Abstract
In bacteria, multiple σs direct RNA polymerase to distinct sets of promoters. Housekeeping σs direct transcription from thousands of promoters, whereas most alternative σs are more selective, recognizing more highly conserved promoter motifs. For σ32 and σ28, two Escherichia coli Group 3 σs, altering a few residues in Region 2.3, the portion of σ implicated in promoter melting, to those universally conserved in housekeeping σs relaxed their stringent promoter requirements and significantly enhanced melting of suboptimal promoters. All Group 3 σs and the more divergent Group 4 σs have nonconserved amino acids at these positions and rarely transcribe >100 promoters. We suggest that the balance of “melting” and “recognition” functions of σs is critical to setting the stringency of promoter recognition. Divergent σs may generally use a nonoptimal Region 2.3 to increase promoter stringency, enabling them to mount a focused response to altered conditions.
Keywords: σ factor, Region 2.3, melting proficiency, promoter stringency
In bacteria, promoter recognition is accomplished primarily by the σ subunit of RNA polymerase (RNAP). A single housekeeping σ directs RNAP to thousands of promoters, whereas alternative σs generally orchestrate transcription from substantially fewer promoters, allowing focused responses to cellular, environmental, and developmental signals (Gruber and Gross 2003; Paget and Helmann 2003; Gama-Castro et al. 2008). This unequal division of labor requires a housekeeping σ with broad, flexible promoter recognition (Hook-Barnard and Hinton 2007), and alternative σs with more restricted recognition (Amaya et al. 2001; Britton et al. 2002; Eichenberger et al. 2003; Nonaka et al. 2006; van Schaik et al. 2007; Zhao et al. 2007; Asayama and Imamura 2008; Koo et al. 2009a,b). More extensive use of activators and repressors by the housekeeping σ than by alternative σs partially explains this distinction (Browning and Busby 2004). Here, we provide evidence for an additional strategy intrinsic to the σs themselves, demonstrating that some alternative σs are specifically constructed to limit their ability to transcribe a wide range of promoters.
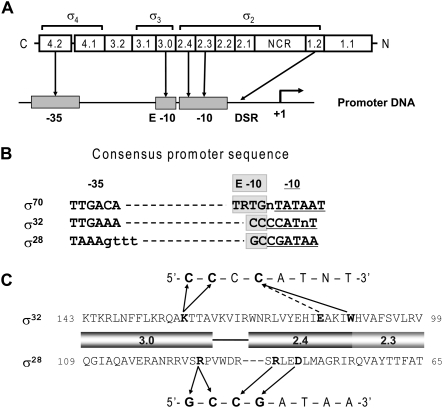
σs are sequence-specific DNA-binding proteins with a modular architecture, consisting of globular domains (Murakami and Darst 2003) subdivided into conserved regions (Fig. 1A). In free σ, DNA-binding determinants are masked by domain interactions (Dombroski et al. 1993; Sorenson and Darst 2006). However, strong interactions between RNAP and domains 2–4 (σ2–4) expose DNA-binding determinants and position domains for interaction with promoter motifs (Fig. 1A; Kuznedelov et al. 2002; Murakami et al. 2002). σ4 recognizes the −35 motif (Gardella et al. 1989; Siegele et al. 1989), σ3 recognizes the extended −10 (E-10) motif (Barne et al. 1997; Koo et al. 2009a,b), and σ2 facilitates strand opening by three sequential activities: (1) recognition of the −10 and discriminator regions (Siegele et al. 1989; Daniels et al. 1990; Waldburger et al. 1990; Tatti et al. 1991; Feklistov et al. 2006; Haugen et al. 2006; Koo et al. 2009a,b), (2) participation in melting (Juang and Helmann 1994; Fenton et al. 2000; Tomsic et al. 2001; Lee and Gralla 2003), and (3) interaction with the −10 region nontemplate strand DNA to stabilize the melted state (Helmann and deHaseth 1999; Schroeder et al. 2009). σ1 (Region 1.1) is unique to housekeeping σs and has regulatory roles (Dombroski et al. 1992, 1993; Hook-Barnard and Hinton 2009).
Figure 1.
Promoter utilization by σ. (A) Domain organization and conserved regions of σs and their functional assignments based on those of σ70. A schematic of a typical promoter with conserved DNA regions −35, −10, and E-10, and discriminator (DSR) region from the transcription start site is shown below the linear representation of σ. Interactions between conserved region of σ and promoter regions are indicated by arrows. In σ2, Region 2.4 is implicated in both duplex recognition and interaction with nontemplate strand, Region 2.3 is implicated in duplex recognition and melting, and Region 1.2 recognizes the discriminator. The crystallized domains of σ are indicated above the linear representation of σ. (B) Consensus promoter sequences of σ70, σ32, and σ28. E-10 motifs of each promoter are boxed in gray, and the −10 motifs of each promoter are underlined. (C) Recognition of the −10 region of promoters by σ32 and σ28. Amino acid sequences of Regions 2.3, 2.4, and 3.0 of σ32 and σ28 are shown. The −10 consensus promoter sequences for each σ are indicated above or below the amino acid sequences. Contacts between σ and promoter are indicated by arrows. Solid arrows indicate direct contacts, and the arrow with the dotted line indicates contact to increase selectivity (Koo et al. 2009a,b).
The σ70 superfamily includes three subfamilies, divided according to phylogenetic relatedness to the essential housekeeping (Group 1) σs (Lonetto et al. 1992; Gruber and Gross 2003; Paget and Helmann 2003). Group 2 σs, most closely related to the Group 1 σs, have σ2–4 but are nonessential; Group 3 σs are less related but also contain σ2–4; Group 4 σs (ECF σs) are the most minimal σs, containing only σ2 and σ4. Here, we examine Group 3 σs. Most bacteria have multiple σs of this type, and Escherichia coli has two: σ28 and σ32 (Gruber and Gross 2003; Paget and Helmann 2003). σ28, the most widely distributed alternative σ, controls flagella-related genes in all motile Gram-negative and Gram-positive bacteria and development in some nonmotile bacteria (Chilcott and Hughes 2000; Yu and Tan 2003; Serizawa et al. 2004; Shen et al. 2006). σ32 controls the heat-shock response and is present in most proteobacteria (Nakahigashi et al. 1995; Guisbert et al. 2008). σ32 and σ28 each direct RNAP to a limited number of promoters (∼50 for σ32 and ∼25 for σ28 in E. coli) (Nonaka et al. 2006; Shen et al. 2006; Wade et al. 2006; Zhao et al. 2007). Expression from these promoters is instituted synchronously in response to a change in the amount and/or activity of the respective σ.
We previously found an important distinction between the promoter recognition properties of the E. coli Group 3 σs and their housekeeping σ, σ70. In σ70, the −35, −10, and E-10 motifs are partially redundant, and functional promoters are constructed from subsets of motifs (Hook-Barnard and Hinton 2007). Most commonly, promoters have −10 and −35 motifs but lack the E-10 (Lisser and Margalit 1993). Conversely, promoters with the E-10 motif do not have good matches to both the −10 and −35 motifs (Mitchell et al. 2003). Most E-10 promoters require neither the −35 motif nor its σ recognition region (4.2) (Ponnambalam et al. 1986; Kumar et al. 1993; Minakhin and Severinov 2003; Young et al. 2004); a few have an excellent −35 and a reduced requirement for −10 (Thouvenot et al. 2004; Hook-Barnard et al. 2006). In sharp contrast, functional σ28 and σ32 promoters require good matches to the −35, −10, and E-10 motifs (Fig. 1B; Koo et al. 2009a,b). Consistent with this, the information content of the core regions of σ32 promoters (18.3 bits) (Nonaka et al. 2006) and σ28 promoters (21.3 bits) (determined from sequences of all E. coli σ28 promoters listed in BioCyc [http://biocyc.org]; V Rhodius, unpubl.; data not shown) is much higher than that for σ70 promoters (9.2 bits) (Shultzaberger et al. 2007). Requiring extensive recognition determinants would limit transcription to a relatively restricted set of promoters, making it important to understand how these σs maintain dependence on all three promoter motifs.
We show that σ32 and σ28 require all three core promoter motifs for function largely because of altered residues in Region 2.3, a 17-amino-acid section of σ2 implicated in promoter melting. Both σs deviate in sequence from critical Region 2.3 residues that are universally conserved in the housekeeping σs and are known to be important in melting. Converting these σ32 and σ28 residues to those present in the housekeeping σs decreases the requirement for extensive recognition motifs and increases melting capacity at nonoptimal promoters. Our results suggest that extensive recognition motifs compensate for nonoptimal Regions 2.3 in σ32 and σ28. All Group 3 and Group 4 σs have nonconsensus amino acids at the Region 2.3 positions we investigated. Decreasing melting capacity may be a general strategy enabling divergent alternative σs to mount a discrete, focused, and structured response to altered conditions.
Results
Phylogenetic analysis identifies elements required to bypass the σ32 requirement for an E-10 motif
σ32s from different proteobacterial groups differ in their stringency of promoter recognition and in their recognition determinants. σ32s from E. coli and other γ-/β-proteobacteria exhibit high dependence on the E-10 motif and recognize it with a universally conserved residue (K130) in σ3 (Koo et al. 2009a). In contrast, α-/δ-/ɛ-proteobacteria show little to no dependence on the E-10 motif, and their comparable σ3 residue is alanine, serine, or glutamine (Green and Donohue 2006; McGrath et al. 2007; Koo et al. 2009a). σ32 of the α-proteobacterium Caulobacter crescentus has little dependence on the E-10 motif even in combination with E. coli core RNAP (Koo et al. 2009a), indicating that the distinction in promoter recognition stringency resided within σ32 itself. This raises the question of how C. crescentus σ32 bypasses the requirement for the E-10 motif.
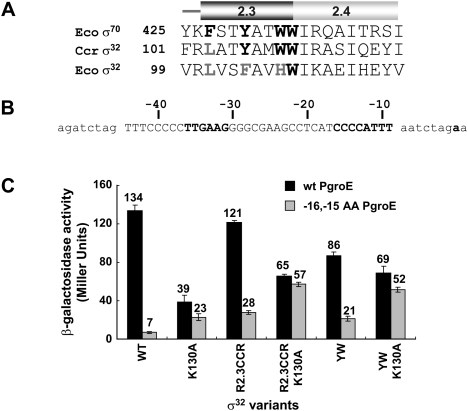
Comparison of E. coli and C. crescentus σ32 revealed that the two differ in important residues in Region 2.3 (Fig. 2A), but not in residues implicated in base-specific recognition of promoters (Fig. 1C) or in Region 2.2, the most important core-binding motif (data not shown). Four Region 2.3 aromatic amino acids are universally conserved in housekeeping σs (bold in Fig. 2A). Of these, F427 is buried and is likely to play a structural role (Murakami et al. 2002), W434 is likely to be involved in promoter recognition (Juang and Helmann 1994), and Y430 and W433 are most directly involved in promoter DNA melting (Juang and Helmann 1994; Fenton et al. 2000; Tomsic et al. 2001). Three of these residues are present in C. crescentus σ32 (and in σ32 of all α-proteobacteria), but only one is present in E. coli σ32 (and in σ32 of all other γ-proteobacteria) (Kourennaia and deHaseth 2007). We tested whether transplantation of C. crescentus Region 2.3 residues (103LATYAMWW110) into E. coli σ32 reduced dependence on the E-10 motif using lacZ reporter assays driven from the groE σ32 promoter (Fig. 2B) or derivatives of this promoter. Whereas authentic E. coli σ32 exhibited a 20-fold dependence (Fig. 2C, columns 1,2), the hybrid σ32 showed only a fourfold dependence (Fig. 2C, columns 5,6); introducing the K130A substitution to more fully mimic authentic C. crescentus σ32 eliminated dependence on the E-10 motif (Fig. 2C, columns 7,8) and reduced the deleterious effect of the K130A substitution alone (Fig. 2C, cf. columns 7,8 and 3,4). Importantly, this effect is recapitulated by simply changing the residues in E. coli σ32 analogous to Y430 and W433 to their counterparts in the housekeeping σs and in C. crescentus σ32 (creating F104YH107Wσ32; YWσ32) (Fig. 2C, cf. columns 9,10 and 5,6, and columns 11,12, and 7,8). Note that Y430 and W433 are the residues most directly implicated in promoter DNA melting (Juang and Helmann 1994; Fenton et al. 2000; Tomsic et al. 2001). Single amino acid substitutions showed a partial effect, and a triple mutant (L101F, F104Y, H107W) is virtually indistinguishable from YWσ32 (data not shown). These results suggest that these YW residues in C. crescentus (and other α-/δ-/ɛ-proteobacteria) enable their σ32s to bypass the requirement for an E-10 motif.
Figure 2.
Identification of elements that bypass the need for the E-10 motif in σ32. (A) Amino acid sequence alignment of a segment of Region 2.3 and Region 2.4 of E. coli σ70, E. coli σ32, and C. crescentus σ32. Bold letters are residues universally conserved in housekeeping and Group 2 σs, and are implicated in promoter melting in σ70; analogous residues that deviate from consensus in σ32s are in gray. The numbers indicate amino acid sequence position. (B) Sequence of σ32-dependent promoter (PgroE) used in this study. Only the wild-type sequence of groE promoter is shown; promoter variants were constructed from this sequence (Koo et al. 2009a). The native sequence of the groE promoter region is shown in capital letters, vector sequence is in lowercase, and −35 and −10 regions and transcription start site are shown in bold. (C) Expression from wild-type or −16,−15 AA mutant groE promoters by E. coli RNAP containing σ32 variants. β-galactosidase activity (Miller units) of promoter∷lacZ fusions for each σ32 variant on the wild-type promoter (black bar) or the −16,−15 AA mutant promoter lacking the extended −10 region (gray bar) are shown. These assays were performed in CAG57101 (ΔrpoH) with plasmids expressing σ32 variants and bearing groE promoter variants∷lacZ. For the β-galactosidase assays shown in this and subsequent figures, all values (indicated above each bar) are averages of at least three independent experiments; error bars indicate one standard deviation. Wild type (WT) is E. coli σ32; variants have substitutions in E. coli σ32 as follows: R2.3CCR, sequence encoding Region 2.3 is replaced with Region 2.3 of C. crescentus σ32; R2.3CCR K130A is R2.3CCR with K130A; YW is F104YH107Wσ32; and YW K130A is YW with K130A.
YWσ32 generally suppresses defects of nonoptimal promoters
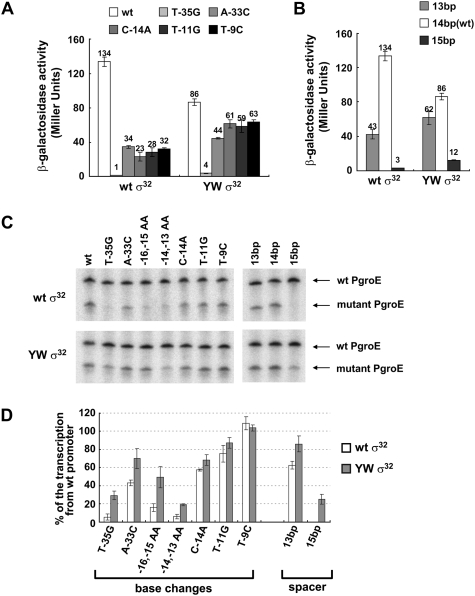
We tested whether YWσ32 was generally proficient in transcribing nonconsensus promoters (Fig. 3). YWσ32 significantly suppressed the two −35 and the three −10 mutations in vivo: Whereas most mutations resulted in a fourfold to fivefold decrease in expression with wild-type σ32, they showed only a 1.5-fold to twofold decrease in expression with YWσ32 (Fig. 3A). Likewise, YWσ32 showed reduced dependence on the length of the spacer DNA separating the −35 and −10 elements (Fig. 3B). This effect was reproduced in vitro. All mutant promoters with a significant transcription defect (≤40% of the wild-type promoter) showed significantly higher transcription with YWσ32 than with wild-type σ32, indicating that effects were direct (Fig. 3C,D). We conclude that YWσ32 generally exhibited higher activity than authentic σ32 on weak promoters.
Figure 3.
Suppression of nonoptimal promoter mutations by YWσ32 in vivo and in vitro. Expression from wild-type and mutant groE promoters (A) or promoters with nonoptimal spacing between −10 and −35 (B) by wild-type and YW σ32 in vivo as assayed by β-galactosidase activities (Miller units) for each σ32 variant. Assays were performed as described in Figure 2C. Numbers above each bar indicate average values of Miller units. (C) Effect of YW substitution on transcription from nonoptimal groE promoter in vitro. Single-round run-off transcriptions were performed as described in the Materials and Methods; representative gels of each transcription reaction are shown. The top band in each lane is from the wild-type groE promoter (internal control); the bottom band originates from either the wild-type groE promoter or the mutant to be tested. (D) Quantification of the in vitro transcripts. The bars indicate the relative transcription from mutant promoters as a percentage of transcript from wild-type promoter for each σ32 variant. Each value was calculated as follows: (1) Each short transcript (lower band) was expressed as a percentage of the long transcript in the same lane to obtain the normalized short transcript; and (2) normalized short transcripts from each promoter variant (shown in C, lanes 2–11) were divided by normalized short transcript from the wild-type promoter (shown in C, lane 1). All values are averages of three independent experiments.
Altering Region 2.3 in σ28 also broadens promoter recognition
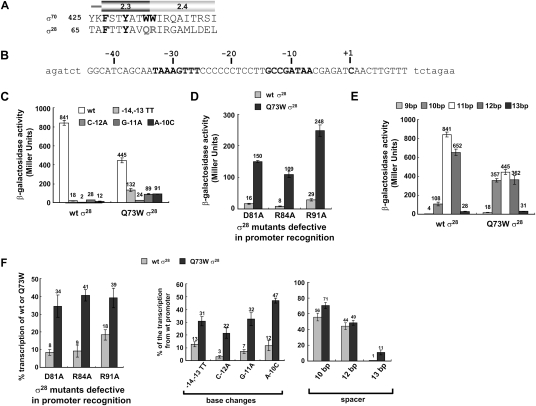
σ28 is highly divergent from σ32 (Gruber and Gross 2003; Paget and Helmann 2003), enabling us to test whether the relationship between Region 2.3 consensus and increased transcription of weak promoters was likely to be generalizable to other Group 3 σs. Comparison with σ70 indicated that only two of the four aromatic Region 2.3 residues universally conserved in the housekeeping σs are present in E. coli σ28 (Fig. 4A). R74 (W434 in σ70) was not considered further as it is highly conserved among σ28 orthologs and participates in nonspecific promoter binding (Koo et al. 2009b). Q73 (W433 in σ70) varies in different σ28s but is never W. We investigated whether Q73Wσ28 has broadened promoter recognition.
Figure 4.
Enhanced promoter utilization by Q73W σ28 in vivo and in vitro. (A) Amino acid sequence alignment of Regions 2.3 and 2.4 of E. coli σ70 and σ28. Bold letters are residues implicated in promoter melting in σ70, and the residues not conserved in σ28 are in gray. The numbers indicate amino acid sequence position. (B) Sequence of σ28-dependent promoter (Ptar) used in this study. Only the wild-type sequence of the tar promoter is shown; promoter variants were constructed from this sequence (Koo et al. 2009b). The native sequence of the tar promoter region is shown in capital letters, vector sequence is in lowercase, and −35 and −10 regions and transcription start site are shown in bold. (C) Expression from tar promoter mutants by RNAP containing wild-type or Q73W σ28 in vivo. β-Galactosidase activities (Miller units) of promoter∷lacZ fusions are shown. Assays were performed in CAG57115 (ΔfliA, ΔflgM) with plasmids expressing σ28 variants and bearing tar promoter variants∷lacZ. All values are averages of three independent experiments. (D) In vivo effects of the Q73W substitution on mutations in σ28 that eliminate promoter recognition determinants. β-Galactosidase activities (Miller units) driven by each σ28 variant on tested promoters are shown. All values are averages of three independent experiments. (Gray bar) Activity of each mutation in wild-type background; (black bar) activity of each mutation in Q73W background. (E) Expression from tar promoter mutants with nonoptimal spacers by RNAP containing wild-type or Q73W σ28 in vivo. β-Galactosidase activities (Miller units) are shown. All values are averages of three independent experiments. The spacer length was varied from 9 to 13 base pairs (bp); the wild-type tar promoter has an 11-bp spacer. (F) Effects of the Q73W substitution on mutations in σ28 that eliminate promoter recognition determinants and of tar promoter variants in vitro. Relative transcription determined from single-round run-off transcription assays (Supplemental Fig. S1) is depicted as bar graphs. Each experiment was repeated a minimum of three times, and numbers above each bar indicate average values of relative transcription. (Left graph) Effect of Q73W substitution on mutations eliminating promoter recognition determinants in σ28. Percent transcription was calculated as (intensity of transcript for RNAP with wild-type or Q73W σ28 carrying the specified additional substitutions in σ28)/(intensity of transcript for RNAP with wild-type or Q73W σ28). (Middle and right graphs) Expression from tar promoters with base substitutions (middle) and nonoptimal spacer lengths (right) by RNAP containing wild-type or Q73W σ28, normalized as described in Figure 3D.
σ28 is exceptionally sensitive to substitution in the −10 region of the promoter, and the Q73W substitution dramatically increases the ability of σ28 to recognize a variety of nonoptimal promoters. We examined promoter activity using lacZ reporter assays driven from the tar σ28 promoter (Fig. 4B) and its derivates. Eliminating the E-10 motif (−14G, −13C) or altering any one of three bases in the −10 motif reduces expression by wild-type σ28 ∼30-fold to 400-fold in vivo (Fig. 4C; Koo et al. 2009b). In sharp contrast, Q73Wσ28 exhibits only a threefold to 18-fold decrease on this same set of promoters (Fig. 4C). Importantly, Q73A does not increase expression from these mutant promoters, indicating that the suppressive effect resulted from adding the W residue at position 73, rather than removing the naturally occurring Q residue (data not shown). Likewise, Q73W exhibits 10-fold suppression of the transcription defects resulting from removing each of the three σ28 residues known to participate in base-specific recognition of the σ28 promoter (R91 [E-10 motif] and D81/R84 [−10 motif]) (Fig. 4D; Koo et al. 2009b) and shows enhanced tolerance for variation in the length of the spacer DNA separating the −35 and −10 elements (Fig. 4E). All promoters with significant transcriptional defects in vitro (≤40% of the wild-type promoter) showed significantly increased transcription with Q73Wσ28 as compared with wild-type σ28 (Fig. 4F). Thus, Q73Wσ28 is directly responsible for significantly enhanced transcription of a broad range of nonoptimal promoters.
YWσ32 broadens promoter recognition of natural promoters
Thus far, we examined the effects of YWσ32 on a promoter set, each differing from the groE consensus promoter at only a single position. Natural promoters may have many changes from the consensus and an UP-element (Zhao et al. 2005; Nonaka et al. 2006; Wade et al. 2006), recognized by the C-terminal domain of the α-subunit (Ross et al. 1993; Gourse et al. 2000), which might obscure the effect of YWσ32 on natural promoters. We compared expression of natural σ32 promoters in E. coli fused to a green fluorescent protein (GFP) reporter driven by either wild-type σ32 or YWσ32 (see the Materials and Methods). The promoter library encompassed sequences −65 to +20 relative to the start site of transcription, and therefore included any UP-element present. We found that promoters very weakly transcribed by σ32 RNAP were more strongly transcribed by YWσ32 RNAP; conversely, promoters strongly transcribed by σ32 RNAP have similar or less expression with YWσ32 RNAP. This is illustrated by comparing the ratio of promoter strengths by σ32 or YWσ32 RNAP against the strength of the promoter in the presence of σ32 RNAP (Fig. 5A; see also Supplemental Fig. S2 for the raw data). Therefore, YWσ32 preferentially increases expression from weaker, less conserved natural promoters. We consider the implications of this finding in the Discussion.
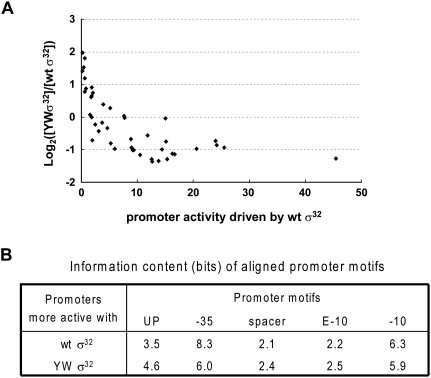
Figure 5.
YWσ32 preferentially increases expression of weak σ32 promoters in vivo. Promoter activities driven by either wild type (wt) or YWσ32 were determined by the expression of GFP from 50 σ32 promoters (Nonaka et al. 2006) as described in the Materials and Methods. (A) Data are displayed as a scatter plot showing the log2 value of the ratio of promoter activities driven by YWσ32 and wild-type σ32 (Y-axis) versus the strength of the promoter when driven by wild-type σ32 (X-axis). Promoter activity was calculated from the slope of the differential plot of OD600 versus GFP fluorescence (RFU); see the Materials and Methods. (B) Information content of promoters more active with either wild type (30 promoters) or YW σ32 (19 promoters). Values were calculated as described in the Materials and Methods. Each motif covers the following sequences: UP element, −60 to −45; −35 motif, −37 to −31; spacer between −35 and −10 motif; extended −10 motif, −16 and −15; and −10 motif, −14 to −9.
Altered Region 2.3 residues exert their effects on a step beyond duplex promoter binding
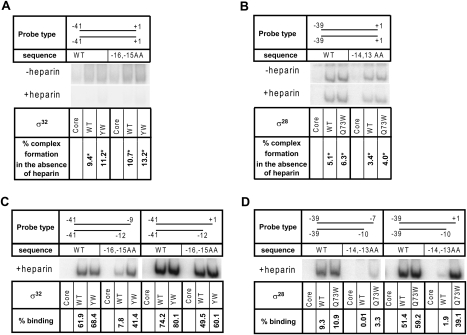
The σ70 residues analogous to those altered in σ28 and/or σ32 (Y430 and W433) have a minor effect on initial recognition of duplex DNA and a major effect on open complex formation (Fenton et al. 2000; Tomsic et al. 2001; Schroeder et al. 2009). The crystal structure of Aquifex aeolicus σ28 complexed to its anti-σ indicates that these Region 2.3 residues in Group 3 σs are roughly in the same position as in the housekeeping σs with respect to the most C-terminal helix of σ2 (Sorenson et al. 2004), suggesting that they might play similar roles in both σ families. We examined whether aromatic amino acid-substituted σ32 and σ28 holoenzymes (Eσ32 and Eσ28) were altered in initial duplex DNA recognition or in open complex formation using assays developed to characterize σ70 holoenzyme (Eσ70). Consistent with their effects in Eσ70, these residues have little or no effect on duplex binding by Eσ32 and Eσ28 (Fig. 6A,B) and exert their major effects on strand opening (Fig. 6C,D).
Figure 6.
The altered Region 2.3 residues in σ32 and σ28 affect open complex formation but not template binding. Electrophoretic mobility shift assay was performed as described in the Materials and Methods (also contains sequences of the DNA probes). End-labeled probe (10 nM) and holoenzyme (25 nM) (both as shown in the figure) were incubated for 10 min at 4°C. In heparin challenge experiments, 100 μg/mL heparin was added and incubation was continued for 5 min. All values shown are averages of at least three independent experiments. Standard deviations are ≤25% of average values. Representative gels are shown. (A,B) Binding to duplex DNA by E-wtσ32 or E-YW σ32 at the groE promoter (A) or by E-wtσ28 or E-Q73W σ28 at the tar promoter (B). Holoenzyme–DNA complexes formed without (top) or with (bottom) heparin challenge are shown. The percent complex formation in the absence of heparin is considered to reflect initial duplex binding. (C,D) Binding of RNAP to fork junction templates by E-wtσ32 or E-YW σ32 at the groE promoter (C) or by E-wtσ28 or E- Q73W σ28 to the tar promoter (D). Percent binding is the extent of formation of heparin-resistant complexes.
We assessed initial recognition by determining Eσ32 and Eσ28 binding to duplex DNA at 4°C, a temperature that prevents strand opening and traps the initial rapidly dissociating complex between RNAP and DNA, as shown for Eσ70 (Fenton et al. 2000). The low-temperature complex is on the pathway to the open complex, and therefore provides an accurate estimate of initial binding (Li and McClure 1998). These trapped complexes are expected to be sensitive to inhibition by heparin, which binds free RNAP irreversibly, thereby inactivating rapidly dissociating RNAP. We assessed binding both to the complete promoter (consensus −35, E-10, and −10 elements) and to a suboptimal promoter (lacking the E-10 motif). YWσ32 holoenzyme (E-YWσ32) and wild-type σ32 holoenzyme (E-wtσ32) show equivalent extents of heparin-sensitive binding (Fig. 6A), providing clear evidence that YWσ32 does not affect initial binding. Likewise, Q73Wσ28 holoenzyme (E-Q73Wσ28) and wild-type σ28 holoenzyme (E-wtσ28) show equivalent extents of binding (Fig. 6B). This binding is partially heparin-resistant, possibly reflecting formation of an “intermediate” complex further down the pathway. In any case, E-Q73Wσ28 and E-wtσ28 do not display any distinction in behavior in this assay, consistent with the idea that initial steps in the process are unaffected. Very similar results were obtained for Eσ32 and Eσ28 using shorter templates truncated just downstream from the −10 regions of each promoter (data not shown). Additionally, we validated that the observed binding of Eσ28 and Eσ32, although weak, is dependent on specific promoter sequences, as it is not observed with random sequence DNA (Supplemental Fig. S3). Taken together, these results support the conclusion that the Region 2.3 alterations in σ28 and σ32 have a minimal effect on duplex recognition.
To examine strand opening, we assessed heparin-resistant binding to “fork junction” templates at 4°C. A fork junction template is one in which the template strand is truncated just upstream of the position of strand opening (T-11 for PgroE with Eσ32 [Mecsas et al. 1991] and T-9 for Ptar with Eσ28 [Givens et al. 2001]), and the nontemplate strand continues as a single-strand overhang. This assay has been validated both kinetically and structurally to be an excellent mimic of open complex formation in Eσ70 (Guo and Gralla 1998; Murakami et al. 2002; Tsujikawa et al. 2002). Indeed, the extent to which a particular combination of fork junction template and holoenzyme is able to form a stable complex (i.e., resistant to heparin challenge) reflects the propensity for open complex formation with the particular set of reactants used (Guo and Gralla 1998; Fenton et al. 2000; Tsujikawa et al. 2002). We observe clear evidence that both YWσ32 and Q73Wσ28 are more proficient than their wild-type counterparts at promoting formation of the open complex, when assayed with the appropriate templates. E-YWσ32 shows fivefold enhancement of open complex formation when assayed on a suboptimal template (no E-10) whose fork extends to −9 (8% E-wtσ32 vs. 41% E-YWσ32) (Fig. 6C, −16,−15 AA). The other templates are almost completely shifted by E-wtσ32 and therefore cannot provide distinction between the two holoenzymes. The effect is even more dramatic for σ28, where E-Q73Wσ28 exhibits ≥20-fold more open complex formation than E-wtσ28 at both the long and short suboptimal fork junction templates (Fig. 6D; −14,−13 AA). We also validated that the observed binding of Eσ28 and Eσ32 to fork junction templates is dependent on specific promoter sequences, as no specific binding to fork junction templates bearing their anti-−10 promoter sequences was observed (Supplemental Fig. S3). Taken together, these results provide strong support for the idea that YWσ32 and Q73Wσ28 significantly promote open complex formation at suboptimal promoter templates.
Discussion
Early “primordial” σ factors have diverged into major subgroups: the housekeeping σs such as σ70, and the alternative σs. This divergence spawned evolution of an important gene expression strategy, allowing differentially regulated σs to recognize discrete classes of promoters. σ factor specialization has another distinguishing feature: Housekeeping σs recognize a large number of diverse promoters (>1000), whereas most alternative σs are much more restrictive in promoter selection, with a tighter requirement for the sequence and spacing of their promoter motifs. In this study, we asked what feature(s) of σs is responsible for this important difference in promoter recognition strategy. Our results suggest the hypothesis that the balance of melting and recognition functions of σs is critical to setting the stringency of promoter recognition.
We investigated the features of σs required for stringent promoter recognition by σ32 and σ28, two highly divergent members of the Group 3 σ subfamily present in E. coli. Sequence variation in Region 2.3 is largely responsible for this requirement: Converting one or two residues in Region 2.3 to their counterpart(s) in the housekeeping σs largely alleviated the stringent requirements. Our results indicated that the consensus variants were fivefold to ≥20-fold more proficient than their wild-type σ counterparts at promoting open complex formation on suboptimal promoters (Fig. 6C,D). Thus, Eσ32 and Eσ28 require highly specified promoters because of their reduced capacity for promoter melting.
σs perform two sequential activities: recognizing promoter motifs (to position RNAP), and facilitating subsequent steps required for melting the −10 region to create the “open complex” poised for transcription initiation. These activities are tightly coupled: The Region 2.3 residues facilitating melting and general duplex recognition are located in the same α-helix as the Region 2.4 residues that recognize specific bases in the −10 region and then stabilize the melted state by interaction with the nontemplate strand. In Eσ70 promoters, strand separation is nucleated within the −10 recognition motif, most likely by flipping out the −11A base (Lim et al. 2001; Schroeder et al. 2009). W433 may “push” the −11A out of the helix (Tomsic et al. 2001). Y430 is believed to interact with and stabilize the conformation of the flipped out −11A (Schroeder et al. 2009). Importantly, it is altering σ32 and σ28 residues at positions analogous to Y430 and W433 to those in housekeeping σs (F104YH107Wσ32; Q73Wσ28) that increases melting and relaxes promoter recognition.
How might a completely consensus promoter decrease the requirements for σ melting functions performed by Y430 and W433? The kinetics of open complex formation by E. coli σ70 holoenzyme at a consensus promoter (consensus −35, E-10, −10) provide a way of thinking about this linkage (Schroeder et al. 2009). It is suggested that the completely consensus promoter greatly stabilizes the transition state of the normally rate-limiting step in open complex formation so that it becomes a kinetically significant intermediate, possibly through contacts between the E-10 region and RNAP. The implication is that base flipping and strand separation, previously coupled kinetically, become two separable steps at the consensus promoter, with the latter being rate-limiting. Thus, the defect in base flipping caused by a suboptimal amino acid sequence in σ Region 2.3 would have no or little effect at consensus promoters but would lead to much reduced expression in weaker promoters. Consistent with this interpretation, Y430 and W433 substitutions do not affect open complex formation at the consensus σ70 promoter, although they have big effects on melting at standard promoters (Tomsic et al. 2001; Schroeder et al. 2009). Thus, the conformational changes driven by the consensus promoter elements obviate the requirement for these Region 2.3 residues. This scenario precisely explains our findings for σ32 and σ28: Their consensus promoters drive melting even though substitutions in residues analogous to Y430 and/or W433 reduce the melting proficiency of σ32 and σ28.
The suboptimal melting capacity of σ32 and σ28 has biological correlates. The necessity of optimal placement and sequence of promoter motifs to create a functional promoter means that deviation from consensus has profound negative effects on promoter activity. This focuses transcription by these σs on their authentic regulons and decreases the possibility that their responses will be diminished because they engage in adventitious transcription of near-match promoters. Likewise, the exquisite sensitivity of transcriptional capacity to promoter sequence also allows promoter strength to be regulated over a broad range so that regulon members are produced in optimal amounts relative to each other. Finally, suboptimal melting allows these σs to maintain the integrity of their recognition across many organisms. σ28 directs synthesis of flagellar components in both Gram-negative and Gram-positive bacteria, separated in evolution by billions of years. Yet the consensus sequence of the promoters recognized by σ28 remains unchanged.
Our results graphically illustrate the extent to which the relative promoter strength is deregulated when the σ factor has consensus melting determinants. Whereas the natural promoters in the σ32 regulon members display an ∼100-fold range in activity when driven by wild-type σ32, only a 20-fold range is seen when they are transcribed by the melting-proficient YWσ32, with the weakest promoters having enhanced activity and the strongest promoters showing decreased transcription by YWσ32 (possibly because of difficulty in promoter clearance) (Fig. 5A; Supplemental Fig. S2). Interestingly, motif comparison indicates that the predominant difference between promoters preferentially transcribed by YWσ32 and by wild-type σ32 is that the former has a less conserved −35 motif (Fig. 5B). This distinction raises the intriguing possibility that wild-type σ32, but not YWσ32, uses the −35 region as a “gatekeeper” both to determine functional promoters and to set promoter strength. A consensus −35 region may be necessary to slow dissociation sufficiently to permit strand opening.
It is interesting to consider why the σ32s of α-protobacteria might have broadened promoter specificity. Interestingly, σ32s in the α-protobacteria often control processes in addition to heat shock, such as development in Myxococcus xanthus (Ueki and Inouye 2001) or other stress responses (e.g., heavy metal stress in C. crescentus) (McGrath et al. 2007). Additionally, groEL, the most important member of the σ32 regulon (Kusukawa and Yura 1988), is regulated by an alternative mechanism in α-proteobacteria (Yura and Nakahigashi 1999). Thus, α-proteobacterial σ32s may transcribe more genes without the necessity of finely controlling the extent of their expression as compared with γ-proteobacterial σ32s.
Ever since Helmann's seminal work (Juang and Helmann 1994) demonstrated the involvement of σ Region 2.3 residues in promoter DNA melting, much effort has been devoted to defining its mechanism (Juang and Helmann 1994; Fenton et al. 2000; Schroeder et al. 2009). However, there has been little consideration of whether σ melting proficiency differs among σ subfamilies and whether this property is used to set the promoter recognition promiscuity of that family. Given that Group 3 σs rarely use >100 promoters in any bacterial species (in marked contrast to the housekeeping σ70) and that all of them share nonconsensus residues in Region 2.3 (Supplemental Fig. S4), we propose the hypothesis that amino acid sequence differences within Region 2.3 are important for the differences in breadth of promoter choice. Importantly, divergent Group 4 σs are also discrepant from the housekeeping σs in some of these important Region 2.3 residues. Melting deficiencies in the more divergent alternative σs may be a universal mechanism to ensure their promoter recognition stringency.
Materials and methods
Details of materials and methods are presented in the Supplemental Material.
Strains, plasmids, and growth conditions
Strains and plasmids used in this study are listed in Supplemental Table S1. Cells were grown at 30°C in Luria-Bertani (LB) media supplemented with appropriate antibiotics such as ampicillin (100 μg/mL), chloramphenicol (30 μg/mL), kanamycin (20 μg/mL), and spectinomycin (50 μg/mL). For the strains lacking rpoH (CAG57101), cells were grown with 0.1% L-(+)-arabinose to induce expression of GroESL (Koo et al. 2009a).
β-galactosidase assay, purification of σs, and in vitro transcription
β-galactosidase assays (used to measure in vivo promoter∷lacZ activities), overproduction and purification of σs, and in vitro single-round transcription assays were performed essentially as described (Koo et al. 2009a,b). Details are in the Supplemental Material.
Promoter activity determined by expression of GFP
The σ32 promoter library was constructed as described previously (Rhodius et al. 2006). Fifty σ32 promoters validated in our previous work (Nonaka et al. 2006) were cloned as XhoI–BamHI fragments into the GFP reporter plasmid, pUA66. Reporter strains were constructed by transforming derivatives of pSAKT32 and promoter library plasmids into CAG57101 sequentially using electroporation.
Promoter assays were performed by direct inoculation of LB broth supplemented with appropriate antibiotics from fresh transformants. Fluorescence and OD600 were measured in a Varioskan spectrofluorometer (Thermo Electron Corporation). σ32-dependent promoter activity was determined as described in the Supplemental Material.
Calculation of information content of promoter motifs
The information content (Iseq) of aligned promoter motifs was calculated using
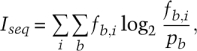 |
where i is the position within the site, b refers to each of the possible bases, fb,i is the observed frequency of each base at that position, and pb is the frequency of base b in the entire genome (in E. coli, taken to be 0.25 for A/G/C/T) (Schneider et al. 1986).
Electrophoretic mobility shift assay
PAGE-purified synthetic oligonucleotides were used for preparing double-strand and fork junction probes. 32P-labeled 10 nM annealed DNA probe and 25 nM holoenzyme were mixed in 10 μL of binding buffer and incubated for 10 min at 4°C. For heparin challenge, 2 μL of 600 μg/mL heparin were added, and the incubation was continued for an additional 5 min. Electrophoresis was performed in a prechilled 5% acrylamide/TBE gel at 4°C. See details in the Supplemental Material.
Acknowledgments
We thank Tania Baker for her critical input in conceptualizing this work, Vivek Mutalik for technical assistance, and members of the Gross laboratory for useful comments. This work was supported by National Institutes of Health Grants GM057755 (to C.A.G.) and GM31808 (to P.L.dH.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1843709.
Supplemental material is available at http://www.genesdev.org.
References
- Amaya E, Khvorova A, Piggot PJ. Analysis of promoter recognition in vivo directed by σF of Bacillus subtilis by using random-sequence oligonucleotides. J Bacteriol. 2001;183:3623–3630. doi: 10.1128/JB.183.12.3623-3630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asayama M, Imamura S. Stringent promoter recognition and autoregulation by the group 3 σ-factor SigF in the cyanobacterium Synechocystis sp. strain PCC 6803. Nucleic Acids Res. 2008;36:5297–5305. doi: 10.1093/nar/gkn453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-wide analysis of the stationary-phase σ factor (σH) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D, Zuber P, Losick R. Two amino acids in an RNA polymerase σ factor involved in the recognition of adjacent base pairs in the −10 region of a cognate promoter. Proc Natl Acad Sci. 1990;87:8075–8079. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Record MT, Jr, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- Dombroski AJ, Walter WA, Gross CA. Amino-terminal amino acids modulate σ-factor DNA-binding activity. Genes & Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, et al. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, et al. A basal promoter element recognized by free RNA polymerase σ subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and −10 recognition: Mutational analysis of σ70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): Gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella T, Moyle H, Susskind MM. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Givens JR, McGovern CL, Dombroski AJ. Formation of intermediate transcription initiation complexes at pfliD and pflgM by σ28 RNA polymerase. J Bacteriol. 2001;183:6244–6252. doi: 10.1128/JB.183.21.6244-6252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: The role of the α subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- Green HA, Donohue TJ. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock σ factor family. J Bacteriol. 2006;188:5712–5721. doi: 10.1128/JB.00405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple σ subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Helmann JD, deHaseth PL. Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: Flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Bio. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM. The promoter spacer influences transcription initiation via σ70 region 1.1 of Escherichia coli RNA polymerase. Proc Natl Acad Sci. 2009;106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard I, Johnson XB, Hinton DM. Escherichia coli RNA polymerase recognition of a σ70-dependent promoter requiring a −35 DNA element and an extended −10 TGn motif. J Bacteriol. 2006;188:8352–8359. doi: 10.1128/JB.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang YL, Helmann JD. A promoter melting region in the primary σ factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- Koo BM, Rhodius VA, Campbell EA, Gross CA. Dissection of recognition determinants of Escherichia coli σ32 suggests a composite −10 region with an ‘extended −10’ motif and a core −10 element. Mol Microbiol. 2009a;72:815–829. doi: 10.1111/j.1365-2958.2009.06690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BM, Rhodius VA, Campbell EA, Gross CA. Mutational analysis of Escherichia coli σ28 and its target promoters reveals recognition of a composite −10 region, comprised of an ‘extended −10 motif’ and a core −10 element. Mol Microbiol. 2009b;72:830–843. doi: 10.1111/j.1365-2958.2009.06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourennaia OV, deHaseth PL. Substitution of a highly conserved histidine in the Escherichia coli heat shock transcription factor, σ32, affects promoter utilization in vitro and leads to overexpression of the biofilm-associated flu protein in vivo. J Bacteriol. 2007;189:8430–8436. doi: 10.1128/JB.01197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Malloch RA, Fujita N, Smillie DA, Ishihama A, Hayward RS. The −35-recognition region of Escherichia coli σ70 is inessential for initiation of transcription at an ‘extended −10’ promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- Kusukawa N, Yura T. Heat shock protein GroE of Escherichia coli: Key protective roles against thermal stress. Genes & Dev. 1988;2:874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the σ subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Gralla JD. Open complex formation in vitro by σ38 (rpoS) RNA polymerase: Roles for region 2 amino acids. J Mol Biol. 2003;329:941–948. doi: 10.1016/s0022-2836(03)00369-3. [DOI] [PubMed] [Google Scholar]
- Li XY, McClure WR. Characterization of the closed complex intermediate formed during transcription initiation by Escherichia coli RNA polymerase. J Biol Chem. 1998;273:23549–23557. doi: 10.1074/jbc.273.36.23549. [DOI] [PubMed] [Google Scholar]
- Lim HM, Lee HJ, Roy S, Adhya S. A ‘master’ in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc Natl Acad Sci. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The σ70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Mecsas J, Cowing DW, Gross CA. Development of RNA polymerase–promoter contacts during open complex formation. J Mol Biol. 1991;220:585–597. doi: 10.1016/0022-2836(91)90102-c. [DOI] [PubMed] [Google Scholar]
- Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase σ70 region 4.2 and α-subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J Biol Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- Mitchell JE, Zheng D, Busby SJ, Minchin SD. Identification and analysis of ‘extended −10’ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: The wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding σ32 homologs from gram negative bacteria: Conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes & Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The σ70 family of σ factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J Biol Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Schroeder LA, Gries TJ, Saecker RM, Record MT, Jr, Harris ME, deHaseth PL. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J Mol Biol. 2009;385:339–349. doi: 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa M, Yamamoto H, Yamaguchi H, Fujita Y, Kobayashi K, Ogasawara N, Sekiguchi J. Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses. Gene. 2004;329:125–136. doi: 10.1016/j.gene.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Shen L, Feng X, Yuan Y, Luo X, Hatch TP, Hughes KT, Liu JS, Zhang YX. Selective promoter recognition by chlamydial σ28 holoenzyme. J Bacteriol. 2006;188:7364–7377. doi: 10.1128/JB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Sorenson MK, Darst SA. Disulfide cross-linking indicates that FlgM-bound and free σ28 adopt similar conformations. Proc Natl Acad Sci. 2006;103:16722–16727. doi: 10.1073/pnas.0606482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MK, Ray SS, Darst SA. Crystal structure of the flagellar σ/anti-σ complex σ28/FlgM reveals an intact σ factor in an inactive conformation. Mol Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- Tatti KM, Jones CH, Moran CP., Jr Genetic evidence for interaction of σE with the spoIIID promoter in Bacillus subtilis. J Bacteriol. 1991;173:7828–7833. doi: 10.1128/jb.173.24.7828-7833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenot B, Charpentier B, Branlant C. The strong efficiency of the Escherichia coli gapA P1 promoter depends on a complex combination of functional determinants. Biochem J. 2004;383:371–382. doi: 10.1042/BJ20040792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli σ70 in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J Biol Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- Tsujikawa L, Tsodikov OV, deHaseth PL. Interaction of RNA polymerase with forked DNA: Evidence for two kinetically significant intermediates on the pathway to the final complex. Proc Natl Acad Sci. 2002;99:3493–3498. doi: 10.1073/pnas.062487299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Inouye S. SigB, SigC, and SigE from Myxococcus xanthus homologous to σ32 are not required for heat shock response but for multicellular differentiation. J Mol Microbiol Biotechnol. 2001;3:287–293. [PubMed] [Google Scholar]
- van Schaik W, van der Voort M, Molenaar D, Moezelaar R, de Vos WM, Abee T. Identification of the σB regulon of Bacillus cereus and conservation of σB-regulated genes in low-GC-content gram-positive bacteria. J Bacteriol. 2007;189:4384–4390. doi: 10.1128/JB.00313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between σ factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- Waldburger C, Gardella T, Wong R, Susskind MM. Changes in conserved region 2 of Escherichia coli σ70 affecting promoter recognition. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Young BA, Gruber TM, Gross CA. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- Yu HH, Tan M. σ28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol Microbiol. 2003;50:577–584. doi: 10.1046/j.1365-2958.2003.03708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Nakahigashi K. Regulation of the heat-shock response. Curr Opin Microbiol. 1999;2:153–158. doi: 10.1016/S1369-5274(99)80027-7. [DOI] [PubMed] [Google Scholar]
- Zhao K, Liu M, Burgess RR. The global transcriptional response of Escherichia coli to induced σ32 protein involves σ32 regulon activation followed by inactivation and degradation of σ32 in vivo. J Biol Chem. 2005;280:17758–17768. doi: 10.1074/jbc.M500393200. [DOI] [PubMed] [Google Scholar]
- Zhao K, Liu M, Burgess RR. Adaptation in bacterial flagellar and motility systems: From regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]