceptor utilization and V3 sequence of HIV env clones from mother-infant pairs.
| Sample | No. of clonesa |
Average RLU in CCR5+ cells |
Average RLU in CXCR4+ cells |
Coreceptor tropismb |
Amino acid sequences of V3 loppc |
|---|---|---|---|---|---|
| Mother 197 | 4 | 71 | 702,609 | X4 | CTRPYEKGTRKSTPIGIGRAWYTMNITGRIRPAHC |
| 4 | 100 | 1,343,705 | X4 | .....R.F........T.........A..AE..Y. | |
| 2 | 114 | 1,164,827 | X4 | .................................Y. | |
| 1 | 91 | 1,448,435 | X4 | .....R.......................AE.... | |
| 1 | 62 | 93,543 | X4 | .....R....R...................E.... | |
| 1 | 83 | 83,578 | X4 | ..........R........................ | |
| Infant 197 | 9 | 95 | 607,949 | X4 | ................................... |
| 2 | 78 | 871,742 | X4 | .....K.......................AE..Y. | |
| 1 | 79 | 1,164,366 | X4 | .....R.F.....................AE.... | |
| 1 | 77 | 513,404 | X4 | .....R.......................AE.... | |
| Mother 185 | 8 | 620,521 | 93 | R5 | CTRPGNNTRKGIHIGPGQAFYAADKIIGDIRQAHC |
| 1 | 15,639 | 69 | R5 | ....N....R.V..........T........... | |
| 1 | 369,812 | 64 | R5 | ....N....R............T.S........Y. | |
| 1 | 58,419 | 4,571 | Dual-R | .................................. | |
| 1 | 9,451 | 419 | Dual-R | ....N....R............T.S........Y. | |
| 2 | 104,675 | 153,727 | Dual-X | .........R..R....RK..............Y. | |
| Infant 185 | 14 | 171,921 | 395,231 | Dual-X | .........R..R....RK..............Y. |
| Mother 632 | 6 | 750,856 | 2,345 | Dual-R | CTRPYNNTRRGTHIGPGRAYFTTNIIGDIRQAHC |
| 4 | 361,183 | 117 | R5 | .................................. | |
| 1 | 383,804 | 98 | R5 | ..................T............... | |
| 1 | 281,590 | 113 | R5 | .........K........................ | |
| 1 | 1,198,350 | 614 | Dual-R | .........T.............S.......... | |
| 1 | 1,321,181 | 17,062 | Dual-R | .....S...T........................ | |
| Infant 632 | 8 | 1,123,855 | 1,547 | Dual-R | ..........S....................... |
| 3 | 245,317 | 113 | R5 | ..........S....................... | |
| 1 | 179,315 | 154 | R5 | ..........S..............V........ | |
| Mother 223 | 11 | 1,132,515 | 123 | R5 | CTRPYNNKRESIHMGPGRALYTKDIIGDIRQAHC |
| 2 | 722,746 | 100 | R5 | .....T..K......................... | |
| 1 | 957,172 | 97 | R5 | ................................Y. | |
| 2 | 105 | 37,191 | X4 | .....I.N.DG..T..R..F..RK.T........ | |
| Infant 223 | 12 | 1,097,648 | 121 | R5 | .................................. |
| 1 | 1,504,316 | 134 | R5 | .....S............................ | |
| 1 | 809,701 | 104 | R5 | .A................................ | |
| 1 | 76 | 340,101 | X4 | .....1.N.DG..T..R..F..RK.T........ | |
| Mother 827 | 5 | 973,307 | 622 | Dual-R | CTRPYSNKRQGIHIGPGRAYYTSEITGNIRQAYC |
| 3 | 1,411,912 | 12,527 | Dual-R | ..............................K... | |
| 1 | 953,950 | 688 | Dual-R | .....T............................ | |
| 1 | 1,012,714 | 1,795 | Dual-R | .........H........................ | |
| 3 | 904,040 | 87 | R5 | .....N.T.K...M...Q....AN...D....H. | |
| 1 | 144,355 | 102 | R5 | .....N.T.K.......Q....AN...D....H. | |
| Infant 827 | 12 | 1,189,705 | 1,912 | Dual-R | .................................. |
| 2 | 70,880 | 88 | R5 | .................................. | |
| 1 | 961,696 | 112 | R5 | .....N.T.K.......Q....AN...D....H. |
Number of clones with the indicated V3 sequence identified in each sample.
Dual-tropic clones were sub-classified as dual-R-tropic and dual-X-tropic [10].
Amino acid residues that differ from the most prevalent sequence in the corresponding mother are indicated. V3 amino acid positions 11 and 25 are highlighted in bold font. Potential N-linked glycosylation sites (PNGS) in V3 at amino acid positions 6–8 are underlined.
The virus populations of both mother and infant 197 were homogenous and comprised solely of X4-tropic variants that generally utilized CXCR4 with equivalent efficiencies. Similarly, R5-tropic and X4-tropic variants within the mixed virus population of infant 223 were representative of the R5-tropic and X4-tropic variants observed in the mother’s virus population. The virus populations of two mother–infant pairs (632 and 827) were comprised of mixed subpopulations of R5-tropic and dual-tropic variants. The infectivity of individual env clones [shown as relative light units (RLU) of luciferase activity in Table 1] indicated that the dual-tropic variants in these mother–infant virus populations utilize CCR5 more efficiently (104 to 106 RLU) than CXCR4 (102 to 104 RLU). In the fifth mother–infant pair (185), the mother’s virus was comprised of an R5-tropic population mixed with two dual-tropic subpopulations that were distinguished based on their abilities to utilize CXCR4 (approximately 103 RLU vs. 105 RLU, respectively). All of the viruses detected in infant 185 displayed the same tropism pattern (dual-tropic with efficient CXCR4 use). No R5-tropic and dual-tropic clones with inefficient CXCR4 use were identified by examining 76 additional functional clones (data not shown), suggesting that the maternal subpopulation of dual-tropic variants with efficient CXCR4 use was selectively transmitted.
We next determined the gp160 sequence of each of the 140 env clones from five mother–infant pairs using conventional chain termination chemistry (Big Dye Terminator Ready Reaction Cycle Sequencing; ABI, Foster City, California, USA). The V3 amino acid sequences of mother and infant env clones derived from nucleotide sequences were correlated with coreceptor tropism results (Table 1). In general, V3 loop sequences of R5-tropic, X4-tropic or dual-tropic env clones from each infant were identical, or similar, to R5-tropic, X4-tropic or dual-tropic env clones from the viruses of the paired mothers. We further sub-classified dual-tropic env clones as dual-X or dual-R based on virus infectivity and the V3 amino acid sequences of individual clones as described previously [10]. Dual-X-tropic clones efficiently utilize CXCR4 and have V3 sequences that are distinct from the R5-tropic clones from the same virus population; dual-R clones utilize CXCR4 less efficiently and have V3 sequences that are closely related or identical to the R5-tropic clones that are present in the same virus population. In infant 197, all env clones were X4-tropic and the V3 sequences were similar to the maternal X4-tropic clones, differing at no more than two amino acid positions. In mother–infant pairs 223 and 185, the infant virus populations contained either X4-tropic clones (223) or dual-X-tropic clones (185) with V3 sequences identical to those of their mother’s X4-tropic or dual-X-tropic clones, but distinct from those of their mother’s R5-tropic clones. In mother–infant pairs 632 and 827, the infants had dual-R-tropic clones with V3 sequences similar to the dual-R-tropic clones from their mothers, and were closely related or identical to the R5-tropic clones. In all five mother–infant pairs, infant V3 sequences matched the maternal V3 sequences of env clones with the same phenotype.
The presence of a charged residue at positions 11 and/or 25 in the V3 loop has been associated with CXCR4 coreceptor use [11–13]. However, in this study, the 11/25 rule correctly predicted CXCR4 use in only the X4-tropic clones for pairs 197 and 223, but not in the dual-R clones for pairs 632 and 827. Furthermore, the charged residue lysine was present at position 25 in R5-tropic and dual-tropic clones for pair 185. The loss of potential N-linked glycosylation site (PNGS) in V3 at amino acid positions 6–8 has also been reported to associate with CXCR4 use [14] and was observed in all X4-tropic clones from pair 197 and pair 223. However, the absence of PNGS was also observed in dual-R clones from pair 827 and in R5-tropic clones from pair 827 and 223. In addition, all clones from pair 185 and all but one clone from pair 632 retained PNGS, including dual-X-tropic, dual-R-tropic and R5-tropic clones. Thus, we observed no clear distinction of 11/25 and PNGS between dual-tropic or R5-tropic viruses in this study.
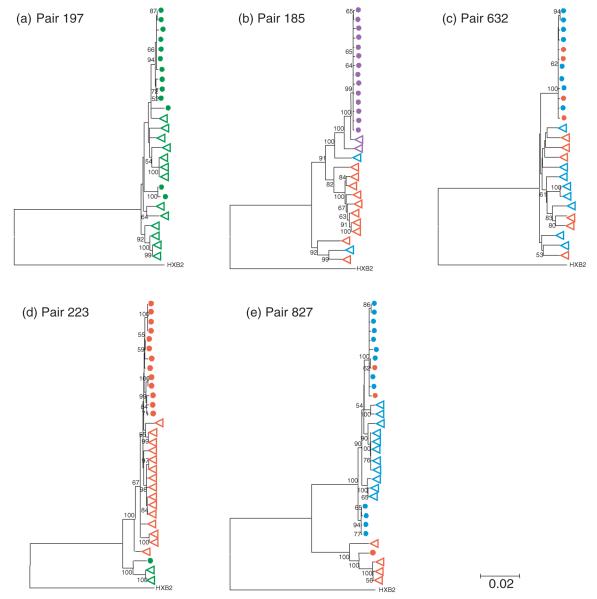
To examine further the genetic relationship between env clones from each mother–infant pair, we performed a phylogenetic analysis of gp160 nucleotide sequences from the full set of 140 env clones isolated from the paired mother and infant samples. Virus subtyping was performed by aligning the gp160 nucleotide sequences of the 140 clones against the HIV-1 subtype reference sequences available from the Los Alamos HIV Sequence Database. All mother–infant pairs were infected with subtype D viruses, with the exception of pair 185 that was infected with subtype A virus. Phylogenetic trees were constructed from gp160 nucleotide sequences using neighbor-joining methods as implemented in MEGA, version 3.1 [15], with 1000 replicate bootstrap resampling. Sequences from each mother–infant pair separated into distinct clusters that were supported by high bootstrap values (>99%) for the nodes of all five mother–infant pairs. We next performed a phylogenetic analysis of the individual env sequences obtained for each mother–infant pair (Fig. 1). The coreceptor tropism result for each clone is indicated on the corresponding branch of the phylogenetic tree (R5-tropic, X4-tropic, dual-X-tropic or dual-R-tropic). In general, sequences from infant viruses clustered tightly within each group of paired mother–infant sequences and exhibited less diversity than the corresponding sequences from the mother’s viruses, irrespective of coreceptor tropism. This observation is consistent with the existence of some degree of selective advantage (or disadvantage) during transmission and is best illustrated by the phylogenetic relationship observed in mother–infant pair 185, which suggests that infection in this infant was established from a minor maternal subpopulation of dual-X-tropic viruses (Fig. 1b). In addition to the relatively homogeneous virus populations from infants 197, 185 and 632 shown in phylogenetic trees (Fig. 1a–c), we also found evidence of oligoclonal infection in infants 223 and 827 (Fig. 1d, e); the phylogenetic trees for these two mother–infant pairs clearly show distinct subpopulations in the infants that were sorted by coreceptor tropism and were most likely derived from the corresponding subpopulations in the mother. Whether the virus populations in these two infants were established by a single oligoclonal transmission event or sequential monoclonal transmission events is uncertain. Taken together, analysis of gp160 sequences from all five mother–infant pairs indicates that the CXCR4-using env clones isolated from each infant virus population are phylogenetically indistinguishable from the corresponding CXCR4-using subpopulations present in the mother at the time of delivery. We found no evidence to suggest that CXCR4-using clones in the infants were the result of rapid evolution of R5-tropic clones following transmission.
Fig. 1. Phylogenetic relationships among env clones from each mother–infant pair.
Phylogenetic trees were constructed from gp160 nucleotide sequences using neighbor-joining methods. Nodes with less than 50% bootstrap support were collapsed. Maternal and infant sequences are indicated with open triangles and filled circles, respectively. Coreceptor tropism of individual env clones is depicted in color: red, R5-tropic; blue, dual-R-tropic; purple, dual-X-tropic; green, X4-tropic. (a) Pair 197; (b) pair 185; (c) pair 632; (d) pair 223; (e) pair 827.  , Infant’s R5-tropic clones;
, Infant’s R5-tropic clones;  , Infant’s X4-tropic clones;
, Infant’s X4-tropic clones;  , Infant’s dual-R clones;
, Infant’s dual-R clones;  , Infant’s dual-X clones;
, Infant’s dual-X clones;  , Mother’s R5-tropic clones;
, Mother’s R5-tropic clones;  , Mother’s X4-tropic clones;
, Mother’s X4-tropic clones;  , Mother’s dual-R clones;
, Mother’s dual-R clones;  , Mother’s dual-X clones.
, Mother’s dual-X clones.
In summary, we demonstrated CXCR4-using viruses were transmitted from mother to infant in five Ugandan mother–infant pairs. Detailed clonal analyses identified different types of CXCR4-using viruses in the viral populations of these newborns, including X4-tropic, dual-X-tropic and dual-R-tropic variants. The phenotypic composition of the virus populations from these infants was strikingly similar to that of their mother’s virus population at delivery. In the single exception (pair 185), a mother transmitted a dual-X-tropic variant to her infant that existed as a minor subpopulation among predominantly R5-tropic variants. Phylogenetic analysis based on gp160 nucleotide sequences further indicated that R5-tropic, X4-tropic and dual-tropic variants from each infant shared a common origin with variants from the infant’s mother. These data support the vertical transmission of CXCR4-using variants from mother to infant during pregnancy in four cases (pairs 185, 197, 632 and 827), and at delivery or shortly after birth in the fifth case (pair 223). Our observations in these mother–infant pairs do not support a model in which CXCR4-using variants evolve from transmitted R5-tropic viruses or the preferential infection by, or outgrowth of R5-tropic viruses in vertically infected infants. The different types of CXCR4-using viruses that we identified previously in pregnant Ugandan women (X4-tropic, dual-X-tropic and dual-R-tropic variants) [10] were also found in the virus populations of the mother–infant pairs evaluated in this study. We can conclude from these two studies that distinct subclasses of CXCR4-using viruses present in pregnant women can be transmitted to their infants. Notably, the two infants infected with either homogeneous X4-tropic (197) or dual-X-tropic (185) viruses died earlier (14.5 and 10.5 months) than the two infants infected with R5-tropic and dual-R-tropic viruses (2 and 4.7 years for 827 and 632) or the one infant (223) infected with a predominantly R5-tropic virus population containing a small subpopulation of X4-tropic variants (2.5 years). However, additional studies are required to adequately address whether infants with infections dominated by X4-tropic or dual-X-tropic viruses are at greater risk for disease progression and death than those infected predominantly with R5-tropic or dual-R-tropic viruses.
Acknowledgements
We thank Jessica Church (Johns Hopkins University) for assistance with sample and data management and for critical reviewing the manuscript; Yolanda Lie for coordinating the study; Sunny Choe and Cynthia Sedik for editorial assistance. We thank Monogram Biosciences Clinical Reference Laboratory for the performance of tropism assays. This study was partially supported by SBIR-AT grant R44-AI-048990 (NIAID, NIH). This work was also supported by the HIV Network for Prevention Trials (HIVNET) and sponsored by the US National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services (DHHS), through contract N01-AI-35173 with Family Health International, contract N01-AI-45200 with Fred Hutchinson Cancer Research Center, and contract N0I-AI-35173-417 with Johns Hopkins University; the HIV Prevention Trials Network (HPTN) sponsored by the NIAID, National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (U01-AI-46745, U01-AI-48054 and U01-AI-068613), and the International Maternal Pediatric Adolescent AIDS Clinical Trials Network of the NIAID (U01-AI-068632).
References
- 1.van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrechtvan Lent N, Scherpbier HJ, Veenstra J, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casper CH, Clevestig P, Carlenor E, Leitner T, Anzen B, Lidman K, et al. Link between the X4 phenotype in human immunodeficiency virus type 1-infected mothers and their children, despite the early presence of R5 in the child. J Infect Dis. 2002;186:914–921. doi: 10.1086/342948. [DOI] [PubMed] [Google Scholar]
- 3.Salvatori F, Scarlatti G. HIV type 1 chemokine receptor usage in mother-to-child transmission. AIDS Res Hum Retroviruses. 2001;17:925–935. doi: 10.1089/088922201750290041. [DOI] [PubMed] [Google Scholar]
- 4.Matala E, Hahn T, Yedavalli VR, Ahmad N. Biological characterization of HIV type 1 envelope V3 regions from mothers and infants associated with perinatal transmission. AIDS Res Hum Retroviruses. 2001;17:1725–1735. doi: 10.1089/08892220152741423. [DOI] [PubMed] [Google Scholar]
- 5.Scarlatti G, Hodara V, Rossi P, Muggiasca L, Bucceri A, Albert J, Fenyo EM. Transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child correlates with viral phenotype. Virology. 1993;197:624–629. doi: 10.1006/viro.1993.1637. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgibbon JE, Gaur S, Gavai M, Gregory P, Frenkel LD, John JF., Jr Effect of the HIV-1 syncytium-inducing phenotype on disease stage in vertically-infected children. J Med Virol. 1998;55:56–63. doi: 10.1002/(sici)1096-9071(199805)55:1<56::aid-jmv10>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 7.Casper C, Naver L, Clevestig P, Belfrage E, Leitner T, Albert J, et al. Coreceptor change appears after immune deficiency is established in children infected with different HIV-1 subtypes. AIDS Res Hum Retroviruses. 2002;18:343–352. doi: 10.1089/088922202753519124. [DOI] [PubMed] [Google Scholar]
- 8.Church JD, Huang W, Mwatha A, Toma J, Stawiski E, Donnell D, et al. HIV-1 tropism and survival in vertically infected Ugandan infants. J Infect Dis. 2008;197:1382–1388. doi: 10.1086/587492. [DOI] [PubMed] [Google Scholar]
- 9.Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchier RA, Brouwer M, Broersen SM, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol. 2002;76:3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polzer S, Dittmar MT, Schmitz H, Schreiber M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glyco-protein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology. 2002;304:70–80. doi: 10.1006/viro.2002.1760. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]