Abstract
A key step in bacterial transcription initiation is melting of the double-stranded promoter DNA by the RNA polymerase holoenzyme. Primary σ factors mediate the melting of thousands of promoters through a conserved set of aromatic amino acids. Alternative σs, which direct transcription of restricted regulons, lack the full set of melting residues. In this issue of Genes & Development, Koo and colleagues (pp. 2426–2436) show that introducing the primary σ melting residues into alternative σs relaxes their promoter specificity, pointing to a trade-off of reduced promoter melting capacity for increased promoter stringency.
Keywords: σ factor, Region 2.3, melting proficiency, promoter stringency
Transcription initiation in bacteria is controlled primarily by σ factors, promoter specificity subunits for the catalytic core RNA polymerase (RNAP) (Gruber and Gross 2003; Paget and Helmann 2003). The primary σ (σ70 in Escherichia coli) directs core RNAP to the majority of promoters active during log-phase growth. Alternative σs control restricted, specialized regulons in response to environmental conditions. Examples in E. coli (Eco) include σS (general stress response, stationary phase) (Klauck et al. 2007), σ32 (also called σH; cytoplasmic heat shock) (Guisbert et al. 2008), σ28 (also called σF; starvation, flagellar synthesis) (Chilcott and Hughes 2000), σE (envelope stress response) (Ades 2008), and FecI (iron deficiency) (Braun et al. 2003). Some bacteria have just one σ factor (Mycoplasma sp.), while some have at least 66 (Streptomyces coelicolor). The vast majority of σ factors, including all those described above, comprise a homologous family of proteins, the σ70 family (Stragier et al. 1985; Gribskov and Burgess 1986), with four regions of conserved amino acid sequence (Lonetto et al. 1992; Gruber and Bryant 1997).
After binding to the core RNAP, the σ factor directs the resulting holoenzyme to a particular set of promoters dictated by sequence-specific recognition of the promoter by σ (Murakami et al. 2002; Gruber and Gross 2003). Promoter motifs recognized by σ70 family members include (from upstream to downstream) the −35 element (Gardella et al. 1989; Siegele et al. 1989; Campbell et al. 2002), the extended −10 element (Keilty and Rosenberg 1987; Barne et al. 1997; Koo et al. 2009a,b), the −10 element (Siegele et al. 1989; Daniels et al. 1990; Waldburger et al. 1990; Tatti et al. 1991), and the discriminator (Feklistov et al. 2006; Haugen et al. 2006). Primary σs, such as σ70, recognize thousands of promoters constructed from subsets of these motifs that are poorly constrained in sequence. In contrast, alternative σs recognize only a handful of promoters—for example, Eco σ32 directs the transcription of ∼50 promoters, while Eco σ28 transcribes ∼25 promoters (Nonaka et al. 2006; Shen et al. 2006)—and these promoters require good matches to the cognate −35, extended −10, and −10 motifs (Koo et al. 2009a,b,c). In other words, primary σs tolerate a great deal of promoter sequence diversity (loose stringency), while alternative σs in general do not (high stringency). What are the sequence determinants of the σ factors that govern this dramatic range of promoter stringency, and what are the mechanistic underpinnings? In this issue of Genes & Development, Koo et al. (2009c) set out to answer these questions.
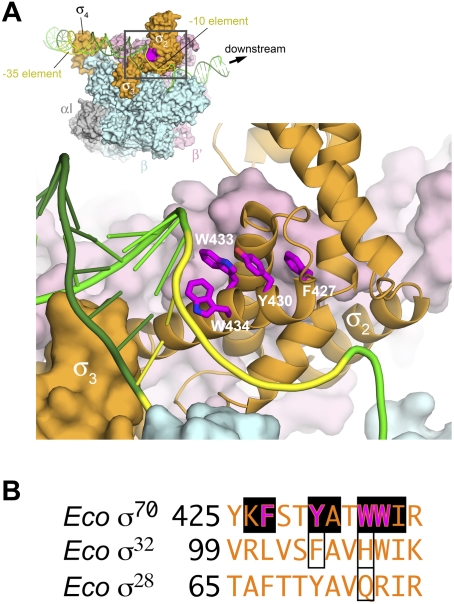
For σ factors belonging to the σ70 family, recognition of the promoter sequence is followed spontaneously by a series of isomerization steps yielding the transcription-competent open promoter complex (Fig. 1A), in which ∼14 base pairs of DNA are melted in a region that includes most of the −10 element and extends downstream to the transcription start site (deHaseth et al. 1998). Studies on primary σs indicate that σ plays a key role in the initiation of the DNA melting process within the promoter −10 element. A set of four aromatic residues that are invariant in primary σs, corresponding to Eco σ70 F427, Y430, W433, and W434 (Gruber and Bryant 1997; Campbell et al. 2002), have been implicated in this promoter melting function (Helmann and Chamberlin 1988; Juang and Helmann 1994, 1995), possibly by interacting with the exposed bases of the nontemplate strand within the −10 element at the upstream edge of the transcription bubble to stabilize the melted state of the DNA (Roberts and Roberts 1996; Huang et al. 1997; Marr and Roberts 1997). Structural studies of an RNAP holoenzyme/promoter fragment complex are consistent with this view (Fig. 1A; Murakami et al. 2002). Despite significant sequence variation, the structures of the conserved domains of primary and alternative σs are essentially identical (Malhotra et al. 1996; Campbell et al. 2002, 2003, 2007; Li et al. 2002; Sorenson et al. 2004), and their interactions with core RNAP and with promoter DNA are expected to be conserved. Nevertheless, the crucial “promoter melting” residues of the primary σs are not shared among the alternative σs (Lonetto et al. 1992).
Figure 1.
Primary σ promoter melting residues in the context of the RNAP holoenzyme open promoter complex. (A) Structural model of the RNAP holoenzyme open promoter complex (after Murakami et al. 2002). The RNAP is shown as a molecular surface, color-coded as follows: (gray) α; (cyan) β; (pink) β′; (orange) σ. The DNA is shown as phosphate backbone ribbons (template strand, dark green; nontemplate strand, light green), with the −35 and −10 elements colored yellow. The direction of transcription (downstream) is to the right. The top left (inset) shows an overview of the entire complex. The region in the gray box is magnified below. In the magnified view, σ domain 2 (σ2) is shown as a backbone ribbon. The side chains of the four, universally conserved “promoter melting” mutants of primary σs (see the text) are shown in magenta. The labels show residue numering according to Eco σ70. (B) Sequence context of the promoter melting residues. The “promoter melting” residues of the primary σ are bold and colored magenta. Absolutely conserved residues of the primary σs are shaded with a solid black box. The corresponding residues of the alternative σs that were substituted by Koo et al. (2009c) are boxed.
The study by Koo et al. (2009c) focuses on two Eco alternative σ factors: σ32 and σ28 (encoded by rpoH and rpoF, respectively). σ32 is induced in response to the presence of unfolded proteins in the cytoplasm, which can result from heat shock or other stressful conditions. σ32 elicits transcription of regulons resulting in the expression of chaperones and proteases helping to refold or degrade damaged proteins (transcription of ∼90 genes is initiated; e.g., dnaK, groESL, and hslU) (Nonaka et al. 2006). σ28 directs transcription from ∼25 promoters controlling flagellin biosynthesis and chemotaxis (Chilcott and Hughes 2000).
The “promiscuous” primary σ, σ70, recognizes an extended −10/−10 element consensus of TGnTATAAT (Pribnow 1975; Shultzaberger et al. 2006). The extended −10 motif (TG) is recognized by σ domain 3 (Barne et al. 1997), while the −10 element is recognized by σ domain 2, which contains the “melting” residues F427, Y430, W433, and W434. σ32 and σ28 recognize −10 elements of CCCCATnT and GCCGATAA, respectively. Previous studies by Koo et al. (2009a,b) showed that these σ32 and σ28 −10 elements are, in fact, composite, and their recognition follows a general bipartite pattern similar to σ70, with an extended −10 element (upstream CC for σ32, GC for σ28) recognized by domain 3, and a −10 element recognized by residues in region 2.
The very first step in promoter opening probably involves flipping out of the consensus adenine residue, dubbed the “master base in promoter opening” (the −11A, the second adenine in the TATAAT sequence), from the DNA duplex into a hydrophobic pocket in σ (Lim et al. 2001). This triggers recognition of the remaining bases of the −10 element, which is followed by extension of the melted region to the transcription start site. The universally conserved primary σ “melting residues” appear to act by promoting the −11A flipping, and in sequence-specific recognition of the −10 element nontemplate strand bases following the melting, with Y430 and W433 being the most crucial for the nucleation process (Tomsic et al. 2001; Schroeder et al. 2009). Using a combined in vivo/in vitro approach, Koo et al. (2009c) investigated the effects of substituting amino acid residues in the alternative σs at positions corresponding to σ70 Y430 and W433 to those present in primary σs (Fig. 1B). These substitutions rendered the alternative σs more promiscuous, in that they were able to tolerate promoters with less than the normally required complete set of promoter motifs (−35/extended −10/−10 elements), with nonoptimal motif sequences, and with nonoptimal spacing between the elements.
The principal finding of the Koo et al. (2009c) study comes from the results of in vitro binding assays comparing σ binding to promoter templates made of dsDNA with so-called fork junction templates (Guo and Gralla 1998). Fork junction DNA, which contains a double-stranded −35 element but mostly only the nontemplate strand of the −10 element, forms stable complexes with the RNAP holoenzyme that mimic many properties of the promoter open complex (Tsujikawa et al. 2001; Murakami et al. 2002). Thus, comparison of RNAP holoenzyme binding to a double-stranded versus a fork junction promoter relates to the stability of the initial, closed promoter complex versus the melted, open promoter complex. Koo et al. (2009c) observed that the mutated alternative σs were much more proficient then their wild-type counterparts in forming fork junction promoter complexes relative to double-stranded promoter complexes. The investigators concluded that the primary σ “melting residues,” placed in the context of the alternative σs, made the mutant alternative σs more efficient in the DNA strand separation (melting) step of promoter utilization, and that this was also the explanation for the increased tolerance toward imperfect promoters.
One can think of an evolutionary trade-off between melting capacity of a σ factor and its promoter specificity. A σ factor with strong melting capability would serve as a global regulator, serving many, poorly conserved promoters. σ Factors with weak melting capability would act as local regulators, confined to a more restricted set of promoters. In a broad sense, the results of Koo et al. (2009c) provide an interesting corollary to the recent observation that transcription factors with high DNA-binding specificity tend to mediate a more focused response on a genome-wide scale, while lower DNA-binding specificity enables them to control a broad range of targets in a global manner (Lozada-Chavez et al. 2008).
The region of σ harboring the “melting residues” (σ70 F427, Y430, W433, and W434), the so-called σ conserved region 2.3 (σ2.3) (Lonetto et al. 1992), is thought to act, at least in part, as a sequence-specific, ssDNA-binding element. In this view, σ2.3 functions by binding the nontemplate strand bases in the melted −10 element, thereby stabilizing the melted state (Roberts and Roberts 1996; Huang et al. 1997; Marr and Roberts 1997). Thinking about σ2.3 function in this way, the results of Koo et al. (2009c) may seem to present a paradox, since substitution of the primary σ melting residues into the alternative σ improves melting function on the cognate promoter of the wild-type alternative σ. However, this can be understood since, in the primary σ cognate −10 element, the −11A (TATAAT) is extremely highly conserved (Shultzaberger et al. 2006) and is thought to be the initial base that flips out of the double helix to interact with σ and nucleate promoter melting (Lim et al. 2001). Alignment of the σ70, σ32, and σ28 promoter sequences indicates that the −11A is retained in each of the promoters (see Fig. 1B of Koo et al. 2009c). The conservation of an A at this promoter position for the primary and alternative σ promoters; the apparent specificity of the primary σ melting residues for this A; and the dominant, nucleating effect of this A on the promoter melting process resolves the paradox.
The ability of the primary σ melting residues to improve the melting capabilities of the mutant alternative σs was inferred by comparing binding of the mutant σs to double-stranded and fork junction promoter fragments. It will be informative to investigate the mechanisms of melting by the alternative σs (both mutant and wild type) using the battery of biophysical and biochemical tools that have been brought to bear on the primary σ (deHaseth et al. 1998).
While primary σs on their own direct transcription from a wide range of poorly conserved promoters, the range of promoters is broadened even further by the influence of trans-acting factors (i.e., transcription activators), which allow transcription from promoters too poor to be used by σ70 on its own. The influence of trans-acting regulatory factors on the promoter selectivity of holoenzymes containing alternative σs has not been investigated to any extent.
While alternative σ factors should clearly know their allowed territory, housekeeping σs can act on behalf of alternative σs in certain situations. For example, bacterial strains lacking σ32 can still transcribe heat-shock genes, possibly through the involvement of σ70 (Zhou et al. 1988). It was also shown that the majority of σ32 promoters can be used by the σ70 holoenzyme in vivo and in vitro. Some σE promoters are recognized by σ70 as well (Wade et al. 2006). The biological role of σ70 in specific responses, normally thought to be the exclusive purview of alternative σs, remains to be understood.
Alternative σs usually mediate responses to environmental stresses, so these responses need to be rapid, focused, and confined to a tightly defined regulon. The work of Koo et al. (2009c) indicates that this is achieved, at least in part, through the strict promoter recognition requirements of the alternative σs. This is governed by the reduced promoter melting capability of the alternative σs due to the loss of “promoter melting” aromatic residues in σ region 2.3, pointing to an evolutionary trade-off between promoter melting capability and promoter stringency.
Acknowledgments
This work was supported by funds from a Merck Post-doctoral Fellowship at The Rockefeller University to A.F., and NIH RO1 GM053759 to S.A.D.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1862609.
References
- Ades SE. Regulation by destruction; design of the σE envelope stress response. Curr Opin Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Barne KA, Bown JA, Busby SJW, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended −10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF σ factor by transmembrane signalling. Curr Opin Microbiol. 2003;6:173–180. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity σ factor. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-σ RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L, Das K, Sofia HJ, Donohue TJ, Darst SA. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol Cell. 2007;27:793–805. doi: 10.1016/j.molcel.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D, Zuber P, Losick R. Two amino acids in an RNA polymerase σ factor involved in the recognition of adjacent base pairs in the −10 region of a cognate promoter. Proc Natl Acad Sci. 1990;87:8075–8079. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHaseth PL, Zupancic ML, Record MTJ. RNA polymerase-promoter interactions: The comings and going of RNA polymerase. J Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkiene E, Stavrovskaya E, Klimasauskas S, et al. A basal promoter element recognized by free RNA polymerase σ subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gardella T, Moyle T, Susskind MM. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gribskov M, Burgess RR. σ Factors from E. coli, B. subtilis, phase SPO1, and phage T4 are homologous proteins. Nucleic Acids Res. 1986;14:6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Bryant DA. Molecular systematic studies of eubacteria, using σ70-type σ factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple σ subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of σ region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ. Structure and function of bacterial σ factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Huang X, Lopez de Saro FJ, Helmann JD. σ Factor mutations affecting the sequence-seletive interaction of RNA polymerase with −10 region single-stranded DNA. Nucleic Acids Res. 1997;25:2603–2609. doi: 10.1093/nar/25.13.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y-L, Helmann JD. A promoter melting region in the primary σ factor of Bacillus subtilis: Identification of functionally important aromatic amino acids. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- Juang Y-L, Helmann JD. Pathway of promoter melting by Bacillus subtilis RNA polymerase at a stable RNA promoter: Effects of temperature, δ protein, and σ factor mutations. Biochemistry. 1995;34:8465–8473. doi: 10.1021/bi00026a030. [DOI] [PubMed] [Google Scholar]
- Keilty S, Rosenberg M. Constituitive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- Klauck E, Typas A, Hengge R. The σS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci Prog. 2007;90:103–127. doi: 10.3184/003685007X215922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-M, Rhodius VA, Campbell EA, Gross CA. Dissection of recognition determinants of Escherichia coli σ32 suggests a composite −10 region with an ‘extended −10’ motif and a core −10 element. Mol Microbiol. 2009a;72:815–829. doi: 10.1111/j.1365-2958.2009.06690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-M, Rhodius VA, Campbell EA, Gross CA. Mutational analysis of Escherichia coli σ28 and its target promoters reveals recognition of a composite −10 regionm, comprised of an ‘extended −10’ motif and a core −10 element. Mol Microbiol. 2009b;72:830–843. doi: 10.1111/j.1365-2958.2009.06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-M, Rhodius VA, Nonaka G, deHaseth PL, Gross CA. Reduced capacity of alternative σs to melt promoters ensures stringent promoter recognition. Genes & Dev. 2009c doi: 10.1101/gad.1843709. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Stevenson CE, Burton N, Jakimowicz P, Paget MS, Buttner MJ, Lawson DM, Kleanthous C. Identification and structure of the anti-σ factor-binding domain of the disulphide-stress regulated σ factor σR from Stretomyces coelicolor. J Mol Biol. 2002;323:225–236. doi: 10.1016/s0022-2836(02)00948-8. [DOI] [PubMed] [Google Scholar]
- Lim HM, Lee HJ, Roy S, Adhya S. A ‘master’ in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc Natl Acad Sci. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The σ70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J Mol Biol. 2008;379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Severinova E, Darst SA. Crystal structure of a σ70 subunit fragment from Escherichia coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- Murakami K, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme/DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-chock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes & Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The σ70 family of σ factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci. 1975;72:784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Schroeder LA, Gries TJ, Saecker RM, Record MTJ, Harris ME, deHaseth PL. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J Mol Biol. 2009;385:339–349. doi: 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Feng X, Yuan Y, Luo X, Hatch TP, Hughes KT, Liu JS, Zhang YX. Selective promoter recognition by chlamydial σ28 holoenzyme. J Bacteriol. 2006;188:7364–7377. doi: 10.1128/JB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2006;35:771–788. doi: 10.1093/nar/gkl956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele DA, Hu JC, Walter WA, Gross CA. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Sorenson MK, Ray SS, Darst SA. Crystal structure of the flagellar σ/anti-σ complex σ28/FlgM reveals an intact σ factor in an inactive conformation. Mol Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- Stragier P, Parsot C, Bouvier J. Two functional domains conserved in major and alternate bacterial σ factors. FEBS Lett. 1985;187:11–15. doi: 10.1016/0014-5793(85)81203-5. [DOI] [PubMed] [Google Scholar]
- Tatti KM, Jones CH, Moran CPJ. Genetic evidence for interaction of σE with the spoIIID promoter in Bacillus subtilis. J Bacteriol. 1991;173:7828–7833. doi: 10.1128/jb.173.24.7828-7833.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli σ70 in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase–promoter complexes. J Biol Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- Tsujikawa L, Tsodikov O, de Haseth P. Interaction of RNA polymerase with forked DNA: Evidence for two kinetically significant intermediates on the pathway to the final complex. Proc Natl Acad Sci. 2001;99:3493–3498. doi: 10.1073/pnas.062487299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between σ factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- Waldburger C, Gardella T, Wong R, Susskind MM. Changes in conserved region 2 of Escherichia coli σ70 affecting promoter recognition. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- Zhou YN, Kusukawa N, Erickson JW, Gross CA, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock σ factor σ32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]